Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
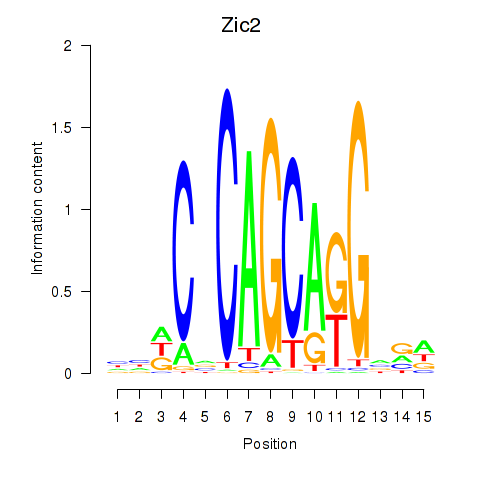
Results for Zic2
Z-value: 1.11
Transcription factors associated with Zic2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic2
|
ENSMUSG00000061524.9 | Zic2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic2 | mm39_v1_chr14_+_122712809_122712847 | -0.33 | 4.4e-03 | Click! |
Activity profile of Zic2 motif
Sorted Z-values of Zic2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_92158054 | 11.57 |
ENSMUST00000071805.4
|
Sprr2a2
|
small proline-rich protein 2A2 |
| chr4_+_40920047 | 8.49 |
ENSMUST00000030122.5
|
Spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr7_-_45315762 | 7.35 |
ENSMUST00000210620.2
ENSMUST00000069800.6 |
Fut2
|
fucosyltransferase 2 |
| chr3_+_92123106 | 6.88 |
ENSMUST00000074449.7
ENSMUST00000090871.3 |
Sprr2a1
Sprr2a2
|
small proline-rich protein 2A1 small proline-rich protein 2A2 |
| chr3_+_92192724 | 5.88 |
ENSMUST00000193337.2
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr6_-_70292451 | 5.20 |
ENSMUST00000103387.3
|
Igkv8-21
|
immunoglobulin kappa variable 8-21 |
| chr15_-_100579813 | 5.17 |
ENSMUST00000230572.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr19_+_58717319 | 5.17 |
ENSMUST00000048644.6
ENSMUST00000236445.2 |
Pnliprp1
|
pancreatic lipase related protein 1 |
| chr8_+_105573693 | 5.00 |
ENSMUST00000055052.6
|
Ces2c
|
carboxylesterase 2C |
| chr19_+_24651362 | 4.90 |
ENSMUST00000057243.6
|
Tmem252
|
transmembrane protein 252 |
| chr6_+_68026941 | 4.88 |
ENSMUST00000103316.2
|
Igkv9-120
|
immunoglobulin kappa chain variable 9-120 |
| chr8_-_106198112 | 4.83 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chr2_+_69210775 | 4.67 |
ENSMUST00000063690.4
|
Dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr11_+_96820220 | 4.53 |
ENSMUST00000062172.6
|
Prr15l
|
proline rich 15-like |
| chr11_-_69696428 | 4.49 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr9_-_14663689 | 4.42 |
ENSMUST00000061498.7
|
Fut4
|
fucosyltransferase 4 |
| chr8_+_93553901 | 4.41 |
ENSMUST00000034187.9
|
Mmp2
|
matrix metallopeptidase 2 |
| chr8_+_105460627 | 4.29 |
ENSMUST00000034346.15
ENSMUST00000164182.3 |
Ces2a
|
carboxylesterase 2A |
| chr8_-_13446769 | 4.26 |
ENSMUST00000033826.4
|
Atp4b
|
ATPase, H+/K+ exchanging, beta polypeptide |
| chr10_+_126836578 | 4.25 |
ENSMUST00000026500.12
ENSMUST00000142698.8 |
Avil
|
advillin |
| chr4_-_58987094 | 4.17 |
ENSMUST00000030069.7
|
Ptgr1
|
prostaglandin reductase 1 |
| chr11_+_96820091 | 4.12 |
ENSMUST00000054311.6
ENSMUST00000107636.4 |
Prr15l
|
proline rich 15-like |
| chr17_-_31363245 | 4.12 |
ENSMUST00000024826.8
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr4_+_129000600 | 4.03 |
ENSMUST00000148979.2
|
Tmem54
|
transmembrane protein 54 |
| chr19_-_4489415 | 3.96 |
ENSMUST00000235680.2
ENSMUST00000117462.2 ENSMUST00000048197.10 |
Rhod
|
ras homolog family member D |
| chr17_+_28988271 | 3.90 |
ENSMUST00000233984.2
ENSMUST00000233460.2 |
Mapk13
|
mitogen-activated protein kinase 13 |
| chr3_-_14843512 | 3.84 |
ENSMUST00000094365.11
|
Car1
|
carbonic anhydrase 1 |
| chr9_+_110848339 | 3.70 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr6_-_68907718 | 3.70 |
ENSMUST00000114212.4
|
Igkv13-85
|
immunoglobulin kappa chain variable 13-85 |
| chr11_+_72851989 | 3.69 |
ENSMUST00000163326.8
ENSMUST00000108485.9 ENSMUST00000021142.8 ENSMUST00000108486.8 ENSMUST00000108484.8 |
Atp2a3
|
ATPase, Ca++ transporting, ubiquitous |
| chr11_-_100012384 | 3.68 |
ENSMUST00000007275.3
|
Krt13
|
keratin 13 |
| chr12_-_80306865 | 3.61 |
ENSMUST00000167327.2
|
Actn1
|
actinin, alpha 1 |
| chr17_-_31383976 | 3.59 |
ENSMUST00000235870.2
|
Tff1
|
trefoil factor 1 |
| chr19_+_11724913 | 3.54 |
ENSMUST00000025585.4
|
Cblif
|
cobalamin binding intrinsic factor |
| chr6_+_17306334 | 3.51 |
ENSMUST00000007799.13
ENSMUST00000115456.6 |
Cav1
|
caveolin 1, caveolae protein |
| chr2_+_24257576 | 3.50 |
ENSMUST00000140547.2
ENSMUST00000102942.8 |
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr6_-_131293361 | 3.39 |
ENSMUST00000121078.2
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr15_+_75027089 | 3.35 |
ENSMUST00000190262.2
|
Ly6g
|
lymphocyte antigen 6 complex, locus G |
| chr4_+_106479624 | 3.33 |
ENSMUST00000047922.3
|
Ttc22
|
tetratricopeptide repeat domain 22 |
| chr11_+_73067909 | 3.30 |
ENSMUST00000040687.12
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr7_+_30475819 | 3.29 |
ENSMUST00000041703.10
|
Dmkn
|
dermokine |
| chr2_+_143757193 | 3.28 |
ENSMUST00000103172.4
|
Dstn
|
destrin |
| chr12_-_108668520 | 3.27 |
ENSMUST00000167978.9
ENSMUST00000021691.6 |
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr11_+_63019799 | 3.24 |
ENSMUST00000108702.8
|
Pmp22
|
peripheral myelin protein 22 |
| chr16_-_56652241 | 3.24 |
ENSMUST00000135672.2
|
Tmem45a
|
transmembrane protein 45a |
| chr11_-_117670430 | 3.23 |
ENSMUST00000143406.8
|
Tmc6
|
transmembrane channel-like gene family 6 |
| chr11_+_73068063 | 3.20 |
ENSMUST00000108477.2
|
Tax1bp3
|
Tax1 (human T cell leukemia virus type I) binding protein 3 |
| chr7_+_45370607 | 3.19 |
ENSMUST00000129507.5
|
Fam83e
|
family with sequence similarity 83, member E |
| chr5_+_72805122 | 3.19 |
ENSMUST00000087212.8
|
Nipal1
|
NIPA-like domain containing 1 |
| chr15_-_100576715 | 3.18 |
ENSMUST00000229869.2
|
Cela1
|
chymotrypsin-like elastase family, member 1 |
| chr1_+_130754413 | 3.18 |
ENSMUST00000027675.14
ENSMUST00000133792.8 |
Pigr
|
polymeric immunoglobulin receptor |
| chr2_+_129854256 | 3.17 |
ENSMUST00000110299.3
|
Tgm3
|
transglutaminase 3, E polypeptide |
| chr15_+_85090150 | 3.16 |
ENSMUST00000057410.14
ENSMUST00000109432.4 |
Fbln1
|
fibulin 1 |
| chr9_+_122980006 | 3.15 |
ENSMUST00000026890.6
|
Clec3b
|
C-type lectin domain family 3, member b |
| chr17_-_26417982 | 3.10 |
ENSMUST00000142410.2
ENSMUST00000120333.8 ENSMUST00000039113.14 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr16_+_92282593 | 3.09 |
ENSMUST00000162181.8
|
Clic6
|
chloride intracellular channel 6 |
| chr3_-_121325887 | 3.03 |
ENSMUST00000039197.9
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr14_+_14091030 | 3.02 |
ENSMUST00000224529.2
|
Oit1
|
oncoprotein induced transcript 1 |
| chr7_+_142030744 | 3.00 |
ENSMUST00000149521.8
|
Lsp1
|
lymphocyte specific 1 |
| chr16_-_10131804 | 2.95 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2 |
| chr4_+_20042045 | 2.94 |
ENSMUST00000098242.4
|
Ggh
|
gamma-glutamyl hydrolase |
| chr2_-_130905428 | 2.87 |
ENSMUST00000147333.2
|
Adam33
|
a disintegrin and metallopeptidase domain 33 |
| chr5_+_93045837 | 2.87 |
ENSMUST00000113051.9
|
Shroom3
|
shroom family member 3 |
| chr19_+_56414114 | 2.86 |
ENSMUST00000238892.2
|
Casp7
|
caspase 7 |
| chr3_-_92922976 | 2.81 |
ENSMUST00000107301.2
ENSMUST00000029521.5 |
Crct1
|
cysteine-rich C-terminal 1 |
| chr12_-_103304573 | 2.79 |
ENSMUST00000149431.2
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr1_+_74448535 | 2.78 |
ENSMUST00000027366.13
|
Vil1
|
villin 1 |
| chr2_-_93283024 | 2.75 |
ENSMUST00000111257.8
ENSMUST00000145553.8 |
Cd82
|
CD82 antigen |
| chr10_+_18720760 | 2.70 |
ENSMUST00000019998.9
|
Perp
|
PERP, TP53 apoptosis effector |
| chr15_-_101621332 | 2.69 |
ENSMUST00000023709.7
|
Krt5
|
keratin 5 |
| chr3_-_83749036 | 2.62 |
ENSMUST00000029623.11
|
Tlr2
|
toll-like receptor 2 |
| chr11_-_117671436 | 2.62 |
ENSMUST00000026659.10
ENSMUST00000127227.2 |
Tmc6
|
transmembrane channel-like gene family 6 |
| chr6_+_36364990 | 2.61 |
ENSMUST00000172278.8
|
Chrm2
|
cholinergic receptor, muscarinic 2, cardiac |
| chr12_-_114286421 | 2.59 |
ENSMUST00000103483.3
|
Ighv3-8
|
immunoglobulin heavy variable V3-8 |
| chr16_-_4950285 | 2.57 |
ENSMUST00000035672.5
|
Ppl
|
periplakin |
| chr11_-_17002344 | 2.56 |
ENSMUST00000020321.13
|
Plek
|
pleckstrin |
| chr6_-_70121150 | 2.53 |
ENSMUST00000197525.2
|
Igkv8-28
|
immunoglobulin kappa variable 8-28 |
| chr12_-_80307110 | 2.53 |
ENSMUST00000021554.16
|
Actn1
|
actinin, alpha 1 |
| chr10_-_76459152 | 2.53 |
ENSMUST00000105413.8
|
Col6a2
|
collagen, type VI, alpha 2 |
| chr16_-_58343789 | 2.51 |
ENSMUST00000137035.8
ENSMUST00000149456.8 |
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr9_-_123045087 | 2.49 |
ENSMUST00000039229.8
|
Cdcp1
|
CUB domain containing protein 1 |
| chr3_+_94249399 | 2.47 |
ENSMUST00000029794.11
|
Them5
|
thioesterase superfamily member 5 |
| chr19_-_40260286 | 2.47 |
ENSMUST00000182432.2
|
Pdlim1
|
PDZ and LIM domain 1 (elfin) |
| chr13_+_52750883 | 2.47 |
ENSMUST00000055087.7
|
Syk
|
spleen tyrosine kinase |
| chr2_+_155593030 | 2.44 |
ENSMUST00000029140.12
ENSMUST00000132608.2 |
Procr
|
protein C receptor, endothelial |
| chr10_+_79726687 | 2.43 |
ENSMUST00000217837.2
ENSMUST00000061653.9 |
Cfd
|
complement factor D (adipsin) |
| chr14_-_55344004 | 2.42 |
ENSMUST00000036041.15
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chr15_-_78449172 | 2.41 |
ENSMUST00000230952.2
|
Rac2
|
Rac family small GTPase 2 |
| chr18_+_20691095 | 2.39 |
ENSMUST00000059787.15
ENSMUST00000120102.8 |
Dsg2
|
desmoglein 2 |
| chr7_-_79492091 | 2.38 |
ENSMUST00000049004.8
|
Anpep
|
alanyl (membrane) aminopeptidase |
| chr1_+_151447124 | 2.37 |
ENSMUST00000148810.8
|
Niban1
|
niban apoptosis regulator 1 |
| chr6_+_17065141 | 2.35 |
ENSMUST00000115467.11
ENSMUST00000154266.3 ENSMUST00000076654.9 |
Tes
|
testin LIM domain protein |
| chr7_-_104114384 | 2.33 |
ENSMUST00000076922.6
|
Trim30a
|
tripartite motif-containing 30A |
| chr15_+_85942928 | 2.32 |
ENSMUST00000138134.8
|
Gramd4
|
GRAM domain containing 4 |
| chr6_-_69877642 | 2.30 |
ENSMUST00000103370.3
|
Igkv5-39
|
immunoglobulin kappa variable 5-39 |
| chr8_+_75599801 | 2.26 |
ENSMUST00000034034.10
|
Isx
|
intestine specific homeobox |
| chr8_+_71858647 | 2.19 |
ENSMUST00000119976.8
ENSMUST00000120725.2 |
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr1_-_128287347 | 2.17 |
ENSMUST00000190495.2
ENSMUST00000027601.11 |
Mcm6
|
minichromosome maintenance complex component 6 |
| chr14_-_52011035 | 2.12 |
ENSMUST00000073860.6
|
Ang4
|
angiogenin, ribonuclease A family, member 4 |
| chr4_-_106657446 | 2.09 |
ENSMUST00000148688.2
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr3_+_94249445 | 2.09 |
ENSMUST00000198083.2
|
Them5
|
thioesterase superfamily member 5 |
| chr5_+_72805153 | 2.08 |
ENSMUST00000197837.3
|
Nipal1
|
NIPA-like domain containing 1 |
| chr7_+_44117444 | 2.06 |
ENSMUST00000206887.2
ENSMUST00000117324.8 ENSMUST00000120852.8 ENSMUST00000134398.3 ENSMUST00000118628.8 |
Josd2
|
Josephin domain containing 2 |
| chr3_+_92223927 | 2.05 |
ENSMUST00000061038.4
|
Sprr2b
|
small proline-rich protein 2B |
| chr6_-_136852792 | 2.05 |
ENSMUST00000032342.3
|
Mgp
|
matrix Gla protein |
| chr14_-_34096574 | 2.02 |
ENSMUST00000023826.5
|
Sncg
|
synuclein, gamma |
| chr5_+_146769915 | 2.00 |
ENSMUST00000075453.9
ENSMUST00000099272.3 |
Rpl21
|
ribosomal protein L21 |
| chr17_-_35293447 | 2.00 |
ENSMUST00000007259.4
|
Ly6g6d
|
lymphocyte antigen 6 complex, locus G6D |
| chr8_-_85740220 | 1.97 |
ENSMUST00000209322.2
|
Best2
|
bestrophin 2 |
| chr10_+_125802084 | 1.94 |
ENSMUST00000074807.8
|
Lrig3
|
leucine-rich repeats and immunoglobulin-like domains 3 |
| chr14_+_51328534 | 1.93 |
ENSMUST00000022428.13
ENSMUST00000171688.9 |
Rnase4
Ang
|
ribonuclease, RNase A family 4 angiogenin, ribonuclease, RNase A family, 5 |
| chr5_-_44259010 | 1.93 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr13_-_100240570 | 1.91 |
ENSMUST00000038104.12
|
Bdp1
|
B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB |
| chr6_+_67873135 | 1.91 |
ENSMUST00000103310.3
|
Igkv14-126
|
immunoglobulin kappa variable 14-126 |
| chr6_-_101176147 | 1.89 |
ENSMUST00000239140.2
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr13_+_30933209 | 1.88 |
ENSMUST00000021784.10
ENSMUST00000110307.3 ENSMUST00000222125.2 |
Irf4
|
interferon regulatory factor 4 |
| chr8_-_93956143 | 1.88 |
ENSMUST00000176282.2
ENSMUST00000034173.14 |
Ces1e
|
carboxylesterase 1E |
| chr7_+_79848138 | 1.87 |
ENSMUST00000205822.2
|
Sema4b
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr5_-_41921834 | 1.87 |
ENSMUST00000060820.8
|
Nkx3-2
|
NK3 homeobox 2 |
| chr7_-_19504446 | 1.86 |
ENSMUST00000003061.14
|
Bcam
|
basal cell adhesion molecule |
| chr4_+_138503046 | 1.85 |
ENSMUST00000030528.9
|
Pla2g2d
|
phospholipase A2, group IID |
| chr13_+_74269554 | 1.85 |
ENSMUST00000036208.7
ENSMUST00000225423.2 ENSMUST00000221703.2 |
Slc9a3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 |
| chr17_+_29333116 | 1.84 |
ENSMUST00000233717.2
ENSMUST00000141239.2 |
Rab44
|
RAB44, member RAS oncogene family |
| chr7_-_140676596 | 1.84 |
ENSMUST00000209199.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr7_+_44117475 | 1.83 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr13_-_74918745 | 1.82 |
ENSMUST00000223033.2
ENSMUST00000222588.2 |
Cast
|
calpastatin |
| chr15_+_102368510 | 1.82 |
ENSMUST00000164688.2
|
Prr13
|
proline rich 13 |
| chr5_-_31102829 | 1.82 |
ENSMUST00000031051.8
|
Cgref1
|
cell growth regulator with EF hand domain 1 |
| chr11_-_118292678 | 1.81 |
ENSMUST00000106290.4
|
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr7_+_120442048 | 1.80 |
ENSMUST00000047875.16
|
Eef2k
|
eukaryotic elongation factor-2 kinase |
| chr2_-_35226981 | 1.80 |
ENSMUST00000028241.7
|
Stom
|
stomatin |
| chr14_-_59602859 | 1.79 |
ENSMUST00000161031.2
|
Phf11d
|
PHD finger protein 11D |
| chr7_-_34353767 | 1.79 |
ENSMUST00000206501.2
ENSMUST00000108069.8 |
Kctd15
|
potassium channel tetramerisation domain containing 15 |
| chr7_-_141241632 | 1.79 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr11_+_5811886 | 1.77 |
ENSMUST00000102923.10
|
Aebp1
|
AE binding protein 1 |
| chr16_+_33338648 | 1.77 |
ENSMUST00000122427.8
ENSMUST00000059056.15 |
Slc12a8
|
solute carrier family 12 (potassium/chloride transporters), member 8 |
| chr5_+_110987839 | 1.76 |
ENSMUST00000200172.2
ENSMUST00000066160.3 |
Chek2
|
checkpoint kinase 2 |
| chr11_+_105183463 | 1.75 |
ENSMUST00000100335.10
ENSMUST00000021038.5 |
Mrc2
|
mannose receptor, C type 2 |
| chr1_-_192883642 | 1.75 |
ENSMUST00000192020.6
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr1_+_132973724 | 1.73 |
ENSMUST00000077730.7
|
Pik3c2b
|
phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta |
| chr15_-_97806142 | 1.73 |
ENSMUST00000023119.15
|
Vdr
|
vitamin D (1,25-dihydroxyvitamin D3) receptor |
| chr19_+_8975249 | 1.73 |
ENSMUST00000236390.2
|
Ahnak
|
AHNAK nucleoprotein (desmoyokin) |
| chr16_+_33338898 | 1.71 |
ENSMUST00000119173.8
|
Slc12a8
|
solute carrier family 12 (potassium/chloride transporters), member 8 |
| chr8_+_81220410 | 1.69 |
ENSMUST00000063359.8
|
Gypa
|
glycophorin A |
| chr1_-_192883743 | 1.69 |
ENSMUST00000043550.11
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr3_-_129126362 | 1.68 |
ENSMUST00000029658.14
|
Enpep
|
glutamyl aminopeptidase |
| chr17_-_27247891 | 1.68 |
ENSMUST00000078691.12
|
Bak1
|
BCL2-antagonist/killer 1 |
| chr11_-_60111391 | 1.66 |
ENSMUST00000020846.8
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr6_-_128803182 | 1.66 |
ENSMUST00000204756.3
ENSMUST00000204394.3 ENSMUST00000204423.3 ENSMUST00000204677.2 ENSMUST00000205130.3 ENSMUST00000174544.2 ENSMUST00000172887.8 ENSMUST00000032472.11 |
Gm44511
Klrb1b
|
predicted gene 44511 killer cell lectin-like receptor subfamily B member 1B |
| chr19_-_6973393 | 1.66 |
ENSMUST00000041686.10
ENSMUST00000180765.2 |
Nudt22
|
nudix (nucleoside diphosphate linked moiety X)-type motif 22 |
| chr8_-_11329656 | 1.65 |
ENSMUST00000208095.2
|
Col4a1
|
collagen, type IV, alpha 1 |
| chr8_-_106660470 | 1.65 |
ENSMUST00000034368.8
|
Ctrl
|
chymotrypsin-like |
| chr19_+_46561801 | 1.65 |
ENSMUST00000026011.8
|
Sfxn2
|
sideroflexin 2 |
| chr7_+_101467512 | 1.64 |
ENSMUST00000008090.11
|
Phox2a
|
paired-like homeobox 2a |
| chrX_-_101232978 | 1.63 |
ENSMUST00000033683.8
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr7_+_141995545 | 1.63 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr12_-_113386312 | 1.61 |
ENSMUST00000177715.8
ENSMUST00000103426.3 |
Ighm
|
immunoglobulin heavy constant mu |
| chr8_+_84393263 | 1.60 |
ENSMUST00000019608.7
|
Ptger1
|
prostaglandin E receptor 1 (subtype EP1) |
| chr6_-_124519240 | 1.60 |
ENSMUST00000159463.8
ENSMUST00000162844.2 ENSMUST00000160505.8 ENSMUST00000162443.8 |
C1s1
|
complement component 1, s subcomponent 1 |
| chr1_-_144124871 | 1.59 |
ENSMUST00000189061.7
|
Rgs1
|
regulator of G-protein signaling 1 |
| chrX_+_21581135 | 1.59 |
ENSMUST00000033414.8
|
Slc6a14
|
solute carrier family 6 (neurotransmitter transporter), member 14 |
| chr1_+_91294133 | 1.59 |
ENSMUST00000086861.12
|
Erfe
|
erythroferrone |
| chr2_-_75534985 | 1.58 |
ENSMUST00000102672.5
|
Nfe2l2
|
nuclear factor, erythroid derived 2, like 2 |
| chr14_+_54701594 | 1.58 |
ENSMUST00000022782.10
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr9_+_51124983 | 1.57 |
ENSMUST00000034554.9
|
Pou2af1
|
POU domain, class 2, associating factor 1 |
| chr5_+_146769700 | 1.54 |
ENSMUST00000035983.12
|
Rpl21
|
ribosomal protein L21 |
| chr15_+_25774070 | 1.54 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr17_+_34311314 | 1.53 |
ENSMUST00000025192.8
|
H2-Oa
|
histocompatibility 2, O region alpha locus |
| chr9_-_44437694 | 1.53 |
ENSMUST00000062215.8
|
Cxcr5
|
chemokine (C-X-C motif) receptor 5 |
| chr8_-_93924426 | 1.52 |
ENSMUST00000034172.8
|
Ces1d
|
carboxylesterase 1D |
| chr4_-_111759951 | 1.52 |
ENSMUST00000102719.8
ENSMUST00000102721.8 |
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr2_+_164611812 | 1.52 |
ENSMUST00000088248.13
ENSMUST00000001439.7 |
Ube2c
|
ubiquitin-conjugating enzyme E2C |
| chr4_-_57301791 | 1.52 |
ENSMUST00000075637.11
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr19_-_11301919 | 1.52 |
ENSMUST00000159269.2
|
Ms4a7
|
membrane-spanning 4-domains, subfamily A, member 7 |
| chr9_-_21710029 | 1.51 |
ENSMUST00000034717.7
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr17_+_37253802 | 1.48 |
ENSMUST00000040498.12
|
Rnf39
|
ring finger protein 39 |
| chr5_+_139408906 | 1.47 |
ENSMUST00000066211.5
|
Gper1
|
G protein-coupled estrogen receptor 1 |
| chr5_-_91550853 | 1.47 |
ENSMUST00000121044.6
|
Btc
|
betacellulin, epidermal growth factor family member |
| chr6_-_142517340 | 1.46 |
ENSMUST00000203945.3
|
Kcnj8
|
potassium inwardly-rectifying channel, subfamily J, member 8 |
| chr9_-_79625800 | 1.46 |
ENSMUST00000071750.13
|
Col12a1
|
collagen, type XII, alpha 1 |
| chr17_+_49735413 | 1.46 |
ENSMUST00000173033.8
|
Mocs1
|
molybdenum cofactor synthesis 1 |
| chr5_-_92426014 | 1.46 |
ENSMUST00000159345.8
ENSMUST00000113102.10 |
Naaa
|
N-acylethanolamine acid amidase |
| chr11_-_11976732 | 1.46 |
ENSMUST00000143915.2
|
Grb10
|
growth factor receptor bound protein 10 |
| chr10_-_76562002 | 1.44 |
ENSMUST00000001147.5
|
Col6a1
|
collagen, type VI, alpha 1 |
| chr4_+_135899678 | 1.44 |
ENSMUST00000061721.6
|
E2f2
|
E2F transcription factor 2 |
| chr15_-_101262452 | 1.44 |
ENSMUST00000230909.2
|
Krt80
|
keratin 80 |
| chr17_-_36440317 | 1.43 |
ENSMUST00000046131.16
ENSMUST00000173322.8 ENSMUST00000172968.2 |
Gm7030
|
predicted gene 7030 |
| chr14_+_34097422 | 1.42 |
ENSMUST00000111908.3
|
Mmrn2
|
multimerin 2 |
| chr12_-_114012399 | 1.42 |
ENSMUST00000103468.3
|
Ighv11-2
|
immunoglobulin heavy variable V11-2 |
| chr11_-_101062111 | 1.42 |
ENSMUST00000164474.8
ENSMUST00000043397.14 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr15_-_76004395 | 1.42 |
ENSMUST00000239552.1
|
EPPK1
|
epiplakin 1 |
| chr7_+_45364822 | 1.42 |
ENSMUST00000210640.2
|
Rpl18
|
ribosomal protein L18 |
| chr3_+_95566082 | 1.41 |
ENSMUST00000037947.15
ENSMUST00000178686.2 |
Mcl1
|
myeloid cell leukemia sequence 1 |
| chr9_+_70114623 | 1.41 |
ENSMUST00000034745.9
|
Myo1e
|
myosin IE |
| chr9_-_79626002 | 1.41 |
ENSMUST00000121227.8
|
Col12a1
|
collagen, type XII, alpha 1 |
| chr16_-_55642491 | 1.40 |
ENSMUST00000114458.8
|
Nfkbiz
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, zeta |
| chr4_-_19570073 | 1.39 |
ENSMUST00000029885.5
|
Cpne3
|
copine III |
| chr5_+_12433344 | 1.39 |
ENSMUST00000030868.11
|
Sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chrX_+_41149264 | 1.39 |
ENSMUST00000224454.2
|
Xiap
|
X-linked inhibitor of apoptosis |
| chr5_-_68004702 | 1.38 |
ENSMUST00000135930.8
|
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 1.4 | 4.3 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 1.2 | 8.4 | GO:0060309 | elastin catabolic process(GO:0060309) |
| 1.1 | 3.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 1.1 | 3.2 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.9 | 3.7 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.9 | 4.4 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.9 | 2.6 | GO:0042495 | positive regulation of interleukin-18 production(GO:0032741) detection of triacyl bacterial lipopeptide(GO:0042495) |
| 0.9 | 2.6 | GO:0030845 | regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010924) positive regulation of inositol-polyphosphate 5-phosphatase activity(GO:0010925) phospholipase C-inhibiting G-protein coupled receptor signaling pathway(GO:0030845) regulation of cell diameter(GO:0060305) |
| 0.8 | 0.8 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.8 | 3.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.8 | 1.6 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.8 | 2.4 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 0.8 | 3.0 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.8 | 2.3 | GO:1901738 | regulation of vitamin A metabolic process(GO:1901738) |
| 0.7 | 2.9 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.7 | 2.2 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.7 | 2.8 | GO:1901376 | mycotoxin metabolic process(GO:0043385) aflatoxin metabolic process(GO:0046222) organic heteropentacyclic compound metabolic process(GO:1901376) |
| 0.7 | 2.8 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.7 | 4.1 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.7 | 2.7 | GO:2001201 | regulation of transforming growth factor-beta secretion(GO:2001201) |
| 0.6 | 1.9 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.6 | 3.2 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.6 | 1.9 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.6 | 2.5 | GO:0045425 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) cellular response to molecule of fungal origin(GO:0071226) |
| 0.6 | 3.5 | GO:1900085 | negative regulation of peptidyl-tyrosine autophosphorylation(GO:1900085) negative regulation of inward rectifier potassium channel activity(GO:1903609) |
| 0.6 | 2.9 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.6 | 5.0 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.5 | 1.6 | GO:0021558 | trochlear nerve development(GO:0021558) |
| 0.5 | 3.3 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.5 | 1.5 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.5 | 1.5 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.5 | 1.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.5 | 1.5 | GO:2000724 | response to mineralocorticoid(GO:0051385) regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.5 | 1.0 | GO:0086098 | angiotensin-activated signaling pathway involved in heart process(GO:0086098) |
| 0.5 | 1.4 | GO:0060935 | cardiac fibroblast cell differentiation(GO:0060935) cardiac fibroblast cell development(GO:0060936) epicardium-derived cardiac fibroblast cell differentiation(GO:0060938) epicardium-derived cardiac fibroblast cell development(GO:0060939) negative regulation of myofibroblast differentiation(GO:1904761) negative regulation of vascular smooth muscle cell differentiation(GO:1905064) |
| 0.4 | 1.8 | GO:0051329 | interphase(GO:0051325) mitotic interphase(GO:0051329) |
| 0.4 | 1.7 | GO:0038183 | bile acid signaling pathway(GO:0038183) |
| 0.4 | 1.7 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.4 | 4.2 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.4 | 1.7 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.4 | 0.8 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.4 | 2.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.4 | 1.2 | GO:0098974 | postsynaptic actin cytoskeleton organization(GO:0098974) |
| 0.4 | 1.8 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.4 | 2.2 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.4 | 1.1 | GO:2000451 | positive regulation of CD8-positive, alpha-beta T cell extravasation(GO:2000451) |
| 0.4 | 1.1 | GO:0072313 | metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.4 | 2.5 | GO:1900226 | negative regulation of NLRP3 inflammasome complex assembly(GO:1900226) |
| 0.4 | 0.7 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.4 | 1.1 | GO:0016132 | brassinosteroid metabolic process(GO:0016131) brassinosteroid biosynthetic process(GO:0016132) |
| 0.4 | 1.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.3 | 3.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
| 0.3 | 1.4 | GO:1902530 | regulation of protein linear polyubiquitination(GO:1902528) positive regulation of protein linear polyubiquitination(GO:1902530) |
| 0.3 | 1.3 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.3 | 1.9 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.3 | 4.5 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.3 | 1.9 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.3 | 0.9 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.3 | 1.2 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.3 | 5.6 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.3 | 3.0 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.3 | 0.9 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) |
| 0.3 | 1.4 | GO:1903378 | positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903378) |
| 0.3 | 1.9 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.3 | 2.7 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.3 | 1.3 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.3 | 1.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.3 | 2.4 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.3 | 1.1 | GO:0035565 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.3 | 0.8 | GO:0010816 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 0.5 | GO:1901252 | regulation of intracellular transport of viral material(GO:1901252) |
| 0.3 | 0.8 | GO:1905063 | detection of oxygen(GO:0003032) regulation of vascular smooth muscle cell differentiation(GO:1905063) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) cardiac jelly development(GO:1905072) |
| 0.3 | 0.8 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.3 | 6.5 | GO:2000009 | negative regulation of protein localization to cell surface(GO:2000009) |
| 0.2 | 1.0 | GO:0032887 | regulation of spindle elongation(GO:0032887) regulation of mitotic spindle elongation(GO:0032888) anastral spindle assembly(GO:0055048) protein localization to spindle pole body(GO:0071988) regulation of protein localization to spindle pole body(GO:1902363) positive regulation of protein localization to spindle pole body(GO:1902365) positive regulation of mitotic spindle elongation(GO:1902846) |
| 0.2 | 2.7 | GO:0001835 | blastocyst hatching(GO:0001835) hatching(GO:0035188) organism emergence from protective structure(GO:0071684) |
| 0.2 | 1.2 | GO:0006231 | dTMP biosynthetic process(GO:0006231) dTMP metabolic process(GO:0046073) |
| 0.2 | 6.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.2 | 0.9 | GO:0003017 | lymph circulation(GO:0003017) |
| 0.2 | 1.7 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.2 | 16.0 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.2 | 0.7 | GO:0036034 | mediator complex assembly(GO:0036034) regulation of mediator complex assembly(GO:2001176) positive regulation of mediator complex assembly(GO:2001178) |
| 0.2 | 1.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.2 | 0.6 | GO:0035622 | intrahepatic bile duct development(GO:0035622) |
| 0.2 | 1.0 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.2 | 1.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.2 | 0.6 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 1.0 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.2 | 1.7 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.2 | 1.5 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.2 | 1.9 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.2 | 1.7 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.2 | 0.7 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.2 | 1.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 1.5 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.2 | 2.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.2 | 1.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.2 | 1.8 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.2 | 0.7 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.2 | 1.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 1.0 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) protein localization to site of double-strand break(GO:1990166) |
| 0.2 | 0.5 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.2 | 0.5 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) activation of protein kinase C activity(GO:1990051) |
| 0.2 | 0.2 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.2 | 1.7 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.2 | 1.8 | GO:0090232 | positive regulation of spindle checkpoint(GO:0090232) |
| 0.2 | 1.2 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 2.6 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 1.8 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.2 | 1.0 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.2 | 1.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.2 | 0.6 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.2 | 1.9 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.2 | 1.8 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 1.1 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.2 | 1.4 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.2 | 0.8 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 1.0 | GO:0006868 | glutamine transport(GO:0006868) |
| 0.1 | 0.4 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 0.1 | 0.9 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.1 | 2.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.7 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 2.5 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.1 | 1.8 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.7 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 4.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.7 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 1.6 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.5 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.1 | 0.5 | GO:0090346 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 0.8 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.1 | 0.4 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.1 | 1.0 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.1 | 1.0 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.1 | 3.7 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 1.9 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.1 | 0.7 | GO:0060369 | positive regulation of natural killer cell cytokine production(GO:0002729) positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.1 | 1.2 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.1 | 0.8 | GO:0038165 | oncostatin-M-mediated signaling pathway(GO:0038165) |
| 0.1 | 0.9 | GO:0090315 | negative regulation of protein targeting to membrane(GO:0090315) |
| 0.1 | 2.4 | GO:0035814 | negative regulation of renal sodium excretion(GO:0035814) |
| 0.1 | 1.4 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.1 | 1.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 1.1 | GO:0051025 | negative regulation of immunoglobulin secretion(GO:0051025) |
| 0.1 | 1.3 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 1.1 | GO:2000535 | entry of bacterium into host cell(GO:0035635) regulation of entry of bacterium into host cell(GO:2000535) |
| 0.1 | 1.8 | GO:0007343 | egg activation(GO:0007343) |
| 0.1 | 0.9 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.3 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.3 | GO:1904155 | DN2 thymocyte differentiation(GO:1904155) DN3 thymocyte differentiation(GO:1904156) |
| 0.1 | 0.4 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.5 | GO:0071224 | cellular response to peptidoglycan(GO:0071224) |
| 0.1 | 2.6 | GO:0031424 | keratinization(GO:0031424) |
| 0.1 | 0.4 | GO:0060112 | positive regulation of luteinizing hormone secretion(GO:0033686) generation of ovulation cycle rhythm(GO:0060112) |
| 0.1 | 1.4 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.1 | 5.6 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.1 | 0.6 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.1 | 0.6 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 0.7 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.1 | 1.2 | GO:1902510 | regulation of apoptotic DNA fragmentation(GO:1902510) |
| 0.1 | 1.2 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.1 | 3.1 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 0.4 | GO:0050822 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-dependent(GO:0002479) peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.1 | 1.0 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.8 | GO:0008228 | opsonization(GO:0008228) |
| 0.1 | 1.5 | GO:0035810 | positive regulation of urine volume(GO:0035810) |
| 0.1 | 0.3 | GO:0045358 | negative regulation of interferon-beta biosynthetic process(GO:0045358) |
| 0.1 | 4.0 | GO:2000249 | regulation of actin cytoskeleton reorganization(GO:2000249) |
| 0.1 | 1.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.3 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.6 | GO:0015871 | choline transport(GO:0015871) |
| 0.1 | 1.0 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.1 | 0.3 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.1 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.2 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 2.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.1 | 0.5 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.1 | 2.1 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 1.6 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.1 | 0.1 | GO:1903660 | regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 3.4 | GO:0033006 | regulation of mast cell activation involved in immune response(GO:0033006) regulation of mast cell degranulation(GO:0043304) |
| 0.1 | 4.8 | GO:0051693 | actin filament capping(GO:0051693) |
| 0.1 | 0.6 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.1 | 5.8 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.6 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.1 | 0.3 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 3.4 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.7 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.4 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.5 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 0.5 | GO:0001887 | selenium compound metabolic process(GO:0001887) |
| 0.1 | 0.6 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.1 | 0.3 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.7 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.1 | 2.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.1 | 0.8 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.6 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.1 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.3 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 0.6 | GO:0046060 | dATP metabolic process(GO:0046060) |
| 0.1 | 0.4 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 15.8 | GO:0006486 | protein glycosylation(GO:0006486) macromolecule glycosylation(GO:0043413) |
| 0.0 | 0.4 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.8 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 3.7 | GO:0002066 | columnar/cuboidal epithelial cell development(GO:0002066) |
| 0.0 | 0.3 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.5 | GO:0045324 | late endosome to vacuole transport(GO:0045324) |
| 0.0 | 0.0 | GO:0010615 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.4 | GO:0051315 | attachment of mitotic spindle microtubules to kinetochore(GO:0051315) |
| 0.0 | 3.4 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.0 | 4.7 | GO:0046785 | microtubule polymerization(GO:0046785) |
| 0.0 | 0.3 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 2.4 | GO:1901385 | regulation of voltage-gated calcium channel activity(GO:1901385) |
| 0.0 | 0.3 | GO:0080182 | histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.3 | GO:0010838 | positive regulation of keratinocyte proliferation(GO:0010838) |
| 0.0 | 0.2 | GO:1904976 | cellular response to cisplatin(GO:0072719) response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.0 | 0.8 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.8 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 5.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 1.1 | GO:0031295 | T cell costimulation(GO:0031295) |
| 0.0 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.3 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 1.8 | GO:0031952 | regulation of protein autophosphorylation(GO:0031952) |
| 0.0 | 8.2 | GO:0002377 | immunoglobulin production(GO:0002377) |
| 0.0 | 2.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.6 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 2.8 | GO:0006970 | response to osmotic stress(GO:0006970) |
| 0.0 | 0.3 | GO:0061052 | negative regulation of cell growth involved in cardiac muscle cell development(GO:0061052) |
| 0.0 | 0.4 | GO:0000022 | mitotic spindle elongation(GO:0000022) |
| 0.0 | 1.0 | GO:0008206 | bile acid metabolic process(GO:0008206) |
| 0.0 | 0.7 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.8 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 3.6 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.5 | GO:0045947 | negative regulation of translational initiation(GO:0045947) |
| 0.0 | 0.8 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.5 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.4 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.0 | 0.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 1.4 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 4.1 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 1.2 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 1.9 | GO:1902476 | chloride transmembrane transport(GO:1902476) |
| 0.0 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.4 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.9 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.5 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 1.0 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 1.0 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 2.8 | GO:0045445 | myoblast differentiation(GO:0045445) |
| 0.0 | 0.1 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.1 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.9 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.4 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.9 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.5 | GO:0043433 | negative regulation of sequence-specific DNA binding transcription factor activity(GO:0043433) |
| 0.0 | 0.7 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.7 | GO:0090307 | mitotic spindle assembly(GO:0090307) |
| 0.0 | 0.2 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.8 | GO:0042035 | regulation of cytokine biosynthetic process(GO:0042035) |
| 0.0 | 0.2 | GO:2000169 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) regulation of peptidyl-cysteine S-nitrosylation(GO:2000169) |
| 0.0 | 0.1 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.7 | GO:0045739 | positive regulation of DNA repair(GO:0045739) |
| 0.0 | 2.5 | GO:0032496 | response to lipopolysaccharide(GO:0032496) |
| 0.0 | 0.2 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.0 | 0.3 | GO:0019184 | nonribosomal peptide biosynthetic process(GO:0019184) |
| 0.0 | 0.5 | GO:0010501 | RNA secondary structure unwinding(GO:0010501) |
| 0.0 | 1.0 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 1.4 | GO:0006937 | regulation of muscle contraction(GO:0006937) |
| 0.0 | 2.1 | GO:0070588 | calcium ion transmembrane transport(GO:0070588) |
| 0.0 | 0.1 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 6.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.9 | 2.6 | GO:0035354 | Toll-like receptor 1-Toll-like receptor 2 protein complex(GO:0035354) |
| 0.6 | 1.9 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.6 | 3.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.6 | 1.9 | GO:1902912 | pyruvate kinase complex(GO:1902912) |
| 0.6 | 3.7 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.6 | 1.7 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.6 | 1.7 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.5 | 1.5 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.5 | 2.9 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.5 | 3.2 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 5.8 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.4 | 4.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.4 | 4.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.4 | 2.8 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.4 | 3.1 | GO:0001652 | granular component(GO:0001652) |
| 0.3 | 2.4 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.3 | 1.2 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.3 | 17.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.3 | 4.1 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.3 | 1.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.3 | 1.0 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.2 | 1.7 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.2 | 3.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.2 | 1.7 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.2 | 0.7 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.2 | 3.5 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.2 | 0.6 | GO:0036398 | TCR signalosome(GO:0036398) |
| 0.2 | 1.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 2.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 1.2 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.2 | 0.9 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.2 | 1.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.2 | 1.9 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.2 | 0.6 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.1 | 5.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.2 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 1.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 1.9 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.3 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 1.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 1.1 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.7 | GO:0031523 | Myb complex(GO:0031523) |
| 0.1 | 5.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.1 | 2.8 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.1 | 2.2 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.5 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 3.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 2.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.1 | 0.8 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 2.4 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.1 | 1.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 2.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.1 | 0.3 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.7 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 5.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 1.9 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.1 | GO:0005605 | basal lamina(GO:0005605) |
| 0.1 | 0.2 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.1 | 1.0 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 2.4 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.3 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.7 | GO:0000782 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.1 | 3.1 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.1 | 0.9 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.0 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.4 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 5.4 | GO:0019814 | immunoglobulin complex(GO:0019814) immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.4 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) TAP complex(GO:0042825) |
| 0.0 | 8.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.5 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0000938 | GARP complex(GO:0000938) EARP complex(GO:1990745) |
| 0.0 | 1.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 1.7 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.5 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 4.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.5 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.6 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 5.2 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 1.0 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.5 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 2.3 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 6.8 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 5.3 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 7.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 1.2 | GO:0097546 | ciliary base(GO:0097546) |
| 0.0 | 0.2 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 3.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.6 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.2 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.5 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.8 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 3.7 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.5 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.4 | GO:0005732 | small nucleolar ribonucleoprotein complex(GO:0005732) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 1.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 3.7 | GO:0042383 | sarcolemma(GO:0042383) |
| 0.0 | 8.9 | GO:0005925 | focal adhesion(GO:0005925) |
| 0.0 | 1.3 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 1.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.9 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 2.2 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.0 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 29.9 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 2.6 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 0.3 | GO:0031672 | A band(GO:0031672) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 3.2 | GO:0005759 | mitochondrial matrix(GO:0005759) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 7.3 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 1.4 | 4.3 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 1.0 | 4.2 | GO:0047522 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 1.0 | 3.1 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.9 | 2.6 | GO:0042497 | triacyl lipopeptide binding(GO:0042497) |
| 0.8 | 3.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.8 | 4.7 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.8 | 4.6 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.7 | 2.6 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.6 | 1.9 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.6 | 3.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.6 | 1.8 | GO:0004686 | elongation factor-2 kinase activity(GO:0004686) |
| 0.6 | 3.5 | GO:0070320 | inward rectifier potassium channel inhibitor activity(GO:0070320) |
| 0.6 | 1.7 | GO:1902121 | lithocholic acid binding(GO:1902121) |
| 0.5 | 1.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.5 | 0.5 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.5 | 1.5 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.5 | 1.5 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.5 | 2.4 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.5 | 3.2 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.5 | 2.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.4 | 1.7 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 0.4 | 4.7 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.4 | 3.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.4 | 1.6 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.4 | 1.2 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.4 | 1.5 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.4 | 1.2 | GO:0098918 | structural constituent of synapse(GO:0098918) structural constituent of postsynaptic actin cytoskeleton(GO:0098973) |
| 0.4 | 3.8 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.4 | 4.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.4 | 1.9 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.4 | 1.1 | GO:0047598 | sterol delta7 reductase activity(GO:0009918) 7-dehydrocholesterol reductase activity(GO:0047598) |
| 0.3 | 1.0 | GO:0008775 | acetate CoA-transferase activity(GO:0008775) |
| 0.3 | 6.4 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.3 | 3.5 | GO:0022820 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.3 | 5.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.3 | 4.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.3 | 5.6 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.3 | 1.7 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.3 | 2.5 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.3 | 3.2 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.3 | 1.8 | GO:0010859 | calcium-dependent cysteine-type endopeptidase inhibitor activity(GO:0010859) |
| 0.3 | 3.1 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 1.2 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.2 | 1.2 | GO:0004174 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.2 | 1.6 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.2 | 0.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.2 | 1.7 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 0.6 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.2 | 2.9 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 1.4 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.2 | 1.8 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 0.7 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 6.4 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.2 | 3.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.2 | 1.6 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.2 | 3.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 1.0 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.2 | 1.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 2.9 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.2 | 1.6 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.2 | 2.1 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.2 | 4.1 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.2 | 3.9 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.2 | 0.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.0 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.1 | 0.4 | GO:0047945 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 0.1 | 1.8 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.1 | 1.4 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.8 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 2.3 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 2.6 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 3.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 4.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.5 | GO:0047726 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 1.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 0.5 | GO:0071796 | K6-linked polyubiquitin binding(GO:0071796) |
| 0.1 | 3.7 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.1 | 1.0 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 1.0 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.1 | 1.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.1 | 1.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.5 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.7 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 1.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 1.5 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.1 | 2.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 1.2 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.1 | 1.7 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 0.7 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.1 | 1.4 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.6 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.3 | GO:0001096 | TFIIF-class transcription factor binding(GO:0001096) |
| 0.1 | 1.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.1 | 1.6 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 1.7 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.1 | 3.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 11.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.1 | 0.5 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 1.0 | GO:0071253 | connexin binding(GO:0071253) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 2.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 2.3 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 1.7 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 1.0 | GO:0019104 | DNA N-glycosylase activity(GO:0019104) |
| 0.1 | 1.5 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 4.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 1.2 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 1.0 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.1 | 2.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 5.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.1 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.7 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.1 | 1.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.1 | 0.4 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.6 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 13.8 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.1 | 7.0 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.3 | GO:0004043 | L-aminoadipate-semialdehyde dehydrogenase activity(GO:0004043) |
| 0.1 | 0.4 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 2.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.6 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 3.1 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 12.0 | GO:0008236 | serine-type peptidase activity(GO:0008236) serine hydrolase activity(GO:0017171) |
| 0.0 | 3.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 2.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.9 | GO:0035615 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 3.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.7 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.9 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 0.6 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.0 | 1.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 2.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 1.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 1.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.0 | 3.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.9 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 1.9 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 1.3 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.4 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 3.5 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 4.0 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 3.5 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 2.4 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.5 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.8 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 1.0 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.7 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 5.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.5 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.7 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.8 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.7 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.6 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 10.1 | GO:0005198 | structural molecule activity(GO:0005198) |
| 0.0 | 4.7 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 1.4 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 1.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.0 | 1.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.2 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 1.4 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 1.2 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 1.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.8 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.7 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 0.3 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 2.5 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.6 | GO:0008376 | acetylgalactosaminyltransferase activity(GO:0008376) |
| 0.0 | 0.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.8 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 1.0 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.4 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 2.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
| 0.0 | 2.0 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.6 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 3.5 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 1.8 | GO:0017124 | SH3 domain binding(GO:0017124) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.9 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.2 | 6.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.2 | 7.5 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.2 | 2.8 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.2 | 9.0 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 6.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.1 | 7.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 2.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.1 | 3.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 3.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 1.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.1 | 3.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.1 | 11.2 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.1 | 1.7 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 3.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.1 | 5.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 2.6 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.1 | 1.4 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.1 | 2.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.1 | 0.5 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 1.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 3.3 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.1 | 1.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 3.8 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.1 | 2.3 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.1 | 4.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.1 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 2.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.1 | 2.0 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 2.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 7.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.6 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.4 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 8.5 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.9 | PID E2F PATHWAY | E2F transcription factor network |
| 0.0 | 1.2 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.5 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 2.2 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.8 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.5 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 6.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.2 | 6.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.2 | 3.9 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.2 | 4.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 5.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.2 | 1.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.2 | 2.4 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.1 | 1.8 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.1 | 2.2 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 9.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 6.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 2.1 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.1 | 2.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 6.6 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 3.7 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.7 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 5.9 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.1 | 4.2 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.1 | 2.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.1 | 1.8 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.1 | 1.8 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.1 | 4.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.9 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.1 | 1.2 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.1 | 1.1 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 3.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.1 | 1.5 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.7 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 1.0 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.1 | 0.6 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 1.7 | REACTOME SHC1 EVENTS IN ERBB4 SIGNALING | Genes involved in SHC1 events in ERBB4 signaling |
| 0.1 | 7.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.1 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.1 | 4.5 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.1 | 0.5 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 1.4 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 2.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 1.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.9 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 1.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 1.7 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 1.2 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 2.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 2.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 2.2 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.0 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.7 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.8 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.8 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 1.2 | REACTOME CROSS PRESENTATION OF SOLUBLE EXOGENOUS ANTIGENS ENDOSOMES | Genes involved in Cross-presentation of soluble exogenous antigens (endosomes) |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.1 | REACTOME AQUAPORIN MEDIATED TRANSPORT | Genes involved in Aquaporin-mediated transport |
| 0.0 | 0.3 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.4 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.7 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.1 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.9 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.3 | REACTOME ABORTIVE ELONGATION OF HIV1 TRANSCRIPT IN THE ABSENCE OF TAT | Genes involved in Abortive elongation of HIV-1 transcript in the absence of Tat |