Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
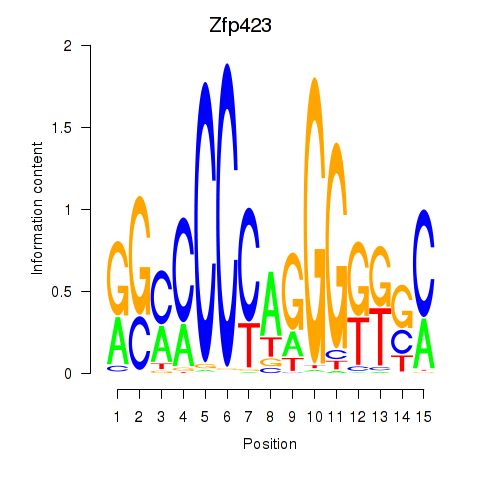
Results for Zfp423
Z-value: 0.40
Transcription factors associated with Zfp423
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp423
|
ENSMUSG00000045333.16 | Zfp423 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp423 | mm39_v1_chr8_-_88686188_88686223 | 0.26 | 2.7e-02 | Click! |
Activity profile of Zfp423 motif
Sorted Z-values of Zfp423 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp423
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_29142965 | 2.78 |
ENSMUST00000155949.2
ENSMUST00000154682.8 ENSMUST00000028141.6 ENSMUST00000071201.5 |
6530402F18Rik
Ntng2
|
RIKEN cDNA 6530402F18 gene netrin G2 |
| chr9_-_37464200 | 2.64 |
ENSMUST00000065668.12
|
Nrgn
|
neurogranin |
| chr16_+_17307503 | 2.28 |
ENSMUST00000023448.15
|
Aifm3
|
apoptosis-inducing factor, mitochondrion-associated 3 |
| chr15_-_66158445 | 1.93 |
ENSMUST00000070256.9
|
Kcnq3
|
potassium voltage-gated channel, subfamily Q, member 3 |
| chr11_+_87469211 | 1.69 |
ENSMUST00000107962.8
ENSMUST00000122067.8 |
Septin4
|
septin 4 |
| chr3_+_114697710 | 1.60 |
ENSMUST00000081752.13
|
Olfm3
|
olfactomedin 3 |
| chr4_+_42917228 | 1.56 |
ENSMUST00000107976.9
ENSMUST00000069184.9 |
Phf24
|
PHD finger protein 24 |
| chr12_+_102915102 | 1.50 |
ENSMUST00000101099.12
|
Unc79
|
unc-79 homolog |
| chr19_-_10847121 | 1.37 |
ENSMUST00000120524.2
ENSMUST00000025645.14 |
Tmem132a
|
transmembrane protein 132A |
| chr18_+_40390013 | 1.32 |
ENSMUST00000096572.2
ENSMUST00000236889.2 |
Kctd16
|
potassium channel tetramerisation domain containing 16 |
| chr6_+_110622533 | 1.27 |
ENSMUST00000071076.13
ENSMUST00000172951.2 |
Grm7
|
glutamate receptor, metabotropic 7 |
| chr17_+_69463786 | 1.27 |
ENSMUST00000112680.8
ENSMUST00000080208.7 ENSMUST00000225977.2 |
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr2_+_38229270 | 1.24 |
ENSMUST00000143783.9
|
Lhx2
|
LIM homeobox protein 2 |
| chr8_+_59365291 | 1.16 |
ENSMUST00000160055.2
|
BC030500
|
cDNA sequence BC030500 |
| chr15_-_78602313 | 1.07 |
ENSMUST00000229441.2
|
Elfn2
|
leucine rich repeat and fibronectin type III, extracellular 2 |
| chr10_+_60185093 | 0.96 |
ENSMUST00000105459.2
|
Vsir
|
V-set immunoregulatory receptor |
| chr19_+_6451667 | 0.91 |
ENSMUST00000113471.3
ENSMUST00000113469.3 |
Rasgrp2
|
RAS, guanyl releasing protein 2 |
| chr2_+_106523532 | 0.87 |
ENSMUST00000111063.8
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr19_-_10079091 | 0.86 |
ENSMUST00000025567.9
|
Fads2
|
fatty acid desaturase 2 |
| chr7_+_29007349 | 0.86 |
ENSMUST00000108230.8
ENSMUST00000065181.12 |
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chrX_-_74174608 | 0.85 |
ENSMUST00000033775.9
|
Mpp1
|
membrane protein, palmitoylated |
| chr11_+_54793569 | 0.83 |
ENSMUST00000082430.11
|
Gpx3
|
glutathione peroxidase 3 |
| chr7_+_30121147 | 0.78 |
ENSMUST00000108176.9
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr4_+_129407374 | 0.73 |
ENSMUST00000062356.7
|
Marcksl1
|
MARCKS-like 1 |
| chr9_+_120321557 | 0.72 |
ENSMUST00000007139.6
|
Eif1b
|
eukaryotic translation initiation factor 1B |
| chr17_+_69463846 | 0.72 |
ENSMUST00000238898.2
|
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr2_+_180141686 | 0.68 |
ENSMUST00000029084.9
|
Ntsr1
|
neurotensin receptor 1 |
| chr5_+_88712840 | 0.66 |
ENSMUST00000196894.5
ENSMUST00000198965.5 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr17_-_11059172 | 0.62 |
ENSMUST00000041463.7
|
Pacrg
|
PARK2 co-regulated |
| chr19_+_7245591 | 0.62 |
ENSMUST00000066646.12
|
Rcor2
|
REST corepressor 2 |
| chrX_-_74174450 | 0.58 |
ENSMUST00000114092.8
ENSMUST00000132501.8 ENSMUST00000153318.8 ENSMUST00000155742.2 |
Mpp1
|
membrane protein, palmitoylated |
| chr11_+_11065782 | 0.57 |
ENSMUST00000056344.5
|
Vwc2
|
von Willebrand factor C domain containing 2 |
| chr12_+_109425769 | 0.57 |
ENSMUST00000173812.2
|
Dlk1
|
delta like non-canonical Notch ligand 1 |
| chr5_-_24628514 | 0.52 |
ENSMUST00000030814.11
|
Cdk5
|
cyclin-dependent kinase 5 |
| chrX_-_74174524 | 0.52 |
ENSMUST00000114091.8
|
Mpp1
|
membrane protein, palmitoylated |
| chr17_-_23864237 | 0.51 |
ENSMUST00000024696.9
|
Mmp25
|
matrix metallopeptidase 25 |
| chr11_+_54793743 | 0.50 |
ENSMUST00000149324.3
|
Gpx3
|
glutathione peroxidase 3 |
| chr10_+_14581325 | 0.50 |
ENSMUST00000191238.7
ENSMUST00000190114.2 |
Nmbr
|
neuromedin B receptor |
| chr19_+_10019023 | 0.49 |
ENSMUST00000237672.2
|
Fads3
|
fatty acid desaturase 3 |
| chr15_-_103242697 | 0.47 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr2_-_30249202 | 0.44 |
ENSMUST00000100215.11
ENSMUST00000113620.10 ENSMUST00000163668.3 ENSMUST00000028214.15 ENSMUST00000113621.10 |
Sh3glb2
|
SH3-domain GRB2-like endophilin B2 |
| chr15_+_78128990 | 0.43 |
ENSMUST00000096357.12
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chrX_+_7688528 | 0.41 |
ENSMUST00000009875.5
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr5_-_24628483 | 0.40 |
ENSMUST00000198990.2
|
Cdk5
|
cyclin-dependent kinase 5 |
| chr10_+_80972089 | 0.40 |
ENSMUST00000048128.15
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr10_-_61946724 | 0.39 |
ENSMUST00000142821.8
ENSMUST00000124615.8 ENSMUST00000064050.5 ENSMUST00000125704.8 ENSMUST00000142796.8 |
Fam241b
|
family with sequence similarity 241, member B |
| chr16_+_49675969 | 0.37 |
ENSMUST00000229101.2
ENSMUST00000230836.2 ENSMUST00000229206.2 ENSMUST00000084838.14 ENSMUST00000230281.2 |
Cd47
|
CD47 antigen (Rh-related antigen, integrin-associated signal transducer) |
| chr8_-_84425724 | 0.37 |
ENSMUST00000005616.16
|
Pkn1
|
protein kinase N1 |
| chr12_-_14202041 | 0.36 |
ENSMUST00000020926.8
|
Lratd1
|
LRAT domain containing 1 |
| chr15_+_78129040 | 0.35 |
ENSMUST00000133618.3
|
Ncf4
|
neutrophil cytosolic factor 4 |
| chr7_-_45519853 | 0.32 |
ENSMUST00000211713.2
|
Grin2d
|
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
| chr2_+_93017887 | 0.31 |
ENSMUST00000150462.8
ENSMUST00000111266.8 |
Trp53i11
|
transformation related protein 53 inducible protein 11 |
| chr2_+_93017916 | 0.30 |
ENSMUST00000090554.11
|
Trp53i11
|
transformation related protein 53 inducible protein 11 |
| chr7_+_127084283 | 0.29 |
ENSMUST00000048896.8
|
Fbrs
|
fibrosin |
| chr19_-_5781503 | 0.29 |
ENSMUST00000162976.2
|
Znrd2
|
zinc ribbon domain containing 2 |
| chr9_+_57037974 | 0.28 |
ENSMUST00000160147.8
ENSMUST00000161663.8 ENSMUST00000034836.16 ENSMUST00000161182.8 |
Man2c1
|
mannosidase, alpha, class 2C, member 1 |
| chr10_-_14581203 | 0.28 |
ENSMUST00000149485.2
ENSMUST00000154132.8 |
Vta1
|
vesicle (multivesicular body) trafficking 1 |
| chr5_-_135963408 | 0.26 |
ENSMUST00000198270.2
ENSMUST00000055808.6 |
Ywhag
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide |
| chr4_+_126915104 | 0.25 |
ENSMUST00000030623.8
|
Sfpq
|
splicing factor proline/glutamine rich (polypyrimidine tract binding protein associated) |
| chr18_+_34892599 | 0.25 |
ENSMUST00000097622.4
|
Fam53c
|
family with sequence similarity 53, member C |
| chr1_+_156386414 | 0.24 |
ENSMUST00000166172.9
ENSMUST00000027888.13 |
Abl2
|
v-abl Abelson murine leukemia viral oncogene 2 (arg, Abelson-related gene) |
| chr17_-_23939700 | 0.24 |
ENSMUST00000201734.4
|
1520401A03Rik
|
RIKEN cDNA 1520401A03 gene |
| chr5_+_67418137 | 0.21 |
ENSMUST00000161369.3
|
Tmem33
|
transmembrane protein 33 |
| chr4_-_148711453 | 0.21 |
ENSMUST00000165113.8
ENSMUST00000172073.8 ENSMUST00000105702.9 ENSMUST00000084125.10 |
Tardbp
|
TAR DNA binding protein |
| chr11_-_99228756 | 0.21 |
ENSMUST00000100482.3
|
Krt26
|
keratin 26 |
| chr19_+_10018914 | 0.21 |
ENSMUST00000115995.4
|
Fads3
|
fatty acid desaturase 3 |
| chr7_-_19192672 | 0.20 |
ENSMUST00000239292.2
ENSMUST00000085715.7 |
Mark4
|
MAP/microtubule affinity regulating kinase 4 |
| chr4_-_116982804 | 0.20 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr8_-_120904179 | 0.18 |
ENSMUST00000048786.13
|
Cibar2
|
CBY1 interacting BAR domain containing 2 |
| chr7_-_128062916 | 0.18 |
ENSMUST00000205835.2
|
Tial1
|
Tia1 cytotoxic granule-associated RNA binding protein-like 1 |
| chr2_+_164627913 | 0.16 |
ENSMUST00000152471.2
|
Snx21
|
sorting nexin family member 21 |
| chr2_+_164627743 | 0.16 |
ENSMUST00000174070.8
ENSMUST00000172577.8 ENSMUST00000056181.7 |
Snx21
|
sorting nexin family member 21 |
| chr19_+_41818409 | 0.14 |
ENSMUST00000087155.5
|
Frat1
|
frequently rearranged in advanced T cell lymphomas |
| chr13_+_41403317 | 0.14 |
ENSMUST00000165561.4
|
Smim13
|
small integral membrane protein 13 |
| chr17_+_11059248 | 0.12 |
ENSMUST00000191124.7
|
Prkn
|
parkin RBR E3 ubiquitin protein ligase |
| chr11_+_102727122 | 0.10 |
ENSMUST00000021302.15
ENSMUST00000107072.2 |
Higd1b
|
HIG1 domain family, member 1B |
| chr6_-_124391994 | 0.10 |
ENSMUST00000035861.6
ENSMUST00000112532.8 ENSMUST00000080557.12 |
Pex5
|
peroxisomal biogenesis factor 5 |
| chr4_-_148711211 | 0.10 |
ENSMUST00000186947.7
ENSMUST00000105699.8 |
Tardbp
|
TAR DNA binding protein |
| chr3_+_94882038 | 0.08 |
ENSMUST00000072287.12
|
Pi4kb
|
phosphatidylinositol 4-kinase beta |
| chr7_+_18737944 | 0.08 |
ENSMUST00000053713.5
|
Irf2bp1
|
interferon regulatory factor 2 binding protein 1 |
| chr17_-_25652750 | 0.07 |
ENSMUST00000159610.8
ENSMUST00000159048.8 ENSMUST00000078496.12 |
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr7_-_79443536 | 0.05 |
ENSMUST00000032760.6
|
Mesp1
|
mesoderm posterior 1 |
| chr7_-_25454177 | 0.05 |
ENSMUST00000206832.2
|
Hnrnpul1
|
heterogeneous nuclear ribonucleoprotein U-like 1 |
| chr9_-_99758200 | 0.03 |
ENSMUST00000054819.10
|
Sox14
|
SRY (sex determining region Y)-box 14 |
| chrX_+_57076359 | 0.01 |
ENSMUST00000088631.11
ENSMUST00000088629.4 |
Zic3
|
zinc finger protein of the cerebellum 3 |
| chr15_-_98676007 | 0.00 |
ENSMUST00000226655.2
ENSMUST00000228546.2 ENSMUST00000023732.12 ENSMUST00000226610.2 |
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.2 | 2.0 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.1 | 1.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.1 | 0.5 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.7 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.1 | 0.9 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.1 | 1.9 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 1.5 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.2 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 1.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 0.8 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.1 | 0.5 | GO:0060022 | hard palate development(GO:0060022) |
| 0.1 | 2.2 | GO:1900273 | positive regulation of long-term synaptic potentiation(GO:1900273) |
| 0.1 | 1.9 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.0 | 2.0 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) |
| 0.0 | 0.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.0 | 1.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0016561 | protein import into peroxisome matrix, translocation(GO:0016561) |
| 0.0 | 0.4 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.3 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.3 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.3 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.8 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.7 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 2.3 | GO:0097194 | execution phase of apoptosis(GO:0097194) |
| 0.0 | 1.3 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.2 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.6 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.2 | 2.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 0.7 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 1.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 2.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 2.2 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.8 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 1.9 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 2.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.7 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.3 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.3 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.0 | 0.3 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.3 | GO:0070905 | serine binding(GO:0070905) |
| 0.2 | 0.7 | GO:0016492 | G-protein coupled neurotensin receptor activity(GO:0016492) |
| 0.2 | 0.5 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.1 | 0.9 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.9 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 2.6 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 1.3 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 2.3 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 0.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 1.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.8 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 1.9 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.6 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.9 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.9 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.9 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.9 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |