Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zic3
Z-value: 1.02
Transcription factors associated with Zic3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic3
|
ENSMUSG00000067860.12 | Zic3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic3 | mm39_v1_chrX_+_57076359_57076378 | -0.33 | 4.7e-03 | Click! |
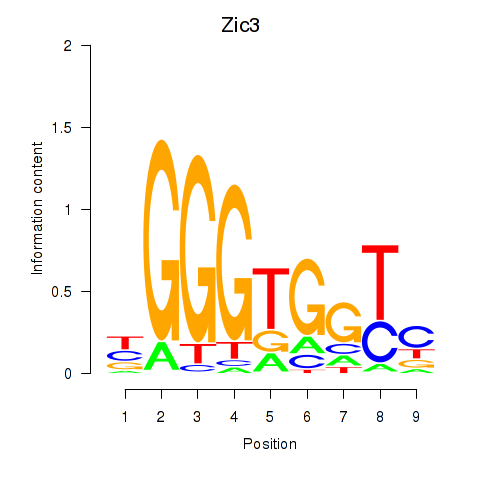
Activity profile of Zic3 motif
Sorted Z-values of Zic3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_87357657 | 9.99 |
ENSMUST00000020241.17
|
Pah
|
phenylalanine hydroxylase |
| chr10_+_87357782 | 9.36 |
ENSMUST00000219813.2
|
Pah
|
phenylalanine hydroxylase |
| chr10_+_87357816 | 9.10 |
ENSMUST00000218573.2
|
Pah
|
phenylalanine hydroxylase |
| chr7_+_51528715 | 7.49 |
ENSMUST00000051912.13
|
Gas2
|
growth arrest specific 2 |
| chr7_+_51528788 | 6.44 |
ENSMUST00000107591.9
|
Gas2
|
growth arrest specific 2 |
| chr11_-_75329726 | 5.84 |
ENSMUST00000108437.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr4_+_108022645 | 5.11 |
ENSMUST00000116309.10
ENSMUST00000116307.8 |
Echdc2
|
enoyl Coenzyme A hydratase domain containing 2 |
| chr4_+_108022628 | 5.00 |
ENSMUST00000052999.13
|
Echdc2
|
enoyl Coenzyme A hydratase domain containing 2 |
| chr12_-_103739847 | 4.75 |
ENSMUST00000078869.6
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr15_-_78687216 | 4.64 |
ENSMUST00000164826.8
|
Card10
|
caspase recruitment domain family, member 10 |
| chr7_+_127399789 | 4.34 |
ENSMUST00000125188.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr2_+_31840340 | 4.23 |
ENSMUST00000148056.4
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr2_+_31840151 | 4.18 |
ENSMUST00000001920.13
ENSMUST00000151276.3 |
Aif1l
|
allograft inflammatory factor 1-like |
| chr7_+_127399776 | 4.04 |
ENSMUST00000046863.12
ENSMUST00000206674.2 ENSMUST00000106272.8 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr8_+_127790772 | 3.93 |
ENSMUST00000079777.12
ENSMUST00000160272.8 ENSMUST00000162907.8 ENSMUST00000162536.8 ENSMUST00000026921.13 ENSMUST00000162665.8 ENSMUST00000162602.8 ENSMUST00000160581.8 ENSMUST00000161355.8 ENSMUST00000162531.8 ENSMUST00000160766.8 ENSMUST00000159537.8 |
Pard3
|
par-3 family cell polarity regulator |
| chr3_+_14545751 | 3.85 |
ENSMUST00000037321.8
ENSMUST00000120484.8 ENSMUST00000120801.2 |
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr1_+_192835414 | 3.85 |
ENSMUST00000076521.7
|
Irf6
|
interferon regulatory factor 6 |
| chr7_+_127399848 | 3.84 |
ENSMUST00000139068.8
|
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr14_-_31362909 | 3.51 |
ENSMUST00000022437.16
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr14_-_31362835 | 3.50 |
ENSMUST00000167066.8
ENSMUST00000127204.9 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr2_+_91087668 | 3.44 |
ENSMUST00000111349.9
ENSMUST00000131711.8 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr11_-_70120503 | 3.44 |
ENSMUST00000153449.2
ENSMUST00000000326.12 |
Bcl6b
|
B cell CLL/lymphoma 6, member B |
| chr6_-_52211882 | 3.34 |
ENSMUST00000125581.2
|
Hoxa10
|
homeobox A10 |
| chr3_+_118355778 | 3.33 |
ENSMUST00000039177.12
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr14_-_30665232 | 3.29 |
ENSMUST00000006704.17
ENSMUST00000163118.2 |
Itih1
|
inter-alpha trypsin inhibitor, heavy chain 1 |
| chr2_-_170339061 | 3.28 |
ENSMUST00000038824.6
|
Cyp24a1
|
cytochrome P450, family 24, subfamily a, polypeptide 1 |
| chr4_-_106349480 | 3.03 |
ENSMUST00000054472.4
|
Bsnd
|
barttin CLCNK type accessory beta subunit |
| chr15_+_7159038 | 2.99 |
ENSMUST00000067190.12
ENSMUST00000164529.9 |
Lifr
|
LIF receptor alpha |
| chr2_+_70305267 | 2.97 |
ENSMUST00000100043.3
|
Sp5
|
trans-acting transcription factor 5 |
| chr10_-_127358231 | 2.97 |
ENSMUST00000219239.2
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr6_-_52168675 | 2.90 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr14_+_33662976 | 2.82 |
ENSMUST00000100720.2
|
Gdf2
|
growth differentiation factor 2 |
| chr2_+_124994425 | 2.81 |
ENSMUST00000110494.9
ENSMUST00000110495.3 ENSMUST00000028630.9 |
Slc12a1
|
solute carrier family 12, member 1 |
| chr10_-_127358300 | 2.79 |
ENSMUST00000026470.6
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr2_+_78699360 | 2.77 |
ENSMUST00000028398.14
|
Ube2e3
|
ubiquitin-conjugating enzyme E2E 3 |
| chr11_-_7163897 | 2.72 |
ENSMUST00000020702.11
ENSMUST00000135887.3 |
Igfbp3
|
insulin-like growth factor binding protein 3 |
| chr6_+_24597724 | 2.72 |
ENSMUST00000031694.8
|
Lmod2
|
leiomodin 2 (cardiac) |
| chr9_-_96900876 | 2.66 |
ENSMUST00000055433.5
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr8_+_120163857 | 2.63 |
ENSMUST00000152420.8
ENSMUST00000212112.2 ENSMUST00000098365.4 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr9_-_50663571 | 2.60 |
ENSMUST00000042790.5
|
Hspb2
|
heat shock protein 2 |
| chr14_+_34097422 | 2.48 |
ENSMUST00000111908.3
|
Mmrn2
|
multimerin 2 |
| chr11_+_96920956 | 2.45 |
ENSMUST00000153482.2
|
Scrn2
|
secernin 2 |
| chr19_+_56862681 | 2.43 |
ENSMUST00000026068.8
|
Vwa2
|
von Willebrand factor A domain containing 2 |
| chr10_-_107321938 | 2.41 |
ENSMUST00000000445.2
|
Myf5
|
myogenic factor 5 |
| chr9_+_50663171 | 2.39 |
ENSMUST00000214609.2
|
Cryab
|
crystallin, alpha B |
| chr11_+_96920751 | 2.39 |
ENSMUST00000021249.11
|
Scrn2
|
secernin 2 |
| chr10_-_95159933 | 2.33 |
ENSMUST00000053594.7
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr3_+_118355811 | 2.30 |
ENSMUST00000149101.3
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr16_-_30086317 | 2.19 |
ENSMUST00000064856.9
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr12_-_69771604 | 2.19 |
ENSMUST00000021370.10
|
L2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr14_-_64654397 | 2.12 |
ENSMUST00000210428.2
|
Msra
|
methionine sulfoxide reductase A |
| chr13_+_74269554 | 2.07 |
ENSMUST00000036208.7
ENSMUST00000225423.2 ENSMUST00000221703.2 |
Slc9a3
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 |
| chr6_+_82029288 | 2.05 |
ENSMUST00000149023.2
|
Eva1a
|
eva-1 homolog A (C. elegans) |
| chr14_+_31363004 | 2.04 |
ENSMUST00000090147.7
|
Btd
|
biotinidase |
| chr1_-_105284383 | 2.02 |
ENSMUST00000058688.7
|
Rnf152
|
ring finger protein 152 |
| chr13_-_25121568 | 2.00 |
ENSMUST00000037615.7
|
Aldh5a1
|
aldhehyde dehydrogenase family 5, subfamily A1 |
| chr11_+_69729340 | 1.98 |
ENSMUST00000133967.8
ENSMUST00000094065.5 |
Tmem256
|
transmembrane protein 256 |
| chr7_+_28524627 | 1.97 |
ENSMUST00000066264.13
|
Ech1
|
enoyl coenzyme A hydratase 1, peroxisomal |
| chr12_-_119202527 | 1.96 |
ENSMUST00000026360.9
|
Itgb8
|
integrin beta 8 |
| chr7_+_100966289 | 1.96 |
ENSMUST00000163799.9
ENSMUST00000164479.9 |
Stard10
|
START domain containing 10 |
| chr11_+_3981769 | 1.95 |
ENSMUST00000019512.8
|
Sec14l4
|
SEC14-like lipid binding 4 |
| chr9_+_44584523 | 1.93 |
ENSMUST00000034609.11
ENSMUST00000071219.12 |
Treh
|
trehalase (brush-border membrane glycoprotein) |
| chr11_-_70590923 | 1.93 |
ENSMUST00000108543.4
ENSMUST00000108542.8 ENSMUST00000108541.9 ENSMUST00000126114.9 ENSMUST00000073625.8 |
Inca1
|
inhibitor of CDK, cyclin A1 interacting protein 1 |
| chr19_-_4889284 | 1.90 |
ENSMUST00000236451.2
ENSMUST00000236178.2 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr3_-_57483330 | 1.89 |
ENSMUST00000120977.2
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr15_-_89310060 | 1.89 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr1_-_105284407 | 1.88 |
ENSMUST00000172299.2
|
Rnf152
|
ring finger protein 152 |
| chr1_-_132067404 | 1.87 |
ENSMUST00000027697.12
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr19_-_4889314 | 1.85 |
ENSMUST00000235245.2
ENSMUST00000037246.7 |
Ccs
|
copper chaperone for superoxide dismutase |
| chr9_+_21746785 | 1.85 |
ENSMUST00000058777.8
|
Angptl8
|
angiopoietin-like 8 |
| chrX_+_72030945 | 1.85 |
ENSMUST00000164800.8
ENSMUST00000114546.9 |
Zfp185
|
zinc finger protein 185 |
| chr3_+_19698631 | 1.80 |
ENSMUST00000029139.9
|
Trim55
|
tripartite motif-containing 55 |
| chr9_+_90045109 | 1.80 |
ENSMUST00000113059.8
ENSMUST00000167122.8 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr9_-_108141105 | 1.79 |
ENSMUST00000166905.8
ENSMUST00000191899.6 |
Dag1
|
dystroglycan 1 |
| chr16_-_85347305 | 1.78 |
ENSMUST00000175700.8
ENSMUST00000114174.3 |
Cyyr1
|
cysteine and tyrosine-rich protein 1 |
| chr3_-_57483175 | 1.77 |
ENSMUST00000029380.14
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr11_+_106265645 | 1.76 |
ENSMUST00000106816.8
|
Prr29
|
proline rich 29 |
| chr18_+_24842493 | 1.76 |
ENSMUST00000037097.9
ENSMUST00000234526.2 ENSMUST00000234834.2 |
Fhod3
|
formin homology 2 domain containing 3 |
| chr8_-_11528615 | 1.75 |
ENSMUST00000033900.7
|
Rab20
|
RAB20, member RAS oncogene family |
| chr9_+_90045219 | 1.75 |
ENSMUST00000147250.8
ENSMUST00000113060.3 |
Adamts7
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 7 |
| chr2_+_131075965 | 1.75 |
ENSMUST00000041362.12
ENSMUST00000110199.3 |
Mavs
|
mitochondrial antiviral signaling protein |
| chr2_-_92201311 | 1.71 |
ENSMUST00000176774.8
|
Large2
|
LARGE xylosyl- and glucuronyltransferase 2 |
| chr15_-_101833160 | 1.71 |
ENSMUST00000023797.8
|
Krt4
|
keratin 4 |
| chr2_+_172392911 | 1.69 |
ENSMUST00000170744.2
|
Tfap2c
|
transcription factor AP-2, gamma |
| chr1_-_132067343 | 1.68 |
ENSMUST00000112362.3
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr13_-_34529157 | 1.68 |
ENSMUST00000040336.12
|
Slc22a23
|
solute carrier family 22, member 23 |
| chr15_+_78912642 | 1.68 |
ENSMUST00000180086.3
|
H1f0
|
H1.0 linker histone |
| chr11_-_69696428 | 1.59 |
ENSMUST00000051025.5
|
Tmem102
|
transmembrane protein 102 |
| chr19_+_44744484 | 1.59 |
ENSMUST00000174490.9
|
Pax2
|
paired box 2 |
| chr2_+_30061469 | 1.59 |
ENSMUST00000015481.6
|
Endog
|
endonuclease G |
| chr9_+_37439367 | 1.57 |
ENSMUST00000002011.14
|
Esam
|
endothelial cell-specific adhesion molecule |
| chr10_+_108168520 | 1.57 |
ENSMUST00000218332.2
|
Pawr
|
PRKC, apoptosis, WT1, regulator |
| chr11_+_60428788 | 1.56 |
ENSMUST00000044250.4
|
Alkbh5
|
alkB homolog 5, RNA demethylase |
| chr6_-_52195663 | 1.55 |
ENSMUST00000134367.4
|
Hoxa7
|
homeobox A7 |
| chr7_+_45522551 | 1.54 |
ENSMUST00000211234.2
|
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr11_+_67689094 | 1.53 |
ENSMUST00000168612.8
|
Dhrs7c
|
dehydrogenase/reductase (SDR family) member 7C |
| chr5_-_98178811 | 1.50 |
ENSMUST00000031281.14
|
Antxr2
|
anthrax toxin receptor 2 |
| chr8_-_117720198 | 1.49 |
ENSMUST00000040484.6
|
Gcsh
|
glycine cleavage system protein H (aminomethyl carrier) |
| chr15_-_7427815 | 1.47 |
ENSMUST00000096494.5
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr15_-_7427759 | 1.47 |
ENSMUST00000058593.10
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr16_-_3895642 | 1.46 |
ENSMUST00000006137.9
|
Trap1
|
TNF receptor-associated protein 1 |
| chr14_+_67470735 | 1.44 |
ENSMUST00000022637.14
|
Ebf2
|
early B cell factor 2 |
| chr5_-_53370761 | 1.44 |
ENSMUST00000031090.8
|
Sel1l3
|
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr17_-_85097945 | 1.43 |
ENSMUST00000112308.9
|
Lrpprc
|
leucine-rich PPR-motif containing |
| chr8_-_71964379 | 1.43 |
ENSMUST00000048452.6
|
Plvap
|
plasmalemma vesicle associated protein |
| chr7_+_127400016 | 1.42 |
ENSMUST00000106271.2
ENSMUST00000138432.2 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr3_+_121220146 | 1.42 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr3_+_145694400 | 1.40 |
ENSMUST00000200546.2
|
Syde2
|
synapse defective 1, Rho GTPase, homolog 2 (C. elegans) |
| chr2_-_33530492 | 1.40 |
ENSMUST00000176067.2
ENSMUST00000239476.2 ENSMUST00000041730.12 |
Lmx1b
|
LIM homeobox transcription factor 1 beta |
| chr12_+_111780604 | 1.36 |
ENSMUST00000021714.9
ENSMUST00000223211.2 ENSMUST00000222843.2 ENSMUST00000221375.2 |
Zfyve21
|
zinc finger, FYVE domain containing 21 |
| chr14_+_55798517 | 1.34 |
ENSMUST00000117701.8
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr4_+_43631935 | 1.33 |
ENSMUST00000030191.15
|
Npr2
|
natriuretic peptide receptor 2 |
| chr9_-_44632680 | 1.33 |
ENSMUST00000148929.2
ENSMUST00000123406.8 |
Phldb1
|
pleckstrin homology like domain, family B, member 1 |
| chr11_+_95010935 | 1.32 |
ENSMUST00000092768.7
|
Dlx3
|
distal-less homeobox 3 |
| chr3_+_94249445 | 1.32 |
ENSMUST00000198083.2
|
Them5
|
thioesterase superfamily member 5 |
| chr4_-_144190326 | 1.31 |
ENSMUST00000105749.2
|
Aadacl3
|
arylacetamide deacetylase like 3 |
| chr11_+_98337655 | 1.31 |
ENSMUST00000019456.5
|
Grb7
|
growth factor receptor bound protein 7 |
| chr17_-_56312555 | 1.29 |
ENSMUST00000043785.8
|
Stap2
|
signal transducing adaptor family member 2 |
| chr11_+_6241606 | 1.29 |
ENSMUST00000003461.15
|
Ogdh
|
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
| chr14_+_55797934 | 1.28 |
ENSMUST00000121622.8
ENSMUST00000143431.2 ENSMUST00000150481.8 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr5_-_66309244 | 1.27 |
ENSMUST00000167950.8
|
Rbm47
|
RNA binding motif protein 47 |
| chr2_-_104647041 | 1.25 |
ENSMUST00000117237.2
ENSMUST00000231375.2 |
Qser1
|
glutamine and serine rich 1 |
| chr10_-_95159410 | 1.25 |
ENSMUST00000217809.2
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr19_+_24853039 | 1.24 |
ENSMUST00000073080.7
|
Gm10053
|
predicted gene 10053 |
| chr4_+_140688514 | 1.24 |
ENSMUST00000010007.9
|
Sdhb
|
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chr1_-_182109773 | 1.22 |
ENSMUST00000133052.2
|
Degs1
|
delta(4)-desaturase, sphingolipid 1 |
| chr2_+_172392678 | 1.21 |
ENSMUST00000099058.10
|
Tfap2c
|
transcription factor AP-2, gamma |
| chr6_-_92920466 | 1.20 |
ENSMUST00000113438.8
|
Adamts9
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 9 |
| chr14_+_55798362 | 1.18 |
ENSMUST00000072530.11
ENSMUST00000128490.9 |
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr1_+_135768409 | 1.17 |
ENSMUST00000189826.7
|
Tnnt2
|
troponin T2, cardiac |
| chrX_+_19925772 | 1.16 |
ENSMUST00000044138.8
|
Chst7
|
carbohydrate (N-acetylglucosamino) sulfotransferase 7 |
| chr6_-_40521911 | 1.15 |
ENSMUST00000089490.3
|
Olfr461
|
olfactory receptor 461 |
| chr11_-_113600838 | 1.14 |
ENSMUST00000018871.8
|
Cpsf4l
|
cleavage and polyadenylation specific factor 4-like |
| chr6_+_48819523 | 1.13 |
ENSMUST00000204309.2
|
Tmem176a
|
transmembrane protein 176A |
| chr9_-_108140904 | 1.13 |
ENSMUST00000194698.2
|
Dag1
|
dystroglycan 1 |
| chr10_-_128237087 | 1.12 |
ENSMUST00000042666.13
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr7_-_111379170 | 1.12 |
ENSMUST00000049430.15
ENSMUST00000106663.2 |
Galnt18
|
polypeptide N-acetylgalactosaminyltransferase 18 |
| chr17_+_29077385 | 1.11 |
ENSMUST00000056866.8
|
Pnpla1
|
patatin-like phospholipase domain containing 1 |
| chr5_+_135754568 | 1.11 |
ENSMUST00000127096.2
|
Por
|
P450 (cytochrome) oxidoreductase |
| chr15_-_89309998 | 1.11 |
ENSMUST00000168376.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr2_-_155772110 | 1.11 |
ENSMUST00000109636.11
ENSMUST00000109631.8 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr11_+_102727122 | 1.10 |
ENSMUST00000021302.15
ENSMUST00000107072.2 |
Higd1b
|
HIG1 domain family, member 1B |
| chr17_-_68311073 | 1.10 |
ENSMUST00000024840.12
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr6_+_73225616 | 1.10 |
ENSMUST00000203632.2
|
Suclg1
|
succinate-CoA ligase, GDP-forming, alpha subunit |
| chr18_-_84969601 | 1.09 |
ENSMUST00000025547.4
|
Timm21
|
translocase of inner mitochondrial membrane 21 |
| chr9_+_106331111 | 1.09 |
ENSMUST00000185779.7
|
Pcbp4
|
poly(rC) binding protein 4 |
| chr9_-_50663648 | 1.09 |
ENSMUST00000217159.2
|
Hspb2
|
heat shock protein 2 |
| chr11_-_72380730 | 1.08 |
ENSMUST00000045303.10
|
Spns2
|
spinster homolog 2 |
| chr5_-_66308421 | 1.08 |
ENSMUST00000200775.4
ENSMUST00000094756.11 |
Rbm47
|
RNA binding motif protein 47 |
| chr17_-_28736483 | 1.07 |
ENSMUST00000114792.8
ENSMUST00000177939.8 |
Fkbp5
|
FK506 binding protein 5 |
| chr13_-_3854307 | 1.07 |
ENSMUST00000077698.5
|
Calml3
|
calmodulin-like 3 |
| chr11_+_115790768 | 1.06 |
ENSMUST00000152171.8
|
Smim5
|
small integral membrane protein 5 |
| chr11_+_86375441 | 1.05 |
ENSMUST00000020827.7
|
Rnft1
|
ring finger protein, transmembrane 1 |
| chr1_-_136876902 | 1.05 |
ENSMUST00000195428.2
|
Nr5a2
|
nuclear receptor subfamily 5, group A, member 2 |
| chr16_+_48662894 | 1.05 |
ENSMUST00000238847.2
ENSMUST00000023329.7 |
Retnla
|
resistin like alpha |
| chr9_-_45817666 | 1.04 |
ENSMUST00000161187.8
|
Rnf214
|
ring finger protein 214 |
| chr7_+_139695707 | 1.04 |
ENSMUST00000211757.2
|
Paox
|
polyamine oxidase (exo-N4-amino) |
| chr9_+_106331041 | 1.04 |
ENSMUST00000024260.14
ENSMUST00000216379.2 ENSMUST00000215656.2 ENSMUST00000214252.2 |
Pcbp4
|
poly(rC) binding protein 4 |
| chr8_-_65471175 | 1.02 |
ENSMUST00000078409.5
ENSMUST00000048565.9 |
Trim61
Trim60
|
tripartite motif-containing 61 tripartite motif-containing 60 |
| chr5_-_31453206 | 1.02 |
ENSMUST00000041266.11
ENSMUST00000172435.8 ENSMUST00000201417.2 |
Fndc4
|
fibronectin type III domain containing 4 |
| chr3_-_92393193 | 1.02 |
ENSMUST00000054599.8
|
Sprr1a
|
small proline-rich protein 1A |
| chr2_-_155771938 | 1.01 |
ENSMUST00000152766.8
ENSMUST00000139232.8 ENSMUST00000109632.8 ENSMUST00000006036.13 ENSMUST00000142655.2 ENSMUST00000159238.2 |
Uqcc1
|
ubiquinol-cytochrome c reductase complex assembly factor 1 |
| chr2_-_35995283 | 0.98 |
ENSMUST00000112960.8
ENSMUST00000112967.12 ENSMUST00000112963.8 |
Lhx6
|
LIM homeobox protein 6 |
| chr11_+_70431063 | 0.98 |
ENSMUST00000018429.12
ENSMUST00000108557.10 ENSMUST00000108556.2 |
Pld2
|
phospholipase D2 |
| chr2_+_90865958 | 0.98 |
ENSMUST00000111445.10
ENSMUST00000111446.10 ENSMUST00000050323.6 |
Rapsn
|
receptor-associated protein of the synapse |
| chr13_-_21257469 | 0.97 |
ENSMUST00000058168.2
|
Olfr1370
|
olfactory receptor 1370 |
| chr7_+_45522196 | 0.97 |
ENSMUST00000002855.14
ENSMUST00000211716.2 |
Kdelr1
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 |
| chr5_-_37874461 | 0.97 |
ENSMUST00000094836.6
|
Stk32b
|
serine/threonine kinase 32B |
| chr17_-_74354844 | 0.94 |
ENSMUST00000043458.9
|
Srd5a2
|
steroid 5 alpha-reductase 2 |
| chr6_-_39702381 | 0.94 |
ENSMUST00000002487.15
|
Braf
|
Braf transforming gene |
| chr15_+_102178975 | 0.93 |
ENSMUST00000181801.2
|
Gm9918
|
predicted gene 9918 |
| chr1_+_187341225 | 0.92 |
ENSMUST00000110939.8
|
Esrrg
|
estrogen-related receptor gamma |
| chr11_+_106265660 | 0.91 |
ENSMUST00000188561.7
ENSMUST00000190795.7 ENSMUST00000185986.7 ENSMUST00000190268.2 |
Prr29
|
proline rich 29 |
| chr7_-_89166781 | 0.91 |
ENSMUST00000041761.7
|
Prss23
|
protease, serine 23 |
| chr8_+_93687561 | 0.90 |
ENSMUST00000072939.8
|
Slc6a2
|
solute carrier family 6 (neurotransmitter transporter, noradrenalin), member 2 |
| chr14_-_104081119 | 0.90 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr3_+_138058139 | 0.89 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V) |
| chr8_-_71308229 | 0.89 |
ENSMUST00000212086.2
|
Kcnn1
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
| chr2_-_102903680 | 0.88 |
ENSMUST00000132449.8
ENSMUST00000111183.2 ENSMUST00000011058.9 |
Pdhx
|
pyruvate dehydrogenase complex, component X |
| chr17_-_74017410 | 0.88 |
ENSMUST00000112591.3
ENSMUST00000024858.12 |
Galnt14
|
polypeptide N-acetylgalactosaminyltransferase 14 |
| chr11_+_115790951 | 0.88 |
ENSMUST00000142089.2
ENSMUST00000131566.2 |
Smim5
|
small integral membrane protein 5 |
| chr3_+_63203235 | 0.88 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr14_+_30853010 | 0.88 |
ENSMUST00000227096.2
|
Nt5dc2
|
5'-nucleotidase domain containing 2 |
| chr15_-_85466009 | 0.87 |
ENSMUST00000023015.15
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr5_+_53748323 | 0.86 |
ENSMUST00000201883.4
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr2_+_121287444 | 0.86 |
ENSMUST00000126764.2
|
Hypk
|
huntingtin interacting protein K |
| chr1_+_187340952 | 0.86 |
ENSMUST00000127489.8
|
Esrrg
|
estrogen-related receptor gamma |
| chr7_+_44117475 | 0.85 |
ENSMUST00000118493.8
|
Josd2
|
Josephin domain containing 2 |
| chr4_+_106418224 | 0.85 |
ENSMUST00000047973.4
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr10_+_106306122 | 0.84 |
ENSMUST00000029404.17
ENSMUST00000217854.2 |
Ppfia2
|
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr6_-_124790029 | 0.84 |
ENSMUST00000149610.3
|
Tpi1
|
triosephosphate isomerase 1 |
| chrX_+_139857640 | 0.84 |
ENSMUST00000112971.2
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr2_+_102904014 | 0.83 |
ENSMUST00000011055.7
|
Apip
|
APAF1 interacting protein |
| chr11_+_102556397 | 0.83 |
ENSMUST00000100378.4
|
Meioc
|
meiosis specific with coiled-coil domain |
| chr7_+_43079512 | 0.83 |
ENSMUST00000004732.7
|
Lim2
|
lens intrinsic membrane protein 2 |
| chr7_-_140530194 | 0.83 |
ENSMUST00000026562.6
|
Ifitm5
|
interferon induced transmembrane protein 5 |
| chr16_-_59459486 | 0.83 |
ENSMUST00000149797.3
ENSMUST00000023405.16 |
Arl6
|
ADP-ribosylation factor-like 6 |
| chr7_-_49286594 | 0.83 |
ENSMUST00000032717.7
|
Dbx1
|
developing brain homeobox 1 |
| chr2_+_19450443 | 0.82 |
ENSMUST00000028068.3
|
Ptf1a
|
pancreas specific transcription factor, 1a |
| chr4_+_156300325 | 0.81 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 28.5 | GO:0009095 | tyrosine biosynthetic process(GO:0006571) aromatic amino acid family biosynthetic process(GO:0009073) aromatic amino acid family biosynthetic process, prephenate pathway(GO:0009095) |
| 1.9 | 5.8 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 1.9 | 5.6 | GO:0006212 | uracil catabolic process(GO:0006212) uracil metabolic process(GO:0019860) |
| 1.2 | 13.6 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 1.0 | 7.0 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 1.0 | 2.9 | GO:0021682 | nerve maturation(GO:0021682) |
| 0.7 | 5.8 | GO:0003072 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.7 | 2.9 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.7 | 4.3 | GO:1902512 | positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.6 | 2.4 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.6 | 1.7 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.6 | 3.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.6 | 2.8 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.5 | 1.6 | GO:0035566 | optic nerve formation(GO:0021634) regulation of metanephros size(GO:0035566) optic chiasma development(GO:0061360) regulation of optic nerve formation(GO:2000595) positive regulation of optic nerve formation(GO:2000597) |
| 0.5 | 3.7 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.5 | 1.6 | GO:0060450 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) positive regulation of hindgut contraction(GO:0060450) regulation of relaxation of smooth muscle(GO:1901080) positive regulation of relaxation of smooth muscle(GO:1901082) |
| 0.5 | 3.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.5 | 1.5 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.5 | 1.5 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.4 | 1.3 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.4 | 2.1 | GO:1902163 | negative regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902163) |
| 0.4 | 2.0 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.4 | 1.9 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.4 | 2.5 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.4 | 1.4 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.3 | 1.4 | GO:0002930 | trabecular meshwork development(GO:0002930) |
| 0.3 | 1.0 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.3 | 3.7 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.3 | 1.3 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.3 | 1.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.3 | 0.8 | GO:0019405 | alditol catabolic process(GO:0019405) glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.3 | 1.1 | GO:0043602 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.3 | 1.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.3 | 1.6 | GO:0035553 | oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.3 | 1.8 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 1.0 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.2 | 2.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 1.2 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.2 | 2.4 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.2 | 3.9 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.2 | 1.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 1.6 | GO:0051697 | protein delipidation(GO:0051697) |
| 0.2 | 1.8 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.2 | 2.9 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.2 | 0.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 2.4 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.2 | 4.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.2 | 0.8 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.2 | 2.7 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.2 | 2.7 | GO:0070131 | positive regulation of mitochondrial translation(GO:0070131) |
| 0.2 | 0.6 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.2 | 3.6 | GO:0006977 | DNA damage response, signal transduction by p53 class mediator resulting in cell cycle arrest(GO:0006977) |
| 0.2 | 2.0 | GO:0035360 | positive regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035360) |
| 0.2 | 1.4 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.2 | 3.3 | GO:0033280 | response to vitamin D(GO:0033280) |
| 0.2 | 0.5 | GO:1903778 | protein localization to vacuolar membrane(GO:1903778) |
| 0.2 | 0.7 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.2 | 0.9 | GO:0046449 | creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.2 | 2.1 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.2 | 3.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 0.5 | GO:1903412 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.2 | 0.8 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.2 | 1.6 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.2 | 2.5 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.2 | 0.5 | GO:0072076 | nephrogenic mesenchyme development(GO:0072076) |
| 0.1 | 0.9 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.1 | 0.4 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 1.7 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.1 | 0.4 | GO:0043181 | vacuolar sequestering(GO:0043181) |
| 0.1 | 1.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 1.6 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.1 | 1.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.1 | 0.2 | GO:0046881 | positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.1 | 1.0 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 3.5 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.1 | 0.6 | GO:0021775 | smoothened signaling pathway involved in ventral spinal cord interneuron specification(GO:0021775) smoothened signaling pathway involved in spinal cord motor neuron cell fate specification(GO:0021776) |
| 0.1 | 1.2 | GO:0010764 | negative regulation of fibroblast migration(GO:0010764) |
| 0.1 | 3.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0050883 | negative regulation of sodium:proton antiporter activity(GO:0032416) musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.1 | 1.3 | GO:1904259 | regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904259) positive regulation of basement membrane assembly involved in embryonic body morphogenesis(GO:1904261) basement membrane assembly involved in embryonic body morphogenesis(GO:2001197) |
| 0.1 | 1.5 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 0.3 | GO:1903401 | mitochondrial ornithine transport(GO:0000066) L-lysine transport(GO:1902022) L-ornithine transmembrane transport(GO:1903352) L-arginine transmembrane transport(GO:1903400) L-lysine transmembrane transport(GO:1903401) arginine transmembrane transport(GO:1903826) |
| 0.1 | 0.4 | GO:1904046 | negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.1 | 0.5 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.1 | 1.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 3.3 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 2.5 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 0.7 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.1 | 10.1 | GO:0006635 | fatty acid beta-oxidation(GO:0006635) |
| 0.1 | 1.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.1 | 0.3 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.1 | 0.8 | GO:0061074 | regulation of neural retina development(GO:0061074) |
| 0.1 | 0.3 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.1 | 0.3 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.1 | 3.9 | GO:0060065 | uterus development(GO:0060065) |
| 0.1 | 0.4 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.9 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 1.5 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.1 | 0.6 | GO:1900109 | regulation of histone H3-K9 dimethylation(GO:1900109) |
| 0.1 | 1.1 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.1 | 1.1 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.1 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.1 | 0.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.7 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.4 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.1 | 3.8 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.1 | 1.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.1 | 2.8 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.1 | 0.7 | GO:0036309 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.9 | GO:0061370 | testosterone biosynthetic process(GO:0061370) |
| 0.1 | 3.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.1 | 3.4 | GO:0042092 | type 2 immune response(GO:0042092) |
| 0.1 | 2.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 1.0 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 1.9 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.1 | 1.1 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) |
| 0.1 | 2.8 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.1 | 0.8 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.4 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.2 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.1 | 0.4 | GO:0061525 | hindgut development(GO:0061525) |
| 0.1 | 1.4 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 0.2 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.1 | 2.0 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.1 | 1.0 | GO:0002031 | G-protein coupled receptor internalization(GO:0002031) |
| 0.1 | 9.4 | GO:0007050 | cell cycle arrest(GO:0007050) |
| 0.1 | 0.7 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.3 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 1.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.1 | 1.4 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.1 | 0.3 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.1 | 1.6 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.1 | 0.4 | GO:1903011 | negative regulation of bone development(GO:1903011) negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 1.5 | GO:0010880 | regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum(GO:0010880) |
| 0.0 | 0.5 | GO:0070814 | homoserine metabolic process(GO:0009092) transsulfuration(GO:0019346) hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.1 | GO:0014908 | myotube differentiation involved in skeletal muscle regeneration(GO:0014908) |
| 0.0 | 0.5 | GO:0044829 | positive regulation by host of viral genome replication(GO:0044829) |
| 0.0 | 0.3 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 3.0 | GO:0015909 | long-chain fatty acid transport(GO:0015909) |
| 0.0 | 1.2 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.9 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.6 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.8 | GO:0006656 | phosphatidylcholine biosynthetic process(GO:0006656) |
| 0.0 | 0.2 | GO:0043654 | recognition of apoptotic cell(GO:0043654) |
| 0.0 | 0.8 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 1.1 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 1.8 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.4 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.3 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.2 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.2 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 1.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.6 | GO:0048485 | sympathetic nervous system development(GO:0048485) |
| 0.0 | 0.2 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.5 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.0 | 1.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.3 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.4 | GO:0045717 | negative regulation of fatty acid biosynthetic process(GO:0045717) |
| 0.0 | 0.8 | GO:0045669 | positive regulation of osteoblast differentiation(GO:0045669) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 1.7 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.4 | GO:0030157 | pancreatic juice secretion(GO:0030157) |
| 0.0 | 0.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 2.2 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 0.2 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.1 | GO:0042631 | cellular response to water deprivation(GO:0042631) cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.5 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 1.0 | GO:0030500 | regulation of bone mineralization(GO:0030500) |
| 0.0 | 2.6 | GO:0030308 | negative regulation of cell growth(GO:0030308) |
| 0.0 | 1.4 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.6 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) |
| 0.0 | 0.4 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.3 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.8 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 3.2 | GO:0006631 | fatty acid metabolic process(GO:0006631) |
| 0.0 | 0.2 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 5.8 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.6 | 5.8 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.5 | 1.5 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.5 | 3.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.5 | 2.0 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.3 | 6.1 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.3 | 1.2 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.3 | 2.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.2 | 2.9 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.2 | 1.8 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.2 | 1.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.6 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) |
| 0.1 | 0.4 | GO:0060473 | cortical granule(GO:0060473) |
| 0.1 | 1.3 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 0.7 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.1 | 1.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.1 | 0.9 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.1 | 2.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 2.0 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 3.8 | GO:0080008 | Cul4-RING E3 ubiquitin ligase complex(GO:0080008) |
| 0.1 | 0.4 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.1 | 1.3 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 2.9 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.0 | GO:0000786 | nucleosome(GO:0000786) |
| 0.1 | 0.7 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.1 | 1.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.1 | 0.9 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.3 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.4 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 1.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 9.5 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.0 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 1.9 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 2.4 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 4.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.6 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 5.3 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 1.4 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.2 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 4.9 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 2.1 | GO:1990204 | oxidoreductase complex(GO:1990204) |
| 0.0 | 0.5 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 1.5 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.4 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 5.3 | GO:0005578 | proteinaceous extracellular matrix(GO:0005578) |
| 0.0 | 4.5 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 0.2 | GO:0031597 | cytosolic proteasome complex(GO:0031597) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.9 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 2.8 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 5.3 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 1.1 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 7.3 | GO:0005667 | transcription factor complex(GO:0005667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.7 | 28.5 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 3.4 | 13.6 | GO:0047016 | cholest-5-ene-3-beta,7-alpha-diol 3-beta-dehydrogenase activity(GO:0047016) |
| 1.9 | 5.8 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) |
| 1.9 | 5.6 | GO:0002061 | uracil binding(GO:0002058) pyrimidine nucleobase binding(GO:0002061) |
| 0.8 | 10.1 | GO:0004300 | enoyl-CoA hydratase activity(GO:0004300) |
| 0.7 | 2.8 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.7 | 8.3 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.6 | 3.8 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.6 | 3.0 | GO:0004923 | leukemia inhibitory factor receptor activity(GO:0004923) |
| 0.5 | 2.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.5 | 2.0 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.5 | 2.5 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.5 | 3.0 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.4 | 1.2 | GO:0051538 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) 3 iron, 4 sulfur cluster binding(GO:0051538) |
| 0.4 | 1.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.4 | 2.9 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.3 | 1.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.3 | 0.9 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.3 | 0.9 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.3 | 1.1 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.3 | 1.6 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.2 | 1.0 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 0.9 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 0.7 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 0.9 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.2 | 3.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.2 | 4.8 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.2 | 1.0 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.2 | 2.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.2 | 1.7 | GO:0042285 | xylosyltransferase activity(GO:0042285) |
| 0.2 | 1.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.2 | 1.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.2 | 1.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.2 | 0.5 | GO:0016784 | 3-mercaptopyruvate sulfurtransferase activity(GO:0016784) |
| 0.2 | 1.9 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.2 | 0.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.6 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 0.9 | GO:0034604 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.1 | 0.4 | GO:0070002 | glutamic-type peptidase activity(GO:0070002) |
| 0.1 | 2.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 2.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 3.0 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.1 | 1.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.1 | 1.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 3.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.1 | 0.3 | GO:0005292 | high-affinity basic amino acid transmembrane transporter activity(GO:0005287) high-affinity arginine transmembrane transporter activity(GO:0005289) high-affinity lysine transmembrane transporter activity(GO:0005292) |
| 0.1 | 0.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.6 | GO:0089720 | death effector domain binding(GO:0035877) caspase binding(GO:0089720) |
| 0.1 | 1.9 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 1.6 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 1.8 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.8 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.1 | 13.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 0.7 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 1.5 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.1 | 3.5 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.1 | 0.5 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.1 | 0.4 | GO:0005223 | intracellular cAMP activated cation channel activity(GO:0005222) intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.7 | GO:0005347 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.1 | 1.5 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.6 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 3.4 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.1 | 1.7 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 1.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.3 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 0.4 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.7 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.1 | 3.4 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 1.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.1 | 1.1 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.5 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.1 | 1.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.3 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 1.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.4 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) syndecan binding(GO:0045545) |
| 0.0 | 1.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 5.4 | GO:0000980 | RNA polymerase II distal enhancer sequence-specific DNA binding(GO:0000980) |
| 0.0 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 2.1 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.8 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.8 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 1.1 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 2.4 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.4 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 3.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 3.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 1.9 | GO:0051287 | NAD binding(GO:0051287) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 1.9 | GO:0015179 | L-amino acid transmembrane transporter activity(GO:0015179) |
| 0.0 | 0.4 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.5 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 3.2 | GO:0042826 | histone deacetylase binding(GO:0042826) |
| 0.0 | 0.9 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.0 | 1.2 | GO:0008237 | metallopeptidase activity(GO:0008237) |
| 0.0 | 1.1 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.5 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 1.1 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.1 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.7 | GO:0005487 | nucleocytoplasmic transporter activity(GO:0005487) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 2.1 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 1.6 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 1.5 | GO:0051082 | unfolded protein binding(GO:0051082) |
| 0.0 | 1.5 | GO:0016614 | oxidoreductase activity, acting on CH-OH group of donors(GO:0016614) |
| 0.0 | 0.8 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 16.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 4.0 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 3.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 2.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 1.5 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 2.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.1 | 4.8 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.9 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 14.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 3.4 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 3.0 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 4.6 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 1.3 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.7 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 1.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 1.2 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.7 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.9 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 1.1 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 13.6 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.5 | 1.9 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.3 | 5.6 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.3 | 9.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.2 | 1.7 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 3.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 3.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 4.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 1.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 2.7 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 3.1 | REACTOME PYRUVATE METABOLISM AND CITRIC ACID TCA CYCLE | Genes involved in Pyruvate metabolism and Citric Acid (TCA) cycle |
| 0.1 | 4.2 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.1 | 28.7 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.1 | 3.0 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.1 | 1.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.2 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.1 | 1.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 3.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 1.1 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.9 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 4.9 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.4 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 4.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.8 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 5.3 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 2.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.7 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.4 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.7 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |