Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
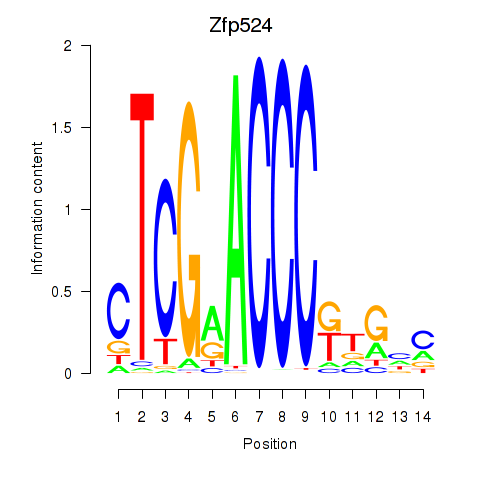
Results for Zfp524
Z-value: 1.56
Transcription factors associated with Zfp524
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp524
|
ENSMUSG00000051184.8 | Zfp524 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp524 | mm39_v1_chr7_+_5017414_5017453 | 0.03 | 7.7e-01 | Click! |
Activity profile of Zfp524 motif
Sorted Z-values of Zfp524 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp524
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_149671012 | 13.43 |
ENSMUST00000039144.7
|
Clstn1
|
calsyntenin 1 |
| chr1_-_136161850 | 9.91 |
ENSMUST00000120339.8
|
Inava
|
innate immunity activator |
| chr2_+_118603247 | 8.26 |
ENSMUST00000061360.4
ENSMUST00000130293.8 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chr12_+_36042899 | 8.10 |
ENSMUST00000020898.12
|
Agr2
|
anterior gradient 2 |
| chr19_+_54033681 | 7.18 |
ENSMUST00000237285.2
|
Adra2a
|
adrenergic receptor, alpha 2a |
| chr12_-_76869282 | 7.08 |
ENSMUST00000021459.14
|
Rab15
|
RAB15, member RAS oncogene family |
| chr2_+_151414524 | 7.02 |
ENSMUST00000028950.9
|
Sdcbp2
|
syndecan binding protein (syntenin) 2 |
| chr4_-_4138432 | 6.63 |
ENSMUST00000070375.8
|
Penk
|
preproenkephalin |
| chr8_+_124059414 | 6.53 |
ENSMUST00000010298.7
|
Spire2
|
spire type actin nucleation factor 2 |
| chr17_+_69382694 | 6.42 |
ENSMUST00000225062.2
|
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr7_-_126651847 | 6.06 |
ENSMUST00000205424.2
|
Zg16
|
zymogen granule protein 16 |
| chr8_+_124138163 | 6.05 |
ENSMUST00000071134.4
ENSMUST00000212743.2 |
Tubb3
|
tubulin, beta 3 class III |
| chr14_+_14090981 | 6.05 |
ENSMUST00000022269.7
|
Oit1
|
oncoprotein induced transcript 1 |
| chr7_-_126651120 | 5.93 |
ENSMUST00000051122.7
|
Zg16
|
zymogen granule protein 16 |
| chr7_+_29991101 | 5.91 |
ENSMUST00000150892.2
ENSMUST00000126216.2 ENSMUST00000014065.16 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr2_-_119590776 | 5.80 |
ENSMUST00000082130.13
ENSMUST00000028759.13 |
Ltk
|
leukocyte tyrosine kinase |
| chr13_-_6686686 | 5.75 |
ENSMUST00000136585.2
|
Pfkp
|
phosphofructokinase, platelet |
| chr9_+_22011488 | 5.69 |
ENSMUST00000213607.2
|
Cnn1
|
calponin 1 |
| chr10_-_62258195 | 5.68 |
ENSMUST00000020277.9
|
Hkdc1
|
hexokinase domain containing 1 |
| chr12_-_76869510 | 5.64 |
ENSMUST00000154765.8
|
Rab15
|
RAB15, member RAS oncogene family |
| chr9_-_75518585 | 5.54 |
ENSMUST00000098552.10
ENSMUST00000064433.11 |
Tmod2
|
tropomodulin 2 |
| chrX_-_7884688 | 5.53 |
ENSMUST00000033503.3
|
Glod5
|
glyoxalase domain containing 5 |
| chr2_-_113588983 | 5.37 |
ENSMUST00000099575.4
|
Grem1
|
gremlin 1, DAN family BMP antagonist |
| chr3_-_98247237 | 5.35 |
ENSMUST00000065793.12
|
Phgdh
|
3-phosphoglycerate dehydrogenase |
| chr19_+_10366753 | 5.32 |
ENSMUST00000169121.9
ENSMUST00000076968.11 ENSMUST00000235479.2 ENSMUST00000223586.2 ENSMUST00000235784.2 ENSMUST00000224135.3 ENSMUST00000225452.3 ENSMUST00000237366.2 |
Syt7
|
synaptotagmin VII |
| chr4_+_43059028 | 5.30 |
ENSMUST00000163653.8
ENSMUST00000107952.9 ENSMUST00000107953.9 |
Unc13b
|
unc-13 homolog B |
| chr11_+_70396070 | 5.20 |
ENSMUST00000019063.3
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr9_+_15150341 | 5.17 |
ENSMUST00000034413.8
|
Vstm5
|
V-set and transmembrane domain containing 5 |
| chr17_+_48037758 | 5.00 |
ENSMUST00000024782.12
ENSMUST00000144955.2 |
Pgc
|
progastricsin (pepsinogen C) |
| chr9_+_75213570 | 4.87 |
ENSMUST00000213990.2
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5 |
| chr19_-_46315543 | 4.85 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chrX_+_7504913 | 4.72 |
ENSMUST00000128890.2
|
Syp
|
synaptophysin |
| chr4_+_62398262 | 4.49 |
ENSMUST00000030088.12
ENSMUST00000107449.4 |
Bspry
|
B-box and SPRY domain containing |
| chr7_+_140658101 | 4.43 |
ENSMUST00000106039.9
|
Pkp3
|
plakophilin 3 |
| chr13_-_92268156 | 4.42 |
ENSMUST00000151408.9
ENSMUST00000216219.2 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr1_-_135846937 | 4.41 |
ENSMUST00000027667.13
|
Pkp1
|
plakophilin 1 |
| chr14_+_14091030 | 4.31 |
ENSMUST00000224529.2
|
Oit1
|
oncoprotein induced transcript 1 |
| chr1_+_93613405 | 4.26 |
ENSMUST00000201863.4
|
Bok
|
BCL2-related ovarian killer |
| chr15_-_79048674 | 4.24 |
ENSMUST00000230261.2
ENSMUST00000040019.5 |
Sox10
|
SRY (sex determining region Y)-box 10 |
| chr1_+_93613295 | 4.23 |
ENSMUST00000027499.13
|
Bok
|
BCL2-related ovarian killer |
| chr13_-_10410857 | 4.18 |
ENSMUST00000187510.7
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chrX_+_72716756 | 4.14 |
ENSMUST00000033752.14
ENSMUST00000114467.9 |
Slc6a8
|
solute carrier family 6 (neurotransmitter transporter, creatine), member 8 |
| chr13_-_96807346 | 4.05 |
ENSMUST00000022176.15
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr17_+_29312737 | 4.05 |
ENSMUST00000023829.8
ENSMUST00000233296.2 |
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr13_-_96807326 | 4.00 |
ENSMUST00000169196.8
|
Hmgcr
|
3-hydroxy-3-methylglutaryl-Coenzyme A reductase |
| chr5_-_44256528 | 3.94 |
ENSMUST00000196178.2
ENSMUST00000197750.5 |
Prom1
|
prominin 1 |
| chr1_+_55445033 | 3.93 |
ENSMUST00000042986.10
|
Plcl1
|
phospholipase C-like 1 |
| chr17_+_35286293 | 3.91 |
ENSMUST00000173478.2
ENSMUST00000174876.2 |
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr17_-_57394718 | 3.90 |
ENSMUST00000071135.6
|
Tubb4a
|
tubulin, beta 4A class IVA |
| chr8_+_27513839 | 3.82 |
ENSMUST00000209563.2
ENSMUST00000209520.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr7_-_44318710 | 3.76 |
ENSMUST00000208131.2
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr9_-_75518387 | 3.75 |
ENSMUST00000215462.2
ENSMUST00000217233.2 ENSMUST00000215614.2 |
Tmod2
|
tropomodulin 2 |
| chr8_-_106198112 | 3.75 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr12_-_36092475 | 3.74 |
ENSMUST00000020896.17
|
Tspan13
|
tetraspanin 13 |
| chr11_+_9068507 | 3.66 |
ENSMUST00000164791.8
ENSMUST00000130522.8 |
Upp1
|
uridine phosphorylase 1 |
| chr2_-_179976458 | 3.64 |
ENSMUST00000015771.3
|
Gata5
|
GATA binding protein 5 |
| chr16_+_33338648 | 3.63 |
ENSMUST00000122427.8
ENSMUST00000059056.15 |
Slc12a8
|
solute carrier family 12 (potassium/chloride transporters), member 8 |
| chr11_+_9068134 | 3.61 |
ENSMUST00000170444.8
|
Upp1
|
uridine phosphorylase 1 |
| chr10_-_78554104 | 3.60 |
ENSMUST00000005488.9
|
Casp14
|
caspase 14 |
| chr11_+_9068098 | 3.56 |
ENSMUST00000020677.8
ENSMUST00000101525.9 |
Upp1
|
uridine phosphorylase 1 |
| chr4_+_43058938 | 3.46 |
ENSMUST00000207569.2
ENSMUST00000079978.13 |
Unc13b
|
unc-13 homolog B |
| chr5_+_76805520 | 3.45 |
ENSMUST00000128112.8
|
Cracd
|
capping protein inhibiting regulator of actin |
| chr6_+_48881913 | 3.45 |
ENSMUST00000162948.7
ENSMUST00000167529.3 |
Aoc1
|
amine oxidase, copper-containing 1 |
| chr11_+_69917640 | 3.42 |
ENSMUST00000135916.9
ENSMUST00000232659.2 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr15_+_103411461 | 3.33 |
ENSMUST00000023132.5
|
Pde1b
|
phosphodiesterase 1B, Ca2+-calmodulin dependent |
| chr11_-_20282684 | 3.28 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr2_-_24653059 | 3.23 |
ENSMUST00000100348.10
ENSMUST00000041342.12 ENSMUST00000114447.8 ENSMUST00000102939.9 ENSMUST00000070864.14 |
Cacna1b
|
calcium channel, voltage-dependent, N type, alpha 1B subunit |
| chr16_+_17945471 | 3.19 |
ENSMUST00000059589.6
|
Rtn4r
|
reticulon 4 receptor |
| chr16_-_97412169 | 3.17 |
ENSMUST00000232141.2
ENSMUST00000000395.8 |
Tmprss2
|
transmembrane protease, serine 2 |
| chr5_-_44256562 | 3.16 |
ENSMUST00000165909.8
|
Prom1
|
prominin 1 |
| chr3_+_96069271 | 3.15 |
ENSMUST00000054356.16
|
Mtmr11
|
myotubularin related protein 11 |
| chr5_-_131645437 | 3.15 |
ENSMUST00000161804.9
|
Auts2
|
autism susceptibility candidate 2 |
| chr4_+_33924632 | 3.15 |
ENSMUST00000057188.7
|
Cnr1
|
cannabinoid receptor 1 (brain) |
| chr13_-_97235745 | 3.12 |
ENSMUST00000071118.7
|
Gm6169
|
predicted gene 6169 |
| chr17_+_69569184 | 3.12 |
ENSMUST00000224951.2
|
Epb41l3
|
erythrocyte membrane protein band 4.1 like 3 |
| chr1_-_135846858 | 3.09 |
ENSMUST00000163260.8
|
Pkp1
|
plakophilin 1 |
| chr4_-_129015027 | 3.08 |
ENSMUST00000030572.10
|
Hpca
|
hippocalcin |
| chr10_-_62067026 | 3.08 |
ENSMUST00000047883.11
|
Tspan15
|
tetraspanin 15 |
| chr12_+_113115632 | 3.07 |
ENSMUST00000006523.12
ENSMUST00000200553.2 |
Crip1
|
cysteine-rich protein 1 (intestinal) |
| chr10_+_75409282 | 3.06 |
ENSMUST00000006508.10
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr2_+_71617266 | 3.06 |
ENSMUST00000112101.8
ENSMUST00000028522.10 |
Itga6
|
integrin alpha 6 |
| chr3_+_124114504 | 3.05 |
ENSMUST00000058994.6
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr8_+_120840883 | 2.99 |
ENSMUST00000108948.8
ENSMUST00000034281.13 ENSMUST00000108951.8 |
6430548M08Rik
|
RIKEN cDNA 6430548M08 gene |
| chr15_+_76763431 | 2.98 |
ENSMUST00000023179.7
|
Zfp7
|
zinc finger protein 7 |
| chr17_+_23879448 | 2.97 |
ENSMUST00000062967.10
|
Bicdl2
|
BICD family like cargo adaptor 2 |
| chr9_+_99494550 | 2.94 |
ENSMUST00000042553.8
|
A4gnt
|
alpha-1,4-N-acetylglucosaminyltransferase |
| chr13_-_34119937 | 2.94 |
ENSMUST00000170991.8
ENSMUST00000171252.8 ENSMUST00000164627.8 ENSMUST00000017188.14 ENSMUST00000167163.8 ENSMUST00000043552.17 |
Serpinb6a
|
serine (or cysteine) peptidase inhibitor, clade B, member 6a |
| chr3_-_84167119 | 2.93 |
ENSMUST00000107691.8
|
Trim2
|
tripartite motif-containing 2 |
| chr2_-_144369261 | 2.84 |
ENSMUST00000163701.2
ENSMUST00000081982.12 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr7_+_16678568 | 2.84 |
ENSMUST00000094807.6
|
Pnmal2
|
PNMA-like 2 |
| chr18_+_67221287 | 2.84 |
ENSMUST00000025402.15
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr9_+_75922137 | 2.83 |
ENSMUST00000008052.13
ENSMUST00000183425.8 ENSMUST00000183979.8 ENSMUST00000117981.3 |
Hmgcll1
|
3-hydroxymethyl-3-methylglutaryl-Coenzyme A lyase-like 1 |
| chr13_-_34119816 | 2.81 |
ENSMUST00000168350.8
ENSMUST00000167260.8 |
Serpinb6a
|
serine (or cysteine) peptidase inhibitor, clade B, member 6a |
| chr2_-_180776920 | 2.81 |
ENSMUST00000197015.5
ENSMUST00000103050.10 ENSMUST00000081528.13 ENSMUST00000049792.15 ENSMUST00000103048.10 ENSMUST00000103047.10 ENSMUST00000149964.9 |
Kcnq2
|
potassium voltage-gated channel, subfamily Q, member 2 |
| chrX_+_135723420 | 2.81 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr5_+_37495801 | 2.78 |
ENSMUST00000056365.9
|
Evc2
|
EvC ciliary complex subunit 2 |
| chr9_-_66032134 | 2.76 |
ENSMUST00000034946.15
|
Snx1
|
sorting nexin 1 |
| chr9_-_99302205 | 2.76 |
ENSMUST00000123771.2
|
Mras
|
muscle and microspikes RAS |
| chr17_+_27160203 | 2.73 |
ENSMUST00000194598.6
|
Syngap1
|
synaptic Ras GTPase activating protein 1 homolog (rat) |
| chr7_-_120581535 | 2.71 |
ENSMUST00000033169.9
|
Cdr2
|
cerebellar degeneration-related 2 |
| chr2_-_144173615 | 2.71 |
ENSMUST00000103171.10
|
Ovol2
|
ovo like zinc finger 2 |
| chr2_+_149672760 | 2.70 |
ENSMUST00000109934.2
ENSMUST00000140870.8 |
Syndig1
|
synapse differentiation inducing 1 |
| chr7_-_110581652 | 2.66 |
ENSMUST00000005751.13
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr3_-_122413361 | 2.65 |
ENSMUST00000239148.2
ENSMUST00000162409.8 ENSMUST00000162947.3 |
Fnbp1l
|
formin binding protein 1-like |
| chr3_-_92564232 | 2.54 |
ENSMUST00000029531.4
|
Lce1b
|
late cornified envelope 1B |
| chr18_-_64794338 | 2.54 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr7_+_99184858 | 2.53 |
ENSMUST00000032995.15
ENSMUST00000162404.8 |
Arrb1
|
arrestin, beta 1 |
| chr2_-_148574353 | 2.49 |
ENSMUST00000028926.13
|
Napb
|
N-ethylmaleimide sensitive fusion protein attachment protein beta |
| chr15_+_98532866 | 2.48 |
ENSMUST00000230490.2
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr2_+_14393127 | 2.48 |
ENSMUST00000114731.8
ENSMUST00000082290.8 |
Slc39a12
|
solute carrier family 39 (zinc transporter), member 12 |
| chr2_-_173117936 | 2.38 |
ENSMUST00000139306.2
|
Pmepa1
|
prostate transmembrane protein, androgen induced 1 |
| chr11_+_83300481 | 2.37 |
ENSMUST00000175848.8
ENSMUST00000108140.10 |
Rasl10b
|
RAS-like, family 10, member B |
| chr5_-_118382926 | 2.37 |
ENSMUST00000117177.8
ENSMUST00000133372.2 ENSMUST00000154786.8 ENSMUST00000121369.8 |
Rnft2
|
ring finger protein, transmembrane 2 |
| chr2_-_168583670 | 2.33 |
ENSMUST00000029060.11
|
Atp9a
|
ATPase, class II, type 9A |
| chr5_+_140491305 | 2.32 |
ENSMUST00000043050.9
ENSMUST00000124142.2 |
Chst12
|
carbohydrate sulfotransferase 12 |
| chr9_+_108708939 | 2.32 |
ENSMUST00000192235.2
|
Celsr3
|
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr10_+_13376745 | 2.32 |
ENSMUST00000060212.13
ENSMUST00000121465.3 |
Fuca2
|
fucosidase, alpha-L- 2, plasma |
| chr7_-_133378410 | 2.31 |
ENSMUST00000130182.2
ENSMUST00000106139.8 |
Dhx32
|
DEAH (Asp-Glu-Ala-His) box polypeptide 32 |
| chr14_+_115329676 | 2.31 |
ENSMUST00000176912.8
ENSMUST00000175665.8 |
Gpc5
|
glypican 5 |
| chr16_-_55659194 | 2.29 |
ENSMUST00000096026.9
ENSMUST00000036273.13 ENSMUST00000114457.8 |
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr10_+_81093110 | 2.28 |
ENSMUST00000117488.8
ENSMUST00000105328.10 ENSMUST00000121205.8 |
Matk
|
megakaryocyte-associated tyrosine kinase |
| chr4_-_152402915 | 2.28 |
ENSMUST00000170820.3
ENSMUST00000076183.12 |
Rnf207
|
ring finger protein 207 |
| chr17_+_6307123 | 2.26 |
ENSMUST00000232383.2
|
Tmem181a
|
transmembrane protein 181A |
| chr5_-_124939428 | 2.25 |
ENSMUST00000036206.14
|
Ccdc92
|
coiled-coil domain containing 92 |
| chr3_+_131270529 | 2.24 |
ENSMUST00000029666.14
|
Papss1
|
3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| chr7_-_19363280 | 2.23 |
ENSMUST00000094762.10
ENSMUST00000049912.15 ENSMUST00000098754.5 |
Relb
|
avian reticuloendotheliosis viral (v-rel) oncogene related B |
| chr11_-_109188892 | 2.22 |
ENSMUST00000106706.8
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr7_+_4467730 | 2.21 |
ENSMUST00000086372.8
ENSMUST00000169820.8 ENSMUST00000163893.8 ENSMUST00000170635.2 |
Eps8l1
|
EPS8-like 1 |
| chr5_+_137551774 | 2.21 |
ENSMUST00000136088.8
ENSMUST00000139395.8 |
Actl6b
|
actin-like 6B |
| chr10_+_40759468 | 2.19 |
ENSMUST00000019975.14
|
Wasf1
|
WASP family, member 1 |
| chr17_+_35284315 | 2.19 |
ENSMUST00000173207.8
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr13_-_49301407 | 2.19 |
ENSMUST00000162581.8
ENSMUST00000110097.9 ENSMUST00000049265.15 ENSMUST00000035538.13 ENSMUST00000110096.8 ENSMUST00000091623.10 |
Wnk2
|
WNK lysine deficient protein kinase 2 |
| chr11_-_109188947 | 2.18 |
ENSMUST00000020920.10
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr17_-_46455082 | 2.17 |
ENSMUST00000024762.3
|
Rsph9
|
radial spoke head 9 homolog (Chlamydomonas) |
| chr7_+_16625679 | 2.17 |
ENSMUST00000078182.6
|
Gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr2_+_149672708 | 2.17 |
ENSMUST00000109935.8
|
Syndig1
|
synapse differentiation inducing 1 |
| chr12_-_115916604 | 2.14 |
ENSMUST00000196991.2
|
Ighv1-82
|
immunoglobulin heavy variable 1-82 |
| chr5_-_114518520 | 2.12 |
ENSMUST00000001125.6
|
Kctd10
|
potassium channel tetramerisation domain containing 10 |
| chr4_+_127062924 | 2.12 |
ENSMUST00000046659.14
|
Dlgap3
|
DLG associated protein 3 |
| chr2_-_102282844 | 2.10 |
ENSMUST00000099678.5
|
Fjx1
|
four jointed box 1 |
| chr19_-_57107413 | 2.07 |
ENSMUST00000111528.8
ENSMUST00000111529.8 ENSMUST00000104902.9 |
Ablim1
|
actin-binding LIM protein 1 |
| chr12_-_107969673 | 2.07 |
ENSMUST00000109887.8
ENSMUST00000109891.3 |
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr19_+_45006552 | 2.03 |
ENSMUST00000237043.2
ENSMUST00000178087.3 |
Lzts2
|
leucine zipper, putative tumor suppressor 2 |
| chr7_+_73025243 | 2.01 |
ENSMUST00000119206.3
ENSMUST00000094312.12 |
Rgma
|
repulsive guidance molecule family member A |
| chr12_+_24881582 | 1.96 |
ENSMUST00000221952.2
ENSMUST00000078902.8 ENSMUST00000110942.11 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr18_+_67221355 | 1.96 |
ENSMUST00000237445.2
ENSMUST00000237985.2 ENSMUST00000236551.2 |
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr7_+_29991366 | 1.95 |
ENSMUST00000144508.2
|
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr7_-_110581376 | 1.94 |
ENSMUST00000154466.2
|
Irag1
|
inositol 1,4,5-triphosphate receptor associated 1 |
| chr3_+_65573644 | 1.94 |
ENSMUST00000099075.4
ENSMUST00000177434.3 |
Lekr1
|
leucine, glutamate and lysine rich 1 |
| chr5_-_25200745 | 1.93 |
ENSMUST00000076306.12
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr1_+_135074520 | 1.92 |
ENSMUST00000027684.11
|
Arl8a
|
ADP-ribosylation factor-like 8A |
| chr17_-_36220924 | 1.91 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr1_-_98023321 | 1.90 |
ENSMUST00000058762.15
ENSMUST00000097625.10 |
Pam
|
peptidylglycine alpha-amidating monooxygenase |
| chr11_-_100650768 | 1.90 |
ENSMUST00000107363.3
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr7_+_127345909 | 1.90 |
ENSMUST00000033081.14
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr2_+_29855572 | 1.89 |
ENSMUST00000113719.9
ENSMUST00000113717.8 ENSMUST00000113741.8 ENSMUST00000100225.9 ENSMUST00000095083.11 ENSMUST00000046257.14 |
Sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr1_-_5140504 | 1.87 |
ENSMUST00000147158.2
ENSMUST00000118000.8 |
Rgs20
|
regulator of G-protein signaling 20 |
| chr8_-_111629074 | 1.85 |
ENSMUST00000041382.7
|
Fcsk
|
fucose kinase |
| chr2_+_80447389 | 1.85 |
ENSMUST00000028384.5
|
Dusp19
|
dual specificity phosphatase 19 |
| chr5_+_143636977 | 1.84 |
ENSMUST00000110727.2
|
Cyth3
|
cytohesin 3 |
| chr3_+_30910163 | 1.83 |
ENSMUST00000108258.8
ENSMUST00000147697.2 |
Gpr160
|
G protein-coupled receptor 160 |
| chr19_-_46314945 | 1.83 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr2_+_154498917 | 1.83 |
ENSMUST00000044277.10
|
Chmp4b
|
charged multivesicular body protein 4B |
| chr10_+_24099415 | 1.82 |
ENSMUST00000095784.3
|
Moxd1
|
monooxygenase, DBH-like 1 |
| chr2_+_71617402 | 1.82 |
ENSMUST00000238991.2
|
Itga6
|
integrin alpha 6 |
| chr4_-_135221926 | 1.82 |
ENSMUST00000102549.10
|
Nipal3
|
NIPA-like domain containing 3 |
| chr10_+_125802084 | 1.81 |
ENSMUST00000074807.8
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr5_+_81169430 | 1.81 |
ENSMUST00000121707.8
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr4_+_28813152 | 1.80 |
ENSMUST00000108194.9
ENSMUST00000108191.2 |
Epha7
|
Eph receptor A7 |
| chr1_+_185064339 | 1.79 |
ENSMUST00000027916.13
ENSMUST00000210277.2 ENSMUST00000151769.2 ENSMUST00000110965.2 |
Bpnt1
|
3'(2'), 5'-bisphosphate nucleotidase 1 |
| chr11_-_20781009 | 1.78 |
ENSMUST00000047028.9
|
Lgalsl
|
lectin, galactoside binding-like |
| chr11_-_120472022 | 1.76 |
ENSMUST00000067936.6
|
Arhgdia
|
Rho GDP dissociation inhibitor (GDI) alpha |
| chr17_+_56277451 | 1.75 |
ENSMUST00000044216.8
ENSMUST00000225145.2 |
Shd
|
src homology 2 domain-containing transforming protein D |
| chrX_+_105070907 | 1.74 |
ENSMUST00000055941.7
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr16_-_84970580 | 1.74 |
ENSMUST00000227737.2
ENSMUST00000226801.2 |
App
|
amyloid beta (A4) precursor protein |
| chr19_-_6947112 | 1.71 |
ENSMUST00000025912.10
|
Plcb3
|
phospholipase C, beta 3 |
| chr8_-_13940234 | 1.68 |
ENSMUST00000033839.9
|
Coprs
|
coordinator of PRMT5, differentiation stimulator |
| chr7_-_65020955 | 1.65 |
ENSMUST00000102592.10
|
Tjp1
|
tight junction protein 1 |
| chr4_+_43875524 | 1.65 |
ENSMUST00000030198.7
|
Reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr11_-_97464866 | 1.64 |
ENSMUST00000207653.2
ENSMUST00000107593.8 |
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr9_-_75518357 | 1.63 |
ENSMUST00000213565.2
|
Tmod2
|
tropomodulin 2 |
| chr11_+_42310557 | 1.63 |
ENSMUST00000007797.10
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2 |
| chr5_-_74229021 | 1.62 |
ENSMUST00000119154.8
ENSMUST00000068058.14 |
Usp46
|
ubiquitin specific peptidase 46 |
| chr8_+_27513819 | 1.61 |
ENSMUST00000033873.9
ENSMUST00000211043.2 |
Erlin2
|
ER lipid raft associated 2 |
| chr4_+_28813125 | 1.61 |
ENSMUST00000080934.11
ENSMUST00000029964.12 |
Epha7
|
Eph receptor A7 |
| chr3_-_122910662 | 1.60 |
ENSMUST00000051443.8
|
Synpo2
|
synaptopodin 2 |
| chr15_+_76544058 | 1.60 |
ENSMUST00000230451.2
|
Kifc2
|
kinesin family member C2 |
| chr15_-_79967543 | 1.59 |
ENSMUST00000081650.15
|
Rpl3
|
ribosomal protein L3 |
| chr7_-_118594365 | 1.58 |
ENSMUST00000008878.10
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B |
| chr6_-_136150076 | 1.55 |
ENSMUST00000053880.13
|
Grin2b
|
glutamate receptor, ionotropic, NMDA2B (epsilon 2) |
| chrX_+_105070865 | 1.54 |
ENSMUST00000113557.8
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr7_+_19242634 | 1.54 |
ENSMUST00000002112.15
ENSMUST00000108455.8 |
Trappc6a
|
trafficking protein particle complex 6A |
| chr7_-_45829353 | 1.53 |
ENSMUST00000033123.8
|
Abcc8
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 8 |
| chr4_+_100926170 | 1.53 |
ENSMUST00000106955.2
ENSMUST00000038463.15 |
Raver2
|
ribonucleoprotein, PTB-binding 2 |
| chr2_+_149672814 | 1.53 |
ENSMUST00000137280.2
ENSMUST00000149705.2 |
Syndig1
|
synapse differentiation inducing 1 |
| chr9_-_99450693 | 1.53 |
ENSMUST00000185524.7
ENSMUST00000186049.2 |
Armc8
|
armadillo repeat containing 8 |
| chr2_-_132095146 | 1.53 |
ENSMUST00000028817.7
|
Pcna
|
proliferating cell nuclear antigen |
| chr2_-_121273054 | 1.52 |
ENSMUST00000116432.2
|
Ell3
|
elongation factor RNA polymerase II-like 3 |
| chr13_-_92667321 | 1.52 |
ENSMUST00000022217.9
|
Zfyve16
|
zinc finger, FYVE domain containing 16 |
| chr15_-_96540760 | 1.52 |
ENSMUST00000088452.11
|
Slc38a1
|
solute carrier family 38, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.6 | 10.8 | GO:0046108 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 2.8 | 16.6 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 1.8 | 5.3 | GO:1990926 | short-term synaptic potentiation(GO:1990926) |
| 1.8 | 8.8 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 1.6 | 6.5 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 1.6 | 4.7 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 1.4 | 7.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 1.4 | 4.1 | GO:0015881 | creatine transport(GO:0015881) |
| 1.3 | 5.4 | GO:1900158 | negative regulation of monocyte chemotaxis(GO:0090027) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 1.2 | 3.5 | GO:0015898 | amiloride transport(GO:0015898) cellular response to copper ion starvation(GO:0035874) response to azide(GO:0097184) cellular response to azide(GO:0097185) |
| 1.1 | 3.3 | GO:0051542 | elastin biosynthetic process(GO:0051542) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 1.1 | 3.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 1.1 | 7.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 1.1 | 5.3 | GO:0006566 | threonine metabolic process(GO:0006566) |
| 1.1 | 4.2 | GO:0099541 | trans-synaptic signaling by lipid(GO:0099541) trans-synaptic signaling by endocannabinoid(GO:0099542) |
| 1.0 | 3.1 | GO:0030827 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.0 | 7.2 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 1.0 | 5.0 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.9 | 7.5 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.9 | 2.7 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.9 | 3.6 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.9 | 4.4 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.9 | 7.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.9 | 9.5 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.8 | 10.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.8 | 4.2 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.8 | 3.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.8 | 2.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.8 | 2.3 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.7 | 5.9 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.7 | 2.8 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.7 | 2.1 | GO:0097534 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.7 | 2.0 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.6 | 5.0 | GO:0031179 | peptide modification(GO:0031179) |
| 0.6 | 1.9 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 0.6 | 1.8 | GO:0046333 | octopamine biosynthetic process(GO:0006589) octopamine metabolic process(GO:0046333) |
| 0.6 | 1.8 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.6 | 2.4 | GO:0034036 | purine ribonucleoside bisphosphate biosynthetic process(GO:0034036) 3'-phosphoadenosine 5'-phosphosulfate biosynthetic process(GO:0050428) |
| 0.6 | 1.7 | GO:0071874 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.5 | 2.7 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.5 | 5.7 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.5 | 1.5 | GO:1901082 | regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 0.5 | 0.5 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.5 | 5.8 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.5 | 2.3 | GO:0036515 | serotonergic neuron axon guidance(GO:0036515) |
| 0.5 | 1.4 | GO:0051455 | attachment of spindle microtubules to kinetochore involved in homologous chromosome segregation(GO:0051455) microtubule cytoskeleton organization involved in homologous chromosome segregation(GO:0090172) |
| 0.5 | 0.9 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.4 | 3.6 | GO:0070268 | cornification(GO:0070268) |
| 0.4 | 4.9 | GO:0035878 | nail development(GO:0035878) |
| 0.4 | 3.1 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.4 | 3.9 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.4 | 4.7 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.4 | 1.7 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.4 | 3.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.4 | 2.9 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.4 | 1.6 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.4 | 1.2 | GO:0090149 | mitochondrial membrane fission(GO:0090149) regulation of peroxisome organization(GO:1900063) |
| 0.4 | 12.8 | GO:0010666 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.4 | 1.5 | GO:0006272 | leading strand elongation(GO:0006272) mitotic telomere maintenance via semi-conservative replication(GO:1902990) |
| 0.4 | 3.7 | GO:0099628 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.4 | 1.5 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.4 | 3.6 | GO:1990416 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.4 | 1.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.4 | 6.2 | GO:0002118 | aggressive behavior(GO:0002118) |
| 0.4 | 1.1 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.4 | 4.6 | GO:0036005 | response to macrophage colony-stimulating factor(GO:0036005) cellular response to macrophage colony-stimulating factor stimulus(GO:0036006) |
| 0.4 | 3.2 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.3 | 2.5 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.3 | 5.1 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.3 | 1.5 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.3 | 6.4 | GO:0097091 | synaptic vesicle clustering(GO:0097091) |
| 0.3 | 1.8 | GO:0090611 | ubiquitin-independent protein catabolic process via the multivesicular body sorting pathway(GO:0090611) |
| 0.3 | 2.7 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.3 | 3.2 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.3 | 0.3 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.3 | 3.0 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.3 | 4.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.3 | 2.5 | GO:0010807 | regulation of synaptic vesicle priming(GO:0010807) |
| 0.3 | 0.8 | GO:2000417 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.3 | 13.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.3 | 2.3 | GO:0006004 | fucose metabolic process(GO:0006004) |
| 0.3 | 1.8 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.2 | 3.2 | GO:0001956 | positive regulation of neurotransmitter secretion(GO:0001956) |
| 0.2 | 2.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.2 | 0.7 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.2 | 1.5 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.2 | 1.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.2 | 5.7 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.2 | 1.5 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.2 | 0.9 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.2 | 1.5 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.2 | 7.8 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.2 | 0.8 | GO:0000390 | spliceosomal complex disassembly(GO:0000390) |
| 0.2 | 2.1 | GO:0030397 | membrane disassembly(GO:0030397) nuclear envelope disassembly(GO:0051081) |
| 0.2 | 1.7 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.2 | 2.0 | GO:1903551 | regulation of extracellular exosome assembly(GO:1903551) |
| 0.2 | 0.6 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.2 | 0.8 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 0.2 | GO:0072106 | regulation of ureteric bud formation(GO:0072106) positive regulation of ureteric bud formation(GO:0072107) |
| 0.2 | 0.6 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.2 | 3.9 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.2 | 1.2 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.2 | 0.6 | GO:0030860 | regulation of polarized epithelial cell differentiation(GO:0030860) |
| 0.2 | 2.2 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.2 | 1.1 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.2 | 1.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 1.6 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.2 | 2.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.2 | 1.2 | GO:1901908 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.2 | 3.8 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.2 | 0.8 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.2 | 1.5 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.2 | 10.1 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.2 | 2.7 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.2 | 1.3 | GO:0033058 | directional locomotion(GO:0033058) |
| 0.2 | 3.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.2 | 2.5 | GO:0061577 | calcium ion transmembrane transport via high voltage-gated calcium channel(GO:0061577) |
| 0.2 | 0.8 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.2 | 0.7 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.2 | 4.4 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.2 | 1.7 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 2.3 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 0.7 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.1 | 2.4 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 1.2 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 1.0 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.1 | 0.4 | GO:0006429 | leucyl-tRNA aminoacylation(GO:0006429) |
| 0.1 | 0.3 | GO:1990051 | activation of protein kinase C activity(GO:1990051) |
| 0.1 | 0.8 | GO:0071899 | regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) |
| 0.1 | 2.4 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) cellular response to fluid shear stress(GO:0071498) |
| 0.1 | 2.2 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.1 | 1.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.1 | 2.8 | GO:0032291 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.1 | 2.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.1 | 2.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.1 | 4.9 | GO:0071470 | cellular response to osmotic stress(GO:0071470) |
| 0.1 | 1.1 | GO:0071313 | cellular response to caffeine(GO:0071313) positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 2.8 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 1.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.1 | 0.2 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.1 | 5.6 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.1 | 14.5 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 1.7 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.1 | 1.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 1.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.5 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.1 | 0.7 | GO:0060309 | elastin metabolic process(GO:0051541) elastin catabolic process(GO:0060309) |
| 0.1 | 0.3 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) |
| 0.1 | 0.4 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.1 | 0.5 | GO:0070889 | platelet alpha granule organization(GO:0070889) |
| 0.1 | 1.8 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.8 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 1.0 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.1 | 0.6 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.1 | 2.3 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.1 | 1.6 | GO:0051974 | negative regulation of telomerase activity(GO:0051974) |
| 0.1 | 2.2 | GO:0043584 | nose development(GO:0043584) |
| 0.1 | 2.7 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.1 | 1.7 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.1 | 0.3 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.1 | 0.4 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.1 | 1.6 | GO:0034776 | response to histamine(GO:0034776) cellular response to histamine(GO:0071420) |
| 0.1 | 2.2 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.1 | 1.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 2.7 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.1 | 1.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 4.9 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 1.2 | GO:0046643 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.1 | 1.2 | GO:0097011 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) |
| 0.1 | 2.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 1.2 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.1 | 0.7 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.1 | 0.3 | GO:2000074 | regulation of type B pancreatic cell development(GO:2000074) |
| 0.1 | 2.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.1 | 0.6 | GO:1904672 | regulation of somatic stem cell population maintenance(GO:1904672) |
| 0.1 | 0.4 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 1.4 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 1.0 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.1 | 2.9 | GO:0006493 | protein O-linked glycosylation(GO:0006493) |
| 0.1 | 0.4 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.1 | 0.8 | GO:0046036 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.9 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.6 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 1.8 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.1 | 0.8 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 1.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.6 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.0 | 0.2 | GO:0030026 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.5 | GO:0090500 | endocardial cushion to mesenchymal transition(GO:0090500) |
| 0.0 | 1.6 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.5 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.8 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.9 | GO:2000678 | negative regulation of transcription regulatory region DNA binding(GO:2000678) |
| 0.0 | 1.8 | GO:0071526 | semaphorin-plexin signaling pathway(GO:0071526) |
| 0.0 | 1.5 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.0 | 2.6 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.0 | 0.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.0 | 1.3 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 3.1 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.7 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 1.2 | GO:0032733 | positive regulation of interleukin-10 production(GO:0032733) |
| 0.0 | 0.4 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 1.5 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.4 | GO:0038166 | brain renin-angiotensin system(GO:0002035) angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 1.6 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 2.5 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.8 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.6 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.2 | GO:1900225 | negative regulation of osteoblast proliferation(GO:0033689) regulation of NLRP3 inflammasome complex assembly(GO:1900225) |
| 0.0 | 0.6 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.4 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 1.0 | GO:0090662 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.3 | GO:0051307 | meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.2 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.7 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.0 | 0.6 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 1.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 1.4 | GO:0045921 | positive regulation of exocytosis(GO:0045921) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 4.0 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.1 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.3 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.5 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.4 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.1 | GO:0071639 | positive regulation of monocyte chemotactic protein-1 production(GO:0071639) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.1 | GO:0045338 | farnesyl diphosphate metabolic process(GO:0045338) |
| 0.0 | 3.5 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.9 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.3 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 4.6 | GO:0000375 | RNA splicing, via transesterification reactions(GO:0000375) |
| 0.0 | 0.9 | GO:0050885 | neuromuscular process controlling balance(GO:0050885) |
| 0.0 | 0.0 | GO:0022601 | menstrual cycle phase(GO:0022601) |
| 0.0 | 1.6 | GO:0006333 | chromatin assembly or disassembly(GO:0006333) |
| 0.0 | 1.0 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.7 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.0 | 0.4 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.7 | GO:1902017 | regulation of cilium assembly(GO:1902017) |
| 0.0 | 0.4 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.6 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0046512 | sphingosine biosynthetic process(GO:0046512) |
| 0.0 | 0.1 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.2 | GO:0017004 | cytochrome complex assembly(GO:0017004) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.6 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 1.7 | 6.6 | GO:0032280 | symmetric synapse(GO:0032280) |
| 1.5 | 8.8 | GO:0044305 | calyx of Held(GO:0044305) |
| 1.2 | 5.8 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 1.1 | 12.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 1.0 | 7.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.9 | 2.8 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.9 | 4.4 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.6 | 2.2 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.5 | 2.6 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.5 | 3.6 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.5 | 6.5 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.5 | 1.5 | GO:1903095 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.5 | 3.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 5.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.4 | 26.1 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.4 | 2.5 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.4 | 13.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.4 | 4.5 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.3 | 3.0 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.3 | 7.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.3 | 4.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.3 | 0.9 | GO:0032545 | CURI complex(GO:0032545) UTP-C complex(GO:0034456) |
| 0.3 | 1.6 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.3 | 1.9 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.3 | 3.1 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.2 | 3.6 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.2 | 0.7 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.2 | 1.7 | GO:1990812 | growth cone filopodium(GO:1990812) |
| 0.2 | 13.4 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.2 | 0.8 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.2 | 4.2 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.2 | 5.0 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.2 | 0.8 | GO:0098835 | presynaptic endocytic zone(GO:0098833) presynaptic endocytic zone membrane(GO:0098835) |
| 0.2 | 10.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.2 | 13.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.2 | 5.7 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.2 | 1.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 3.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.2 | 1.3 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.2 | 2.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.2 | 1.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 2.5 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.2 | 0.5 | GO:0032156 | septin cytoskeleton(GO:0032156) |
| 0.2 | 1.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.2 | 1.8 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.2 | 3.8 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.4 | GO:0005712 | chiasma(GO:0005712) |
| 0.1 | 0.8 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.1 | 0.8 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 2.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 2.7 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.1 | 3.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 2.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 3.2 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.9 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 1.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.1 | 0.4 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 1.4 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.1 | 1.2 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.7 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.1 | 0.8 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.1 | 1.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.1 | 0.5 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 3.3 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 3.6 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.1 | 1.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 0.4 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.1 | 1.5 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.1 | 2.5 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.4 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 3.1 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 1.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.1 | 1.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.4 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 0.3 | GO:0097637 | intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 1.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.1 | 2.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.1 | 2.8 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 2.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.1 | 20.2 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.1 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.1 | 1.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 3.9 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.1 | 1.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.3 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.5 | GO:0097546 | ciliary base(GO:0097546) |
| 0.1 | 1.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.1 | 3.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 2.5 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.7 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 4.4 | GO:0005778 | peroxisomal membrane(GO:0005778) microbody membrane(GO:0031903) |
| 0.0 | 1.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 3.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 2.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 11.6 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.0 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 4.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.7 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.1 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.8 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.7 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 1.8 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 1.2 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 1.5 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 2.3 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 5.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.5 | GO:0030430 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 1.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 6.6 | GO:0005681 | spliceosomal complex(GO:0005681) |
| 0.0 | 2.5 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 5.3 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.9 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 2.7 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.3 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 8.0 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.4 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 2.1 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 1.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 2.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.7 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 3.6 | GO:0098857 | membrane raft(GO:0045121) membrane microdomain(GO:0098857) |
| 0.0 | 2.9 | GO:0099572 | postsynaptic density(GO:0014069) postsynaptic specialization(GO:0099572) |
| 0.0 | 3.5 | GO:0043235 | receptor complex(GO:0043235) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.2 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 1.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 0.3 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 7.5 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.5 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.1 | GO:0001674 | female germ cell nucleus(GO:0001674) |
| 0.0 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.9 | 8.8 | GO:0001565 | phorbol ester receptor activity(GO:0001565) non-kinase phorbol ester receptor activity(GO:0001566) |
| 2.7 | 8.1 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 1.8 | 10.8 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 1.8 | 7.2 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 1.6 | 7.9 | GO:0097001 | ceramide binding(GO:0097001) |
| 1.4 | 4.1 | GO:0005308 | creatine transmembrane transporter activity(GO:0005308) |
| 1.4 | 4.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 1.3 | 5.4 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 1.2 | 3.5 | GO:0052598 | diamine oxidase activity(GO:0052597) histamine oxidase activity(GO:0052598) methylputrescine oxidase activity(GO:0052599) propane-1,3-diamine oxidase activity(GO:0052600) |
| 1.2 | 5.8 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.9 | 1.8 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.9 | 7.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.8 | 2.5 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.8 | 4.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.8 | 3.3 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.8 | 3.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.8 | 2.3 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.8 | 2.3 | GO:0004560 | alpha-L-fucosidase activity(GO:0004560) fucosidase activity(GO:0015928) |
| 0.7 | 3.7 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.7 | 5.7 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.7 | 2.8 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.7 | 2.8 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.6 | 1.8 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.6 | 2.4 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.6 | 4.5 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.5 | 3.7 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.5 | 1.5 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) dinucleotide insertion or deletion binding(GO:0032139) |
| 0.5 | 1.5 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.5 | 2.5 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.5 | 1.9 | GO:0004441 | inositol-1,4-bisphosphate 1-phosphatase activity(GO:0004441) |
| 0.4 | 4.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 2.6 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.4 | 10.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.4 | 4.7 | GO:0048101 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.4 | 1.2 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.4 | 2.4 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.4 | 1.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.4 | 1.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.4 | 9.7 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.4 | 1.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.4 | 3.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.4 | 5.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.4 | 8.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.3 | 3.4 | GO:0008046 | GPI-linked ephrin receptor activity(GO:0005004) axon guidance receptor activity(GO:0008046) |
| 0.3 | 3.3 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.3 | 1.9 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.3 | 1.5 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.3 | 2.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.3 | 2.9 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 7.5 | GO:0005521 | lamin binding(GO:0005521) |
| 0.3 | 1.5 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.3 | 3.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.3 | 3.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 2.7 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.2 | 1.0 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.2 | 1.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.2 | 5.6 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.2 | 0.7 | GO:0031755 | endothelial differentiation G-protein coupled receptor binding(GO:0031753) Edg-2 lysophosphatidic acid receptor binding(GO:0031755) |
| 0.2 | 1.7 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 0.6 | GO:0001607 | neuromedin U receptor activity(GO:0001607) |
| 0.2 | 0.8 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 15.5 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.2 | 6.2 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.2 | 1.6 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.2 | 0.8 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.2 | 1.6 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.2 | 2.0 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.2 | 1.8 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.2 | 5.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.2 | 2.8 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.2 | 2.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 7.8 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.2 | 4.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.2 | 1.3 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.2 | 5.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.2 | 0.7 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.2 | 0.5 | GO:0019002 | GMP binding(GO:0019002) |
| 0.2 | 2.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.2 | 1.2 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.2 | 0.8 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 0.8 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.2 | 0.6 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 21.9 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 0.4 | GO:0004823 | leucine-tRNA ligase activity(GO:0004823) |
| 0.1 | 0.8 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.1 | 0.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.7 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.1 | 4.8 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.4 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 1.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.1 | 1.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 2.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 1.3 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 1.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 3.4 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.8 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 2.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.1 | 3.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 6.1 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 7.8 | GO:0042805 | actinin binding(GO:0042805) |
| 0.1 | 3.0 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.2 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.1 | 1.1 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.8 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 6.2 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.1 | 4.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 1.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 2.2 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.3 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 2.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.6 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 6.4 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.1 | 2.4 | GO:0071617 | lysophospholipid acyltransferase activity(GO:0071617) |
| 0.1 | 0.4 | GO:0004452 | isopentenyl-diphosphate delta-isomerase activity(GO:0004452) |
| 0.1 | 1.9 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.1 | 0.5 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.7 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 5.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.1 | 0.7 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 0.2 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 2.3 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.1 | 2.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.6 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 1.5 | GO:0005351 | sugar:proton symporter activity(GO:0005351) |
| 0.1 | 0.4 | GO:0051022 | CTP binding(GO:0002135) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.1 | 11.8 | GO:0005516 | calmodulin binding(GO:0005516) |
| 0.1 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 15.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.1 | 0.2 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.1 | 0.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.9 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.8 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.1 | 1.8 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 0.7 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.8 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 2.5 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 2.3 | GO:0017022 | myosin binding(GO:0017022) |
| 0.0 | 2.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 3.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.9 | GO:0015175 | neutral amino acid transmembrane transporter activity(GO:0015175) |
| 0.0 | 2.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 2.7 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.5 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 1.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 2.8 | GO:0008375 | acetylglucosaminyltransferase activity(GO:0008375) |
| 0.0 | 2.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 12.2 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
| 0.0 | 1.4 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.9 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 2.3 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.8 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 3.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.4 | GO:0008320 | protein transmembrane transporter activity(GO:0008320) |
| 0.0 | 0.3 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.7 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.2 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.5 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 5.9 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 3.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.1 | GO:0008146 | sulfotransferase activity(GO:0008146) |
| 0.0 | 0.1 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) GABA receptor activity(GO:0016917) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004161 | dimethylallyltranstransferase activity(GO:0004161) geranyltranstransferase activity(GO:0004337) |
| 0.0 | 2.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.2 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 4.2 | GO:0015631 | tubulin binding(GO:0015631) |
| 0.0 | 4.6 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.2 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 4.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.3 | 10.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.3 | 4.9 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 6.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 4.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 6.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.7 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 9.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.1 | 5.8 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 3.7 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.1 | 3.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.1 | 2.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.1 | 1.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.1 | 2.7 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.1 | 1.9 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.1 | 1.8 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 3.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.1 | 4.7 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 2.5 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.1 | 1.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.1 | 2.4 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 5.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 3.3 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 2.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.5 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.0 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 5.1 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 1.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 1.9 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.1 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.8 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 1.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.8 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.6 | 11.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.5 | 9.9 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.5 | 5.9 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.4 | 9.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.4 | 17.9 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.3 | 10.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.3 | 4.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 7.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 3.9 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.2 | 2.0 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.2 | 3.6 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 1.9 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.1 | 4.9 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.1 | 5.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.1 | 2.4 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 2.8 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.1 | 3.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 4.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.1 | 2.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 2.6 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 1.5 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.1 | 5.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 5.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 7.4 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 3.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.1 | 6.7 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 5.3 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.1 | 1.5 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.1 | 2.0 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.1 | 0.8 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 2.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.0 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.1 | 1.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.1 | 13.8 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.1 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.1 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.9 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.5 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.1 | 1.7 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 1.4 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 4.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 1.1 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 1.2 | REACTOME APOPTOTIC EXECUTION PHASE | Genes involved in Apoptotic execution phase |
| 0.0 | 0.5 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.9 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.4 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.8 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 1.9 | REACTOME SIGNALING BY ERBB2 | Genes involved in Signaling by ERBB2 |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 1.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.9 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.3 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.4 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 1.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.4 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 1.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.0 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.7 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |