Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
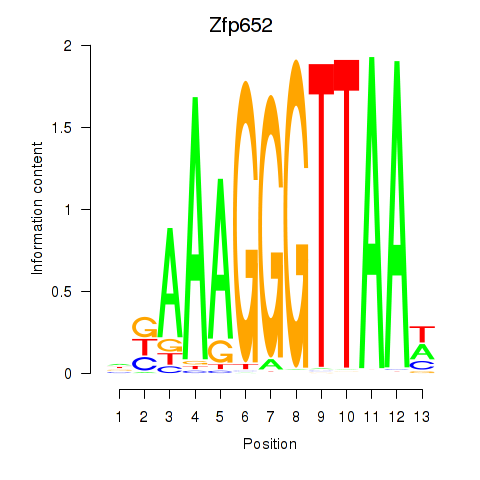
Results for Zfp652
Z-value: 1.50
Transcription factors associated with Zfp652
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp652
|
ENSMUSG00000075595.10 | Zfp652 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp652 | mm39_v1_chr11_+_95603494_95603529 | 0.45 | 8.2e-05 | Click! |
Activity profile of Zfp652 motif
Sorted Z-values of Zfp652 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp652
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_135723531 | 15.93 |
ENSMUST00000113085.2
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr9_-_53882530 | 15.03 |
ENSMUST00000048409.14
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr9_+_50664288 | 14.95 |
ENSMUST00000214962.2
ENSMUST00000216755.2 |
Cryab
|
crystallin, alpha B |
| chr9_+_50664207 | 14.70 |
ENSMUST00000034562.9
|
Cryab
|
crystallin, alpha B |
| chrX_+_135723420 | 14.20 |
ENSMUST00000033800.13
|
Plp1
|
proteolipid protein (myelin) 1 |
| chr15_-_98705791 | 12.28 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chrX_+_158242121 | 11.98 |
ENSMUST00000112470.3
ENSMUST00000043151.12 ENSMUST00000156172.3 |
Map7d2
|
MAP7 domain containing 2 |
| chr2_+_102380357 | 10.93 |
ENSMUST00000028612.8
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr12_-_17226889 | 10.66 |
ENSMUST00000170580.3
|
Kcnf1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr12_+_95658987 | 9.21 |
ENSMUST00000057324.4
|
Flrt2
|
fibronectin leucine rich transmembrane protein 2 |
| chr5_+_146392371 | 8.72 |
ENSMUST00000238592.2
|
Wasf3
|
WASP family, member 3 |
| chr15_-_79897404 | 8.41 |
ENSMUST00000229912.2
ENSMUST00000229795.2 |
Pdgfb
|
platelet derived growth factor, B polypeptide |
| chr12_-_109019507 | 7.77 |
ENSMUST00000185745.2
ENSMUST00000239108.2 |
Begain
|
brain-enriched guanylate kinase-associated |
| chr17_+_44445659 | 7.45 |
ENSMUST00000239215.2
|
Clic5
|
chloride intracellular channel 5 |
| chrX_-_7607527 | 7.37 |
ENSMUST00000033490.13
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr2_+_16361081 | 7.04 |
ENSMUST00000028081.13
|
Plxdc2
|
plexin domain containing 2 |
| chr7_+_92729067 | 6.84 |
ENSMUST00000051179.12
|
Fam181b
|
family with sequence similarity 181, member B |
| chr1_+_143516402 | 6.81 |
ENSMUST00000038252.4
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr11_+_69920542 | 6.76 |
ENSMUST00000232266.2
ENSMUST00000132597.5 |
Dlg4
|
discs large MAGUK scaffold protein 4 |
| chr7_+_4693603 | 6.72 |
ENSMUST00000120836.8
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr11_-_119937970 | 6.63 |
ENSMUST00000103020.8
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr3_+_87878378 | 6.46 |
ENSMUST00000090973.12
|
Nes
|
nestin |
| chr7_+_4693759 | 6.46 |
ENSMUST00000048248.9
|
Brsk1
|
BR serine/threonine kinase 1 |
| chr1_-_189075903 | 6.23 |
ENSMUST00000192723.2
ENSMUST00000110920.7 |
Kcnk2
|
potassium channel, subfamily K, member 2 |
| chr7_-_27146024 | 6.20 |
ENSMUST00000011895.14
|
Sptbn4
|
spectrin beta, non-erythrocytic 4 |
| chr3_+_87878436 | 6.01 |
ENSMUST00000160694.2
|
Nes
|
nestin |
| chr18_+_23886765 | 5.71 |
ENSMUST00000115830.8
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr19_-_21449916 | 5.67 |
ENSMUST00000087600.10
|
Gda
|
guanine deaminase |
| chr18_+_36431732 | 5.60 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein |
| chr1_+_60448703 | 5.44 |
ENSMUST00000052332.15
|
Abi2
|
abl interactor 2 |
| chr8_+_84626715 | 5.41 |
ENSMUST00000141158.8
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr1_+_153541412 | 5.29 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr19_+_8595369 | 5.17 |
ENSMUST00000010250.4
|
Slc22a6
|
solute carrier family 22 (organic anion transporter), member 6 |
| chr8_-_3722290 | 5.17 |
ENSMUST00000159364.3
|
Fcor
|
Foxo1 corepressor |
| chr5_+_139408906 | 5.11 |
ENSMUST00000066211.5
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr4_-_133981387 | 5.08 |
ENSMUST00000060050.6
|
Grrp1
|
glycine/arginine rich protein 1 |
| chr2_+_3119442 | 5.04 |
ENSMUST00000091505.11
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr2_-_32977182 | 5.03 |
ENSMUST00000102810.10
|
Garnl3
|
GTPase activating RANGAP domain-like 3 |
| chr19_+_23881821 | 5.02 |
ENSMUST00000237688.2
|
Apba1
|
amyloid beta (A4) precursor protein binding, family A, member 1 |
| chr17_+_72076678 | 4.98 |
ENSMUST00000230427.2
ENSMUST00000229952.2 ENSMUST00000230333.2 |
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr14_+_56091454 | 4.94 |
ENSMUST00000227465.2
ENSMUST00000168479.3 ENSMUST00000100529.10 |
Nynrin
|
NYN domain and retroviral integrase containing |
| chr1_+_60448813 | 4.76 |
ENSMUST00000188594.7
ENSMUST00000188618.7 ENSMUST00000189980.7 |
Abi2
|
abl interactor 2 |
| chr6_-_85479840 | 4.75 |
ENSMUST00000161546.2
ENSMUST00000161078.8 |
Fbxo41
|
F-box protein 41 |
| chr1_+_135075377 | 4.72 |
ENSMUST00000125774.2
|
Arl8a
|
ADP-ribosylation factor-like 8A |
| chrX_-_47123719 | 4.71 |
ENSMUST00000039026.8
|
Apln
|
apelin |
| chr11_-_119937896 | 4.68 |
ENSMUST00000064307.10
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr1_+_133173826 | 4.59 |
ENSMUST00000105082.9
ENSMUST00000038295.15 |
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr14_-_49763310 | 4.55 |
ENSMUST00000146164.2
ENSMUST00000138884.8 ENSMUST00000074368.11 ENSMUST00000123534.2 |
Slc35f4
|
solute carrier family 35, member F4 |
| chr2_-_45001141 | 4.39 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr1_+_133965228 | 4.28 |
ENSMUST00000162779.2
|
Fmod
|
fibromodulin |
| chr10_-_6930376 | 4.27 |
ENSMUST00000105617.8
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr7_+_127845984 | 4.27 |
ENSMUST00000164710.8
ENSMUST00000070656.12 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr8_-_47742389 | 4.26 |
ENSMUST00000211737.2
|
Stox2
|
storkhead box 2 |
| chr2_+_3119371 | 4.23 |
ENSMUST00000115099.9
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr6_-_54570124 | 4.17 |
ENSMUST00000046520.13
|
Fkbp14
|
FK506 binding protein 14 |
| chr14_-_78774201 | 4.08 |
ENSMUST00000123853.9
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr4_-_134099840 | 4.04 |
ENSMUST00000030643.3
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr17_+_72076728 | 3.99 |
ENSMUST00000230305.2
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr16_-_29360301 | 3.98 |
ENSMUST00000057018.15
ENSMUST00000182627.8 |
Atp13a4
|
ATPase type 13A4 |
| chr2_+_3119558 | 3.97 |
ENSMUST00000062934.7
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chrX_-_72868544 | 3.95 |
ENSMUST00000002080.12
ENSMUST00000114438.3 |
Pdzd4
|
PDZ domain containing 4 |
| chr2_-_84717036 | 3.91 |
ENSMUST00000054514.6
ENSMUST00000151799.8 |
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr6_+_83114020 | 3.88 |
ENSMUST00000121093.8
|
Rtkn
|
rhotekin |
| chr6_+_83114086 | 3.78 |
ENSMUST00000087938.11
|
Rtkn
|
rhotekin |
| chr1_+_153541020 | 3.76 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr6_+_124973752 | 3.72 |
ENSMUST00000162000.4
|
Pianp
|
PILR alpha associated neural protein |
| chr14_-_70864448 | 3.70 |
ENSMUST00000110984.4
|
Dmtn
|
dematin actin binding protein |
| chr5_-_136911969 | 3.67 |
ENSMUST00000057497.13
ENSMUST00000111103.2 |
Col26a1
|
collagen, type XXVI, alpha 1 |
| chr14_-_70864666 | 3.62 |
ENSMUST00000022694.17
|
Dmtn
|
dematin actin binding protein |
| chr8_+_84627332 | 3.55 |
ENSMUST00000045393.15
ENSMUST00000132500.8 ENSMUST00000152978.8 |
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr10_-_44334683 | 3.55 |
ENSMUST00000105490.3
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr11_-_118460736 | 3.51 |
ENSMUST00000136551.3
|
Rbfox3
|
RNA binding protein, fox-1 homolog (C. elegans) 3 |
| chr2_-_179867605 | 3.51 |
ENSMUST00000015791.6
|
Lama5
|
laminin, alpha 5 |
| chr6_-_113478779 | 3.44 |
ENSMUST00000101059.4
ENSMUST00000204268.3 ENSMUST00000205170.2 ENSMUST00000205075.2 ENSMUST00000204134.3 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr6_+_124973644 | 3.39 |
ENSMUST00000032479.11
|
Pianp
|
PILR alpha associated neural protein |
| chr7_+_127846121 | 3.37 |
ENSMUST00000167965.8
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr1_+_86230931 | 3.27 |
ENSMUST00000113306.4
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr1_+_60448931 | 3.22 |
ENSMUST00000189082.7
ENSMUST00000187709.7 |
Abi2
|
abl interactor 2 |
| chr3_+_129326004 | 3.21 |
ENSMUST00000199910.5
ENSMUST00000197070.5 ENSMUST00000071402.7 |
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr4_+_155778767 | 3.21 |
ENSMUST00000099265.3
|
Fndc10
|
fibronectin type III domain containing 10 |
| chr9_-_108329576 | 3.16 |
ENSMUST00000035232.13
ENSMUST00000195435.6 |
Klhdc8b
|
kelch domain containing 8B |
| chr11_-_98620200 | 3.13 |
ENSMUST00000126565.2
ENSMUST00000100500.9 ENSMUST00000017354.13 |
Med24
|
mediator complex subunit 24 |
| chr3_+_129326285 | 3.08 |
ENSMUST00000197235.5
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr15_-_36792649 | 3.08 |
ENSMUST00000126184.2
|
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr18_+_65715460 | 2.90 |
ENSMUST00000169679.8
ENSMUST00000183326.2 |
Zfp532
|
zinc finger protein 532 |
| chr15_+_82031382 | 2.82 |
ENSMUST00000023100.8
ENSMUST00000229336.2 |
Srebf2
|
sterol regulatory element binding factor 2 |
| chr3_-_146476331 | 2.76 |
ENSMUST00000106138.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr3_-_146475974 | 2.71 |
ENSMUST00000106137.8
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta |
| chr1_-_154692678 | 2.67 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr1_-_87322443 | 2.65 |
ENSMUST00000113212.4
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr9_-_56151334 | 2.54 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr10_-_81436671 | 2.53 |
ENSMUST00000151858.8
ENSMUST00000142948.8 ENSMUST00000072020.9 |
Tle6
|
transducin-like enhancer of split 6 |
| chr11_-_102710490 | 2.47 |
ENSMUST00000092567.11
|
Gjc1
|
gap junction protein, gamma 1 |
| chr9_-_50663571 | 2.44 |
ENSMUST00000042790.5
|
Hspb2
|
heat shock protein 2 |
| chr2_-_45000389 | 2.36 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chrX_+_109857866 | 2.25 |
ENSMUST00000078229.5
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr2_-_92876398 | 2.17 |
ENSMUST00000111272.3
ENSMUST00000178666.8 ENSMUST00000147339.3 |
Prdm11
|
PR domain containing 11 |
| chr10_-_7668560 | 2.17 |
ENSMUST00000065124.2
|
Ginm1
|
glycoprotein integral membrane 1 |
| chr1_+_153541339 | 2.07 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr9_-_50663648 | 2.00 |
ENSMUST00000217159.2
|
Hspb2
|
heat shock protein 2 |
| chr5_+_65288418 | 1.98 |
ENSMUST00000101191.10
ENSMUST00000204348.3 |
Klhl5
|
kelch-like 5 |
| chr8_+_112262729 | 1.97 |
ENSMUST00000172856.8
|
Znrf1
|
zinc and ring finger 1 |
| chr5_-_24556602 | 1.90 |
ENSMUST00000036092.10
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr9_+_21077010 | 1.89 |
ENSMUST00000039413.15
|
Pde4a
|
phosphodiesterase 4A, cAMP specific |
| chr13_-_74538899 | 1.83 |
ENSMUST00000223163.3
|
Zfp72
|
zinc finger protein 72 |
| chr10_+_75425197 | 1.76 |
ENSMUST00000189972.2
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr3_-_79053182 | 1.75 |
ENSMUST00000118340.7
|
Rapgef2
|
Rap guanine nucleotide exchange factor (GEF) 2 |
| chrX_+_6690410 | 1.60 |
ENSMUST00000145302.3
|
Dgkk
|
diacylglycerol kinase kappa |
| chr3_-_144738526 | 1.55 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr11_+_96189963 | 1.48 |
ENSMUST00000000704.6
|
Hoxb6
|
homeobox B6 |
| chr7_+_18817767 | 1.48 |
ENSMUST00000032568.14
ENSMUST00000122999.8 ENSMUST00000108473.10 ENSMUST00000108474.2 ENSMUST00000238982.2 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr10_+_53213763 | 1.44 |
ENSMUST00000219491.2
ENSMUST00000163319.9 ENSMUST00000220197.2 ENSMUST00000046221.8 ENSMUST00000218468.2 ENSMUST00000219921.2 |
Pln
|
phospholamban |
| chr1_+_86231208 | 1.38 |
ENSMUST00000188695.2
|
B3gnt7
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 7 |
| chr2_-_90900628 | 1.35 |
ENSMUST00000111436.3
ENSMUST00000073575.12 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr6_-_85479962 | 1.25 |
ENSMUST00000159062.8
|
Fbxo41
|
F-box protein 41 |
| chr2_-_79287095 | 1.24 |
ENSMUST00000041099.5
|
Neurod1
|
neurogenic differentiation 1 |
| chr17_-_27947863 | 1.23 |
ENSMUST00000167489.2
ENSMUST00000138970.3 ENSMUST00000025054.10 ENSMUST00000114870.9 |
Spdef
|
SAM pointed domain containing ets transcription factor |
| chr9_+_108731287 | 1.21 |
ENSMUST00000188557.8
|
Slc26a6
|
solute carrier family 26, member 6 |
| chr5_-_53864595 | 1.21 |
ENSMUST00000200691.4
|
Cckar
|
cholecystokinin A receptor |
| chr2_+_30156523 | 1.17 |
ENSMUST00000091132.13
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr15_-_64254754 | 1.00 |
ENSMUST00000177374.8
ENSMUST00000110114.10 ENSMUST00000110115.9 ENSMUST00000023008.16 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr11_+_108811168 | 1.00 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr10_-_44334711 | 0.92 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr2_+_30156733 | 0.91 |
ENSMUST00000113645.8
ENSMUST00000133877.8 ENSMUST00000139719.8 ENSMUST00000113643.8 ENSMUST00000150695.8 |
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr15_-_98779134 | 0.85 |
ENSMUST00000229348.2
ENSMUST00000229876.2 |
Rhebl1
|
Ras homolog enriched in brain like 1 |
| chr1_+_132128701 | 0.85 |
ENSMUST00000189062.2
|
Lemd1
|
LEM domain containing 1 |
| chr6_+_85888850 | 0.84 |
ENSMUST00000200680.4
|
Tprkb
|
Tp53rk binding protein |
| chr4_+_44756553 | 0.83 |
ENSMUST00000107824.9
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr6_+_125016723 | 0.83 |
ENSMUST00000140131.8
ENSMUST00000032480.14 |
Ing4
|
inhibitor of growth family, member 4 |
| chr11_+_96820091 | 0.77 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr10_-_128334515 | 0.74 |
ENSMUST00000026428.4
|
Myl6b
|
myosin, light polypeptide 6B |
| chr10_+_7668655 | 0.72 |
ENSMUST00000015901.11
|
Ppil4
|
peptidylprolyl isomerase (cyclophilin)-like 4 |
| chr8_+_91681550 | 0.70 |
ENSMUST00000210947.2
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr15_-_98507913 | 0.66 |
ENSMUST00000226500.2
ENSMUST00000227501.2 |
Adcy6
|
adenylate cyclase 6 |
| chr12_+_76593799 | 0.66 |
ENSMUST00000218380.2
ENSMUST00000219751.2 |
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr6_-_126141911 | 0.63 |
ENSMUST00000204542.3
|
Ntf3
|
neurotrophin 3 |
| chr11_-_65160810 | 0.54 |
ENSMUST00000108695.9
|
Myocd
|
myocardin |
| chr9_-_72019053 | 0.53 |
ENSMUST00000183492.8
ENSMUST00000184523.8 ENSMUST00000034755.13 |
Tcf12
|
transcription factor 12 |
| chr9_-_58462720 | 0.50 |
ENSMUST00000165365.3
|
Cd276
|
CD276 antigen |
| chr8_-_49008305 | 0.48 |
ENSMUST00000110346.9
ENSMUST00000211976.2 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr12_-_79054050 | 0.41 |
ENSMUST00000056660.13
ENSMUST00000174721.8 |
Tmem229b
|
transmembrane protein 229B |
| chr1_-_45542442 | 0.40 |
ENSMUST00000086430.5
|
Col5a2
|
collagen, type V, alpha 2 |
| chr11_+_66802807 | 0.38 |
ENSMUST00000123434.3
|
Pirt
|
phosphoinositide-interacting regulator of transient receptor potential channels |
| chr11_-_95201012 | 0.37 |
ENSMUST00000103159.10
ENSMUST00000107734.10 ENSMUST00000107733.10 |
Kat7
|
K(lysine) acetyltransferase 7 |
| chr17_+_35278011 | 0.35 |
ENSMUST00000007255.13
ENSMUST00000174493.8 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr9_-_107482494 | 0.33 |
ENSMUST00000102529.10
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr11_-_109363406 | 0.32 |
ENSMUST00000168740.3
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr16_+_44914397 | 0.31 |
ENSMUST00000061050.6
|
Ccdc80
|
coiled-coil domain containing 80 |
| chr11_+_45946800 | 0.29 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr17_+_23819861 | 0.28 |
ENSMUST00000123866.8
|
Zscan10
|
zinc finger and SCAN domain containing 10 |
| chr10_+_75425171 | 0.26 |
ENSMUST00000072217.9
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr2_+_31649946 | 0.21 |
ENSMUST00000028190.13
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr9_-_65424281 | 0.13 |
ENSMUST00000061766.6
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr4_+_13784749 | 0.09 |
ENSMUST00000098256.4
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr4_-_35845204 | 0.07 |
ENSMUST00000164772.8
ENSMUST00000065173.9 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr3_-_104725535 | 0.07 |
ENSMUST00000002297.12
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr3_-_89245297 | 0.06 |
ENSMUST00000029674.8
|
Efna4
|
ephrin A4 |
| chr11_+_96820220 | 0.06 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr18_-_60881679 | 0.04 |
ENSMUST00000237783.2
|
Ndst1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
| chr2_+_61423469 | 0.03 |
ENSMUST00000112494.2
|
Tank
|
TRAF family member-associated Nf-kappa B activator |
| chr7_-_26928029 | 0.03 |
ENSMUST00000003850.8
|
Itpkc
|
inositol 1,4,5-trisphosphate 3-kinase C |
| chr11_-_65160767 | 0.03 |
ENSMUST00000102635.10
|
Myocd
|
myocardin |
| chr1_-_156546600 | 0.01 |
ENSMUST00000122424.8
ENSMUST00000086153.8 |
Fam20b
|
FAM20B, glycosaminoglycan xylosylkinase |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 5.1 | GO:2000724 | regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 3.0 | 9.0 | GO:1900239 | regulation of phenotypic switching(GO:1900239) |
| 2.7 | 29.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 2.1 | 6.2 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 1.7 | 36.4 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 1.6 | 4.7 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 1.5 | 4.5 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 1.4 | 5.7 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 1.2 | 13.4 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 1.2 | 7.3 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 1.0 | 6.8 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.9 | 11.3 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.8 | 7.6 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.8 | 6.8 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.7 | 11.1 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.7 | 9.2 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.6 | 6.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.6 | 2.5 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.6 | 1.8 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.6 | 5.2 | GO:0070417 | cellular response to cold(GO:0070417) |
| 0.6 | 2.8 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.5 | 3.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.5 | 6.8 | GO:0098953 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.5 | 1.9 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.4 | 4.9 | GO:0015074 | DNA integration(GO:0015074) |
| 0.4 | 1.2 | GO:0046724 | oxalic acid secretion(GO:0046724) |
| 0.4 | 4.4 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.4 | 7.5 | GO:0002024 | diet induced thermogenesis(GO:0002024) |
| 0.3 | 3.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.3 | 5.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.3 | 0.7 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.3 | 1.5 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.3 | 9.0 | GO:0090129 | positive regulation of synapse maturation(GO:0090129) |
| 0.3 | 3.5 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.3 | 2.0 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.2 | 1.2 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.2 | 8.7 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.2 | 13.2 | GO:0031572 | G2 DNA damage checkpoint(GO:0031572) |
| 0.2 | 2.5 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.2 | 5.0 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.2 | 0.7 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.2 | 1.0 | GO:0032423 | regulation of mismatch repair(GO:0032423) regulation of chondrocyte development(GO:0061181) negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.2 | 1.8 | GO:0044789 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.2 | 3.9 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.1 | 1.4 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 6.0 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.1 | 14.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.1 | 0.4 | GO:0072708 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.1 | 2.3 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 0.4 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 1.2 | GO:0060480 | lung goblet cell differentiation(GO:0060480) |
| 0.1 | 3.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.1 | 0.5 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.2 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.1 | 1.6 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.1 | 0.8 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.1 | 0.7 | GO:1901407 | regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901407) |
| 0.1 | 2.6 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.3 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.0 | 10.7 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 3.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 4.2 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 3.1 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 0.4 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.7 | GO:0030049 | muscle filament sliding(GO:0030049) |
| 0.0 | 0.5 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 8.7 | GO:0043413 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 7.1 | GO:0043524 | negative regulation of neuron apoptotic process(GO:0043524) |
| 0.0 | 1.6 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 2.5 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 12.5 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 12.0 | GO:0000226 | microtubule cytoskeleton organization(GO:0000226) |
| 0.0 | 1.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 29.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 1.0 | 7.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 1.0 | 6.2 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.9 | 13.4 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.9 | 3.5 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.8 | 6.2 | GO:0008091 | spectrin(GO:0008091) |
| 0.4 | 5.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.4 | 1.9 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.4 | 2.8 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.3 | 5.5 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.3 | 6.8 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.2 | 11.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.2 | 11.3 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 5.7 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.4 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 2.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 8.7 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.1 | 4.7 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 12.5 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.1 | 2.9 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 10.7 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.1 | 9.0 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 30.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.1 | 18.2 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 0.8 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.1 | 3.1 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 10.1 | GO:0043204 | perikaryon(GO:0043204) |
| 0.1 | 0.7 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 19.2 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.1 | 5.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 19.1 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 6.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 8.7 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.7 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 14.8 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 4.0 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 8.4 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 1.4 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0043195 | terminal bouton(GO:0043195) |
| 0.0 | 1.2 | GO:0000123 | histone acetyltransferase complex(GO:0000123) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.0 | 30.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 1.7 | 5.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 1.7 | 6.8 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 1.3 | 4.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 1.3 | 7.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 1.3 | 7.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 1.0 | 12.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 1.0 | 33.1 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 1.0 | 6.8 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.7 | 13.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.6 | 8.4 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.6 | 2.5 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.6 | 13.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.5 | 6.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.5 | 5.5 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.4 | 9.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.4 | 9.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.4 | 5.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.3 | 1.9 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.3 | 6.2 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) potassium ion leak channel activity(GO:0022841) |
| 0.3 | 1.8 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.2 | 1.4 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 4.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.2 | 1.2 | GO:0015563 | uptake transmembrane transporter activity(GO:0015563) |
| 0.2 | 4.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.2 | 7.3 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.2 | 2.0 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.2 | 11.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.2 | 13.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 4.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 4.7 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 13.3 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.1 | 4.6 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.1 | 1.6 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.1 | 5.7 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 2.7 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 2.3 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 4.5 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.7 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.1 | 1.0 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.1 | 5.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.1 | 9.0 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.1 | 3.1 | GO:0046966 | thyroid hormone receptor binding(GO:0046966) |
| 0.1 | 1.9 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 11.1 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 18.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 10.5 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 5.5 | GO:0016407 | acetyltransferase activity(GO:0016407) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.7 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 3.5 | GO:0044325 | ion channel binding(GO:0044325) |
| 0.0 | 1.2 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 11.2 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 5.7 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 8.7 | GO:0003779 | actin binding(GO:0003779) |
| 0.0 | 10.8 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 1.5 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 8.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.2 | 3.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 16.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.2 | 6.8 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 5.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 13.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.1 | 4.3 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.1 | 4.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 2.8 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.0 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.7 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 6.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 8.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.6 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 2.3 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 6.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.3 | 4.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.3 | 5.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.3 | 5.7 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.2 | 8.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.2 | 12.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.2 | 6.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.2 | 4.6 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.1 | 6.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 4.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.1 | 1.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 9.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.1 | 1.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 11.1 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 2.0 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 6.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |