Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zfp691
Z-value: 0.73
Transcription factors associated with Zfp691
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zfp691
|
ENSMUSG00000045268.14 | Zfp691 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zfp691 | mm39_v1_chr4_-_119031050_119031100 | 0.08 | 5.0e-01 | Click! |
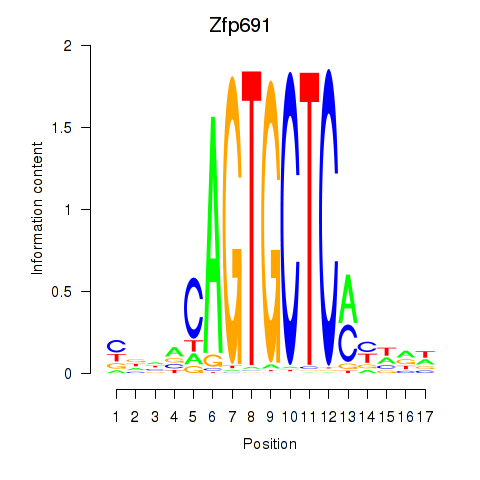
Activity profile of Zfp691 motif
Sorted Z-values of Zfp691 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zfp691
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_120930193 | 6.02 |
ENSMUST00000026159.6
|
Cd7
|
CD7 antigen |
| chrX_-_100311824 | 5.12 |
ENSMUST00000033664.14
|
Il2rg
|
interleukin 2 receptor, gamma chain |
| chr11_-_79414542 | 4.91 |
ENSMUST00000179322.2
|
Evi2b
|
ecotropic viral integration site 2b |
| chr13_+_19528728 | 4.68 |
ENSMUST00000179181.3
|
Trgc4
|
T cell receptor gamma, constant 4 |
| chr19_-_11243530 | 4.10 |
ENSMUST00000169159.3
|
Ms4a1
|
membrane-spanning 4-domains, subfamily A, member 1 |
| chr19_+_11493825 | 3.88 |
ENSMUST00000163078.8
|
Ms4a6b
|
membrane-spanning 4-domains, subfamily A, member 6B |
| chr9_+_32607301 | 3.83 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr8_+_70261323 | 3.63 |
ENSMUST00000036074.15
ENSMUST00000123453.2 |
Gmip
|
Gem-interacting protein |
| chr3_-_15491482 | 3.58 |
ENSMUST00000099201.9
ENSMUST00000194144.3 ENSMUST00000192700.3 |
Sirpb1a
|
signal-regulatory protein beta 1A |
| chr5_+_96104775 | 3.56 |
ENSMUST00000023840.7
|
Cxcl13
|
chemokine (C-X-C motif) ligand 13 |
| chr13_-_100922910 | 3.43 |
ENSMUST00000174038.2
ENSMUST00000091295.14 ENSMUST00000072119.15 |
Ccnb1
|
cyclin B1 |
| chr1_-_181670599 | 3.35 |
ENSMUST00000193030.6
|
Lbr
|
lamin B receptor |
| chr8_+_73072877 | 3.12 |
ENSMUST00000067912.8
|
Klf2
|
Kruppel-like factor 2 (lung) |
| chr2_-_120867529 | 2.90 |
ENSMUST00000102490.10
|
Epb42
|
erythrocyte membrane protein band 4.2 |
| chr2_+_180367056 | 2.87 |
ENSMUST00000094218.4
|
Slc17a9
|
solute carrier family 17, member 9 |
| chr7_-_71956332 | 2.86 |
ENSMUST00000079323.8
|
Mctp2
|
multiple C2 domains, transmembrane 2 |
| chr5_-_130031842 | 2.79 |
ENSMUST00000026613.14
|
Gusb
|
glucuronidase, beta |
| chr12_-_114117264 | 2.78 |
ENSMUST00000103461.5
|
Ighv7-3
|
immunoglobulin heavy variable 7-3 |
| chr8_+_72889073 | 2.63 |
ENSMUST00000003575.11
|
Tpm4
|
tropomyosin 4 |
| chr12_-_113252552 | 2.63 |
ENSMUST00000103416.9
ENSMUST00000195192.2 |
Ighg2c
|
immunoglobulin heavy constant gamma 2C |
| chr3_-_15640045 | 2.62 |
ENSMUST00000192382.6
ENSMUST00000195778.3 ENSMUST00000091319.7 |
Sirpb1b
|
signal-regulatory protein beta 1B |
| chr17_+_8463886 | 2.57 |
ENSMUST00000231545.2
|
Ccr6
|
chemokine (C-C motif) receptor 6 |
| chr12_-_113561594 | 2.42 |
ENSMUST00000103444.3
|
Ighv5-4
|
immunoglobulin heavy variable 5-4 |
| chr7_+_139673300 | 2.37 |
ENSMUST00000026540.9
|
Prap1
|
proline-rich acidic protein 1 |
| chr13_-_55676334 | 2.28 |
ENSMUST00000047877.5
|
Dok3
|
docking protein 3 |
| chr8_+_72889607 | 2.25 |
ENSMUST00000238492.2
|
Tpm4
|
tropomyosin 4 |
| chr3_-_15397325 | 2.20 |
ENSMUST00000108361.2
|
Gm9733
|
predicted gene 9733 |
| chr14_+_52091156 | 2.15 |
ENSMUST00000169070.2
ENSMUST00000074477.7 |
Ear6
|
eosinophil-associated, ribonuclease A family, member 6 |
| chr10_-_62438040 | 2.14 |
ENSMUST00000045866.9
|
Ddx21
|
DExD box helicase 21 |
| chr6_-_128765449 | 2.00 |
ENSMUST00000172601.8
|
Klrb1c
|
killer cell lectin-like receptor subfamily B member 1C |
| chr4_-_118314647 | 1.90 |
ENSMUST00000106375.2
ENSMUST00000168404.9 ENSMUST00000006556.11 |
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr16_+_44632096 | 1.89 |
ENSMUST00000176819.8
ENSMUST00000176321.8 |
Cd200r4
|
CD200 receptor 4 |
| chr17_-_25973288 | 1.88 |
ENSMUST00000075884.8
ENSMUST00000238120.2 ENSMUST00000236137.2 ENSMUST00000237359.2 |
Msln
|
mesothelin |
| chr1_+_51328265 | 1.85 |
ENSMUST00000051572.8
|
Cavin2
|
caveolae associated 2 |
| chr16_+_32427738 | 1.81 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr6_-_87510200 | 1.81 |
ENSMUST00000113637.9
ENSMUST00000071024.7 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr6_-_128599833 | 1.63 |
ENSMUST00000171306.5
ENSMUST00000204819.3 ENSMUST00000032512.15 |
Klrb1a
|
killer cell lectin-like receptor subfamily B member 1A |
| chr3_-_108797022 | 1.57 |
ENSMUST00000180063.8
ENSMUST00000053065.8 |
Fndc7
|
fibronectin type III domain containing 7 |
| chr8_-_23196523 | 1.54 |
ENSMUST00000033939.13
ENSMUST00000063401.10 |
Ikbkb
|
inhibitor of kappaB kinase beta |
| chr1_+_86454511 | 1.51 |
ENSMUST00000188533.2
|
Ptma
|
prothymosin alpha |
| chr4_-_118314707 | 1.50 |
ENSMUST00000102671.10
|
Mpl
|
myeloproliferative leukemia virus oncogene |
| chr17_-_23902992 | 1.50 |
ENSMUST00000085989.8
|
Cldn9
|
claudin 9 |
| chr1_+_86454431 | 1.46 |
ENSMUST00000045897.15
ENSMUST00000186255.7 ENSMUST00000188699.7 |
Ptma
|
prothymosin alpha |
| chr1_-_155022501 | 1.41 |
ENSMUST00000027744.10
|
Mr1
|
major histocompatibility complex, class I-related |
| chr7_-_143203348 | 1.39 |
ENSMUST00000084396.4
ENSMUST00000075588.13 ENSMUST00000146692.8 |
Tnfrsf22
|
tumor necrosis factor receptor superfamily, member 22 |
| chr4_-_137309415 | 1.34 |
ENSMUST00000238941.2
|
Ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr9_+_62765362 | 1.30 |
ENSMUST00000213643.2
ENSMUST00000034777.14 ENSMUST00000163820.3 ENSMUST00000215870.2 ENSMUST00000214633.2 ENSMUST00000215968.2 |
Calml4
|
calmodulin-like 4 |
| chr12_+_69418886 | 1.22 |
ENSMUST00000050063.9
|
Arf6
|
ADP-ribosylation factor 6 |
| chr2_+_118943274 | 1.21 |
ENSMUST00000140939.8
ENSMUST00000028795.10 |
Rad51
|
RAD51 recombinase |
| chr9_-_50528727 | 1.15 |
ENSMUST00000131351.8
ENSMUST00000171462.8 |
Nkapd1
|
NKAP domain containing 1 |
| chr2_+_43445333 | 1.15 |
ENSMUST00000028223.9
ENSMUST00000112826.8 |
Kynu
|
kynureninase |
| chr4_-_141412910 | 1.13 |
ENSMUST00000105782.2
|
Rsc1a1
|
regulatory solute carrier protein, family 1, member 1 |
| chr3_+_98129463 | 1.13 |
ENSMUST00000029469.5
|
Reg4
|
regenerating islet-derived family, member 4 |
| chr14_+_73379930 | 1.13 |
ENSMUST00000170370.8
ENSMUST00000164822.8 ENSMUST00000165429.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr13_-_67480588 | 1.13 |
ENSMUST00000109735.9
ENSMUST00000168892.9 |
Zfp595
|
zinc finger protein 595 |
| chr14_+_73380163 | 1.11 |
ENSMUST00000170368.8
ENSMUST00000171767.8 ENSMUST00000163533.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr2_-_105832353 | 1.11 |
ENSMUST00000155811.2
|
Dnajc24
|
DnaJ heat shock protein family (Hsp40) member C24 |
| chr2_+_26481435 | 1.10 |
ENSMUST00000152988.9
ENSMUST00000149789.2 |
Egfl7
|
EGF-like domain 7 |
| chr1_+_134217727 | 1.08 |
ENSMUST00000027730.6
|
Myog
|
myogenin |
| chr15_+_101308935 | 1.08 |
ENSMUST00000147662.8
|
Krt7
|
keratin 7 |
| chr11_-_78875689 | 1.07 |
ENSMUST00000108269.10
ENSMUST00000108268.10 |
Lgals9
|
lectin, galactose binding, soluble 9 |
| chr10_+_115405891 | 1.07 |
ENSMUST00000173620.2
|
A930009A15Rik
|
RIKEN cDNA A930009A15 gene |
| chr14_+_73380485 | 1.07 |
ENSMUST00000170677.8
ENSMUST00000167401.8 |
Rcbtb2
|
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
| chr4_+_45342069 | 1.06 |
ENSMUST00000155551.8
|
Dcaf10
|
DDB1 and CUL4 associated factor 10 |
| chr7_+_127503812 | 1.03 |
ENSMUST00000151451.3
ENSMUST00000124533.3 ENSMUST00000206745.2 ENSMUST00000206140.2 |
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr6_+_70821481 | 1.03 |
ENSMUST00000034093.15
ENSMUST00000162950.2 |
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chr14_+_54417419 | 1.02 |
ENSMUST00000103703.2
|
Traj39
|
T cell receptor alpha joining 39 |
| chr3_+_102377234 | 1.00 |
ENSMUST00000035952.5
ENSMUST00000198168.5 ENSMUST00000106925.9 |
Ngf
|
nerve growth factor |
| chr7_-_25239229 | 0.96 |
ENSMUST00000044547.10
ENSMUST00000066503.14 ENSMUST00000064862.13 |
Ceacam2
|
carcinoembryonic antigen-related cell adhesion molecule 2 |
| chr10_-_41455203 | 0.91 |
ENSMUST00000095227.10
|
Ccdc162
|
coiled-coil domain containing 162 |
| chr7_-_105386546 | 0.90 |
ENSMUST00000098148.6
|
Rrp8
|
ribosomal RNA processing 8 |
| chrX_-_108056995 | 0.85 |
ENSMUST00000033597.9
|
Hmgn5
|
high-mobility group nucleosome binding domain 5 |
| chr9_+_123921573 | 0.83 |
ENSMUST00000111442.3
ENSMUST00000171499.3 |
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr10_-_88192852 | 0.81 |
ENSMUST00000020249.2
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr1_-_171359228 | 0.79 |
ENSMUST00000168184.2
|
Itln1
|
intelectin 1 (galactofuranose binding) |
| chr7_-_143239600 | 0.79 |
ENSMUST00000208017.2
ENSMUST00000152703.2 |
Tnfrsf23
|
tumor necrosis factor receptor superfamily, member 23 |
| chr14_+_54440591 | 0.78 |
ENSMUST00000103725.2
|
Traj16
|
T cell receptor alpha joining 16 |
| chr3_-_108797306 | 0.78 |
ENSMUST00000102620.10
|
Fndc7
|
fibronectin type III domain containing 7 |
| chr1_-_155627430 | 0.77 |
ENSMUST00000195275.2
|
Lhx4
|
LIM homeobox protein 4 |
| chr7_-_15613769 | 0.76 |
ENSMUST00000172758.3
ENSMUST00000044434.13 |
Crx
|
cone-rod homeobox |
| chr19_-_11637880 | 0.75 |
ENSMUST00000135994.2
ENSMUST00000121793.2 |
Oosp2
|
oocyte secreted protein 2 |
| chr12_-_16850887 | 0.73 |
ENSMUST00000161998.3
|
Greb1
|
gene regulated by estrogen in breast cancer protein |
| chr6_+_28475099 | 0.72 |
ENSMUST00000168362.2
|
Snd1
|
staphylococcal nuclease and tudor domain containing 1 |
| chr13_-_68730207 | 0.70 |
ENSMUST00000221259.2
ENSMUST00000223398.2 ENSMUST00000045827.5 |
Mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr12_-_113236868 | 0.68 |
ENSMUST00000223335.2
ENSMUST00000137336.3 |
Ighe
|
Immunoglobulin heavy constant epsilon |
| chr14_+_43951187 | 0.67 |
ENSMUST00000094051.6
|
Gm7324
|
predicted gene 7324 |
| chr1_+_163979384 | 0.64 |
ENSMUST00000086040.6
|
F5
|
coagulation factor V |
| chr1_-_180823709 | 0.56 |
ENSMUST00000154133.8
|
Ephx1
|
epoxide hydrolase 1, microsomal |
| chr3_+_122305819 | 0.55 |
ENSMUST00000199344.2
|
Bcar3
|
breast cancer anti-estrogen resistance 3 |
| chr2_+_91095597 | 0.54 |
ENSMUST00000028691.7
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr5_+_95136485 | 0.52 |
ENSMUST00000178646.2
|
Gm6367
|
predicted gene 6367 |
| chr14_+_54418031 | 0.51 |
ENSMUST00000103704.2
|
Traj38
|
T cell receptor alpha joining 38 |
| chr7_-_139941566 | 0.50 |
ENSMUST00000215023.2
ENSMUST00000216027.2 ENSMUST00000210932.3 ENSMUST00000211031.3 |
Olfr60
|
olfactory receptor 60 |
| chr4_+_132262853 | 0.49 |
ENSMUST00000094657.10
ENSMUST00000105940.10 ENSMUST00000105939.10 ENSMUST00000150207.8 |
Dnajc8
|
DnaJ heat shock protein family (Hsp40) member C8 |
| chr13_+_30529498 | 0.49 |
ENSMUST00000221743.2
|
Agtr1a
|
angiotensin II receptor, type 1a |
| chr6_+_41279199 | 0.49 |
ENSMUST00000031913.5
|
Try4
|
trypsin 4 |
| chr5_+_75236250 | 0.48 |
ENSMUST00000040477.4
ENSMUST00000160104.3 |
Gsx2
|
GS homeobox 2 |
| chr4_+_32657105 | 0.48 |
ENSMUST00000071642.11
ENSMUST00000178134.2 |
Mdn1
|
midasin AAA ATPase 1 |
| chr9_-_58220469 | 0.47 |
ENSMUST00000061799.10
|
Loxl1
|
lysyl oxidase-like 1 |
| chr10_-_59838815 | 0.47 |
ENSMUST00000182116.8
|
Anapc16
|
anaphase promoting complex subunit 16 |
| chr15_-_82796308 | 0.46 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr11_-_101066266 | 0.45 |
ENSMUST00000062759.4
|
Ccr10
|
chemokine (C-C motif) receptor 10 |
| chr15_-_76235208 | 0.45 |
ENSMUST00000023211.16
|
Sharpin
|
SHANK-associated RH domain interacting protein |
| chr1_-_138784557 | 0.45 |
ENSMUST00000112025.3
|
2310009B15Rik
|
RIKEN cDNA 2310009B15 gene |
| chr7_+_12893495 | 0.44 |
ENSMUST00000236472.2
ENSMUST00000237109.2 |
Vmn1r88
|
vomeronasal 1 receptor, 88 |
| chr10_-_93727003 | 0.44 |
ENSMUST00000180840.8
|
Metap2
|
methionine aminopeptidase 2 |
| chr2_+_181138958 | 0.43 |
ENSMUST00000149163.8
ENSMUST00000000844.15 ENSMUST00000184849.8 ENSMUST00000108800.8 ENSMUST00000069712.9 |
Tpd52l2
|
tumor protein D52-like 2 |
| chr11_+_54582248 | 0.42 |
ENSMUST00000136494.2
|
Rapgef6
|
Rap guanine nucleotide exchange factor (GEF) 6 |
| chr8_-_5155347 | 0.42 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2 |
| chr7_-_37718916 | 0.41 |
ENSMUST00000085513.6
ENSMUST00000206327.2 |
Uri1
|
URI1, prefoldin-like chaperone |
| chr18_-_57108405 | 0.41 |
ENSMUST00000139243.9
ENSMUST00000025488.15 |
C330018D20Rik
|
RIKEN cDNA C330018D20 gene |
| chr7_+_63803583 | 0.41 |
ENSMUST00000085222.12
ENSMUST00000206277.2 |
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr10_-_59838878 | 0.39 |
ENSMUST00000182912.2
ENSMUST00000020307.11 ENSMUST00000182898.8 |
Anapc16
|
anaphase promoting complex subunit 16 |
| chr11_-_75313412 | 0.39 |
ENSMUST00000138661.8
ENSMUST00000000769.14 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chrX_+_105230706 | 0.37 |
ENSMUST00000081593.13
|
Pgk1
|
phosphoglycerate kinase 1 |
| chr6_-_119925387 | 0.36 |
ENSMUST00000162541.8
|
Wnk1
|
WNK lysine deficient protein kinase 1 |
| chr10_+_94412116 | 0.36 |
ENSMUST00000117929.2
|
Tmcc3
|
transmembrane and coiled coil domains 3 |
| chr17_+_19880590 | 0.35 |
ENSMUST00000171741.3
|
Vmn2r102
|
vomeronasal 2, receptor 102 |
| chr4_-_144104503 | 0.30 |
ENSMUST00000121109.2
|
Pramel15
|
PRAME like 15 |
| chr7_+_63803663 | 0.29 |
ENSMUST00000206314.2
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr2_+_181139016 | 0.29 |
ENSMUST00000108799.10
|
Tpd52l2
|
tumor protein D52-like 2 |
| chr17_+_31220910 | 0.29 |
ENSMUST00000235827.2
|
Umodl1
|
uromodulin-like 1 |
| chr18_-_46658957 | 0.26 |
ENSMUST00000036226.6
|
Fem1c
|
fem 1 homolog c |
| chr6_+_83891336 | 0.26 |
ENSMUST00000204751.3
ENSMUST00000204202.3 ENSMUST00000203455.3 ENSMUST00000113836.6 ENSMUST00000113835.10 ENSMUST00000032088.14 |
Zfp638
|
zinc finger protein 638 |
| chr4_+_125940678 | 0.26 |
ENSMUST00000030675.8
|
Mrps15
|
mitochondrial ribosomal protein S15 |
| chr10_+_93983844 | 0.26 |
ENSMUST00000105290.9
|
Nr2c1
|
nuclear receptor subfamily 2, group C, member 1 |
| chr10_-_128204806 | 0.26 |
ENSMUST00000043211.7
ENSMUST00000220227.2 |
Coq10a
|
coenzyme Q10A |
| chr2_-_155571279 | 0.24 |
ENSMUST00000040833.5
|
Edem2
|
ER degradation enhancer, mannosidase alpha-like 2 |
| chr10_+_59839142 | 0.24 |
ENSMUST00000164083.4
|
Ascc1
|
activating signal cointegrator 1 complex subunit 1 |
| chr7_+_103647953 | 0.23 |
ENSMUST00000218325.2
|
Olfr638
|
olfactory receptor 638 |
| chr3_+_92272486 | 0.22 |
ENSMUST00000050397.2
|
Sprr2f
|
small proline-rich protein 2F |
| chr19_+_13608985 | 0.22 |
ENSMUST00000217182.3
|
Olfr1489
|
olfactory receptor 1489 |
| chr8_-_25085654 | 0.21 |
ENSMUST00000110667.8
|
Ido1
|
indoleamine 2,3-dioxygenase 1 |
| chr8_-_78244578 | 0.20 |
ENSMUST00000076316.6
|
Arhgap10
|
Rho GTPase activating protein 10 |
| chr7_+_114318746 | 0.20 |
ENSMUST00000182044.2
|
Calcb
|
calcitonin-related polypeptide, beta |
| chr6_+_145879839 | 0.19 |
ENSMUST00000032383.14
|
Sspn
|
sarcospan |
| chr10_+_4432488 | 0.18 |
ENSMUST00000138112.8
|
Ccdc170
|
coiled-coil domain containing 170 |
| chr9_-_20978389 | 0.18 |
ENSMUST00000115494.3
|
Zglp1
|
zinc finger, GATA-like protein 1 |
| chr9_-_50515089 | 0.17 |
ENSMUST00000000175.6
|
Sdhd
|
succinate dehydrogenase complex, subunit D, integral membrane protein |
| chr8_+_94993453 | 0.17 |
ENSMUST00000212167.2
|
Nup93
|
nucleoporin 93 |
| chr11_-_94864273 | 0.17 |
ENSMUST00000100551.11
ENSMUST00000152042.2 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr2_+_24852409 | 0.16 |
ENSMUST00000028351.9
|
Dph7
|
diphthamine biosynethesis 7 |
| chr2_+_88644840 | 0.16 |
ENSMUST00000214703.2
|
Olfr1202
|
olfactory receptor 1202 |
| chr7_+_102289455 | 0.16 |
ENSMUST00000098221.2
|
Olfr554
|
olfactory receptor 554 |
| chr6_+_88442391 | 0.15 |
ENSMUST00000032165.16
|
Ruvbl1
|
RuvB-like protein 1 |
| chr10_-_77002377 | 0.15 |
ENSMUST00000081654.13
|
Col18a1
|
collagen, type XVIII, alpha 1 |
| chr18_-_80133177 | 0.15 |
ENSMUST00000178391.2
|
Gm21886
|
predicted gene, 21886 |
| chr7_+_140277180 | 0.14 |
ENSMUST00000210357.4
ENSMUST00000209857.2 |
Olfr541
|
olfactory receptor 541 |
| chr6_-_141719536 | 0.14 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr8_-_78244412 | 0.14 |
ENSMUST00000210922.2
ENSMUST00000210519.2 |
Arhgap10
|
Rho GTPase activating protein 10 |
| chr7_-_12853779 | 0.13 |
ENSMUST00000227220.2
ENSMUST00000227700.2 ENSMUST00000226604.2 |
Vmn1r86
|
vomeronasal 1 receptor 86 |
| chr13_-_68730059 | 0.13 |
ENSMUST00000220973.2
|
Mtrr
|
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
| chr11_-_49005701 | 0.12 |
ENSMUST00000060398.3
ENSMUST00000215553.2 ENSMUST00000109201.2 |
Olfr1396
|
olfactory receptor 1396 |
| chr9_-_96407311 | 0.12 |
ENSMUST00000190704.3
|
Gm28729
|
predicted gene 28729 |
| chr2_-_86941996 | 0.11 |
ENSMUST00000213978.2
|
Olfr259
|
olfactory receptor 259 |
| chr7_+_140226365 | 0.11 |
ENSMUST00000084456.6
ENSMUST00000211057.2 ENSMUST00000211399.2 |
Olfr53
|
olfactory receptor 53 |
| chr6_+_41829799 | 0.11 |
ENSMUST00000120605.8
|
Sval2
|
seminal vesicle antigen-like 2 |
| chr1_+_133254950 | 0.11 |
ENSMUST00000178033.5
|
Kiss1
|
KiSS-1 metastasis-suppressor |
| chr9_-_104140099 | 0.11 |
ENSMUST00000035170.13
|
Dnajc13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr2_+_43445359 | 0.09 |
ENSMUST00000050511.7
|
Kynu
|
kynureninase |
| chr5_+_37208198 | 0.07 |
ENSMUST00000043794.11
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr8_-_23295603 | 0.04 |
ENSMUST00000163739.3
ENSMUST00000210656.2 |
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr7_-_108484213 | 0.03 |
ENSMUST00000217803.2
|
Olfr518
|
olfactory receptor 518 |
| chr14_+_26789345 | 0.03 |
ENSMUST00000226105.2
|
Il17rd
|
interleukin 17 receptor D |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0035702 | monocyte homeostasis(GO:0035702) |
| 0.9 | 3.6 | GO:2001027 | negative regulation of endothelial cell chemotaxis(GO:2001027) |
| 0.9 | 3.4 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.9 | 2.6 | GO:1904155 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.9 | 5.1 | GO:0032831 | positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.4 | 3.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.4 | 1.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.4 | 1.2 | GO:1990426 | homologous recombination-dependent replication fork processing(GO:1990426) |
| 0.4 | 1.1 | GO:0014737 | positive regulation of muscle atrophy(GO:0014737) response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.3 | 3.8 | GO:0030578 | PML body organization(GO:0030578) positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.3 | 1.0 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.3 | 1.5 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.2 | 1.1 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.2 | 0.8 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.2 | 0.8 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 1.0 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.2 | 0.8 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.2 | 0.5 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 2.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.8 | GO:0009624 | response to nematode(GO:0009624) |
| 0.1 | 1.8 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.1 | 0.9 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.1 | 3.0 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 6.0 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.1 | 1.0 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.1 | 0.2 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.1 | 2.6 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.4 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.2 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.1 | 1.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.1 | 0.4 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.1 | 1.8 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.1 | 3.0 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.1 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.0 | 1.0 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.3 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 2.0 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 2.2 | GO:1902042 | negative regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902042) |
| 0.0 | 1.4 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 3.6 | GO:0050766 | positive regulation of phagocytosis(GO:0050766) |
| 0.0 | 2.1 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.6 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.0 | 0.2 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.5 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 2.6 | GO:0016126 | sterol biosynthetic process(GO:0016126) |
| 0.0 | 0.4 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.7 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 1.1 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 1.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 3.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.2 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.3 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.0 | 0.4 | GO:0031639 | plasminogen activation(GO:0031639) |
| 0.0 | 1.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.4 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 4.1 | GO:0042113 | B cell activation(GO:0042113) |
| 0.0 | 0.3 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 3.4 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.3 | 4.9 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.2 | 2.6 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.2 | 1.0 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.2 | 3.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.9 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.7 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 1.8 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 0.7 | GO:0097433 | dense body(GO:0097433) |
| 0.1 | 1.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 1.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.1 | 0.4 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.1 | 1.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.5 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 5.7 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.2 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 17.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.6 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 1.1 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.0 | 0.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 1.2 | GO:0035861 | site of double-strand break(GO:0035861) |
| 0.0 | 0.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.2 | GO:0016012 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 2.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.4 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 2.9 | GO:0030863 | cortical cytoskeleton(GO:0030863) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.8 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 1.7 | GO:0001669 | acrosomal vesicle(GO:0001669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.1 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.9 | 3.6 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.7 | 2.8 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.6 | 3.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.4 | 1.8 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.3 | 1.2 | GO:0016823 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.3 | 3.3 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.3 | 2.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.3 | 1.5 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.3 | 1.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.3 | 1.0 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.2 | 3.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.2 | 2.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 0.5 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 4.1 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 1.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 2.2 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 1.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.1 | 0.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.4 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.1 | 0.2 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.1 | 0.4 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.1 | 3.8 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.1 | 1.4 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.1 | 1.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 0.8 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.1 | 3.7 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.8 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.5 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 5.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 2.3 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 1.8 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.4 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.5 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 2.3 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 0.9 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.2 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 3.7 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 6.3 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 1.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.9 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 5.1 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.1 | 4.2 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.1 | 3.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.0 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 3.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.1 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 6.0 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 1.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 4.9 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.7 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.4 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.2 | 2.8 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.2 | 5.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.2 | 0.8 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.2 | 2.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 6.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 3.3 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 1.5 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 4.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.1 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 3.4 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 1.8 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 1.0 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 1.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 5.8 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.1 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |