Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zic4
Z-value: 1.40
Transcription factors associated with Zic4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zic4
|
ENSMUSG00000036972.15 | Zic4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zic4 | mm39_v1_chr9_+_91260688_91260700 | 0.83 | 1.7e-19 | Click! |
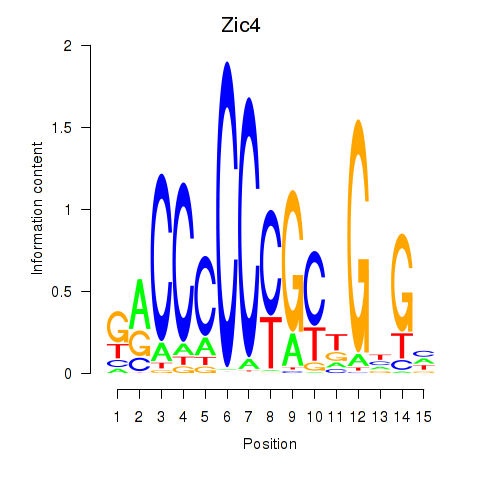
Activity profile of Zic4 motif
Sorted Z-values of Zic4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zic4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_-_107109108 | 19.46 |
ENSMUST00000044532.11
|
Dock3
|
dedicator of cyto-kinesis 3 |
| chr4_-_129015493 | 18.02 |
ENSMUST00000135763.2
ENSMUST00000149763.3 ENSMUST00000164649.8 |
Hpca
|
hippocalcin |
| chr4_-_129015027 | 15.65 |
ENSMUST00000030572.10
|
Hpca
|
hippocalcin |
| chr17_+_25936463 | 13.71 |
ENSMUST00000115108.4
|
Gng13
|
guanine nucleotide binding protein (G protein), gamma 13 |
| chr7_+_139414057 | 12.18 |
ENSMUST00000026548.14
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr9_-_29323500 | 11.86 |
ENSMUST00000115237.8
|
Ntm
|
neurotrimin |
| chr14_-_24054927 | 11.86 |
ENSMUST00000145596.3
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr13_-_69887964 | 10.83 |
ENSMUST00000065118.7
|
Ube2ql1
|
ubiquitin-conjugating enzyme E2Q family-like 1 |
| chr7_-_46783432 | 10.32 |
ENSMUST00000102626.10
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr11_-_102837514 | 10.21 |
ENSMUST00000057849.6
|
C1ql1
|
complement component 1, q subcomponent-like 1 |
| chr15_+_74435587 | 10.09 |
ENSMUST00000185682.7
ENSMUST00000170845.8 ENSMUST00000187599.2 |
Adgrb1
|
adhesion G protein-coupled receptor B1 |
| chr4_+_152423075 | 10.06 |
ENSMUST00000030775.12
ENSMUST00000164662.8 |
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr14_-_55150547 | 9.61 |
ENSMUST00000228495.3
ENSMUST00000228119.3 ENSMUST00000050772.10 ENSMUST00000231305.2 |
Slc22a17
|
solute carrier family 22 (organic cation transporter), member 17 |
| chr19_+_47167259 | 9.55 |
ENSMUST00000111808.11
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr6_+_141470105 | 9.49 |
ENSMUST00000032362.12
ENSMUST00000205214.3 |
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr17_+_25936145 | 9.42 |
ENSMUST00000172002.8
|
Gng13
|
guanine nucleotide binding protein (G protein), gamma 13 |
| chr19_-_4527475 | 9.36 |
ENSMUST00000059295.10
|
Syt12
|
synaptotagmin XII |
| chr2_+_164802729 | 8.99 |
ENSMUST00000202623.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr2_+_164802766 | 8.96 |
ENSMUST00000202223.4
|
Slc12a5
|
solute carrier family 12, member 5 |
| chr4_+_152423344 | 8.45 |
ENSMUST00000005175.5
|
Chd5
|
chromodomain helicase DNA binding protein 5 |
| chr6_-_35285045 | 8.41 |
ENSMUST00000031868.5
|
Slc13a4
|
solute carrier family 13 (sodium/sulfate symporters), member 4 |
| chr4_+_104224774 | 8.30 |
ENSMUST00000106830.9
|
Dab1
|
disabled 1 |
| chr15_+_99122742 | 8.29 |
ENSMUST00000041415.5
|
Kcnh3
|
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
| chr19_+_48194464 | 8.26 |
ENSMUST00000078880.6
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr17_-_29456750 | 8.11 |
ENSMUST00000137727.3
|
Cpne5
|
copine V |
| chr1_-_37758863 | 8.00 |
ENSMUST00000160589.2
|
Cracdl
|
capping protein inhibiting regulator of actin like |
| chr11_-_70924288 | 7.98 |
ENSMUST00000238695.2
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene |
| chr6_+_141470261 | 7.95 |
ENSMUST00000203140.2
|
Slco1c1
|
solute carrier organic anion transporter family, member 1c1 |
| chr4_+_42917228 | 7.93 |
ENSMUST00000107976.9
ENSMUST00000069184.9 |
Phf24
|
PHD finger protein 24 |
| chr14_+_70768257 | 7.91 |
ENSMUST00000047331.8
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr1_-_184731672 | 7.89 |
ENSMUST00000192657.2
ENSMUST00000027929.10 |
Mark1
|
MAP/microtubule affinity regulating kinase 1 |
| chr2_-_152793376 | 7.68 |
ENSMUST00000123121.9
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr19_+_47167554 | 7.63 |
ENSMUST00000235290.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr5_-_113458563 | 7.43 |
ENSMUST00000154248.8
ENSMUST00000112325.8 ENSMUST00000048112.13 |
Sgsm1
|
small G protein signaling modulator 1 |
| chr5_-_49682150 | 7.35 |
ENSMUST00000087395.11
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr2_+_180240015 | 6.99 |
ENSMUST00000103059.2
|
Col9a3
|
collagen, type IX, alpha 3 |
| chr10_+_7465555 | 6.83 |
ENSMUST00000134346.8
ENSMUST00000019931.12 ENSMUST00000130590.8 |
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr14_+_70768289 | 6.77 |
ENSMUST00000226548.2
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr13_-_95359955 | 6.67 |
ENSMUST00000159608.8
|
Pde8b
|
phosphodiesterase 8B |
| chr11_+_92990110 | 6.60 |
ENSMUST00000107863.4
|
Car10
|
carbonic anhydrase 10 |
| chr7_+_49624612 | 6.53 |
ENSMUST00000151721.8
ENSMUST00000081872.13 |
Nell1
|
NEL-like 1 |
| chr8_+_123844090 | 6.52 |
ENSMUST00000037900.9
|
Cpne7
|
copine VII |
| chr2_-_152793469 | 6.45 |
ENSMUST00000037715.7
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr2_-_165076609 | 6.44 |
ENSMUST00000065438.13
|
Cdh22
|
cadherin 22 |
| chr3_+_64856617 | 6.34 |
ENSMUST00000238901.2
|
Kcnab1
|
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr11_+_98632696 | 6.24 |
ENSMUST00000103139.11
|
Thra
|
thyroid hormone receptor alpha |
| chr6_-_126621751 | 6.02 |
ENSMUST00000055168.5
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr1_+_153541412 | 6.00 |
ENSMUST00000111814.8
ENSMUST00000111810.2 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr6_-_115228800 | 5.98 |
ENSMUST00000205131.2
|
Timp4
|
tissue inhibitor of metalloproteinase 4 |
| chr17_+_37356872 | 5.93 |
ENSMUST00000174456.8
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr8_-_112120442 | 5.90 |
ENSMUST00000038475.9
|
Fa2h
|
fatty acid 2-hydroxylase |
| chr7_+_27353331 | 5.67 |
ENSMUST00000008088.9
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr7_+_49624978 | 5.65 |
ENSMUST00000107603.2
|
Nell1
|
NEL-like 1 |
| chr14_-_24053994 | 5.63 |
ENSMUST00000225431.2
ENSMUST00000188210.8 ENSMUST00000224787.2 ENSMUST00000225315.2 ENSMUST00000225556.2 ENSMUST00000223727.2 ENSMUST00000223655.2 ENSMUST00000224077.2 ENSMUST00000224812.2 ENSMUST00000224285.2 ENSMUST00000225471.2 ENSMUST00000224232.2 ENSMUST00000223749.2 ENSMUST00000224025.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr17_-_24752683 | 5.60 |
ENSMUST00000061764.14
|
Rab26
|
RAB26, member RAS oncogene family |
| chr19_-_5040344 | 5.59 |
ENSMUST00000056129.9
|
Npas4
|
neuronal PAS domain protein 4 |
| chr6_-_126621770 | 5.57 |
ENSMUST00000203094.2
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr1_+_153541020 | 5.47 |
ENSMUST00000152114.8
ENSMUST00000111812.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr4_-_24851086 | 5.40 |
ENSMUST00000084781.6
ENSMUST00000108218.10 |
Klhl32
|
kelch-like 32 |
| chr13_+_55097200 | 5.35 |
ENSMUST00000026994.14
ENSMUST00000109994.9 |
Unc5a
|
unc-5 netrin receptor A |
| chr10_-_79473675 | 5.29 |
ENSMUST00000020564.7
|
Shc2
|
SHC (Src homology 2 domain containing) transforming protein 2 |
| chr15_-_103242697 | 5.27 |
ENSMUST00000229373.2
|
Zfp385a
|
zinc finger protein 385A |
| chr3_+_54063459 | 5.11 |
ENSMUST00000029311.11
ENSMUST00000200048.5 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr5_-_49682106 | 4.96 |
ENSMUST00000176191.8
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr10_+_7465845 | 4.92 |
ENSMUST00000135907.8
|
Lrp11
|
low density lipoprotein receptor-related protein 11 |
| chr9_+_87026337 | 4.83 |
ENSMUST00000113149.8
ENSMUST00000049457.14 ENSMUST00000179313.3 |
Mrap2
|
melanocortin 2 receptor accessory protein 2 |
| chr4_+_42158092 | 4.83 |
ENSMUST00000098122.3
|
Gm13306
|
predicted gene 13306 |
| chr17_+_37356854 | 4.78 |
ENSMUST00000025338.16
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr11_-_78388560 | 4.71 |
ENSMUST00000061174.7
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr11_-_78388284 | 4.59 |
ENSMUST00000108287.10
|
Sarm1
|
sterile alpha and HEAT/Armadillo motif containing 1 |
| chr2_-_58247764 | 4.34 |
ENSMUST00000112608.9
ENSMUST00000112607.3 ENSMUST00000028178.14 |
Acvr1c
|
activin A receptor, type IC |
| chr2_+_131751848 | 4.18 |
ENSMUST00000091288.13
ENSMUST00000124100.8 ENSMUST00000136783.2 |
Prnp
Prn
|
prion protein prion protein readthrough transcript |
| chr12_-_31549538 | 4.13 |
ENSMUST00000064240.14
ENSMUST00000185739.8 ENSMUST00000188326.3 ENSMUST00000101499.10 ENSMUST00000085487.12 |
Cbll1
|
Casitas B-lineage lymphoma-like 1 |
| chrX_-_110606766 | 4.06 |
ENSMUST00000113422.9
ENSMUST00000038472.7 |
Hdx
|
highly divergent homeobox |
| chr12_-_67269323 | 4.04 |
ENSMUST00000037181.16
|
Mdga2
|
MAM domain containing glycosylphosphatidylinositol anchor 2 |
| chr10_+_58649181 | 3.80 |
ENSMUST00000135526.9
ENSMUST00000153031.2 |
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr4_+_125384481 | 3.77 |
ENSMUST00000030676.8
|
Grik3
|
glutamate receptor, ionotropic, kainate 3 |
| chr2_-_144173615 | 3.74 |
ENSMUST00000103171.10
|
Ovol2
|
ovo like zinc finger 2 |
| chr7_-_34914675 | 3.68 |
ENSMUST00000118444.3
ENSMUST00000122409.8 |
Lrp3
|
low density lipoprotein receptor-related protein 3 |
| chr2_-_6726417 | 3.65 |
ENSMUST00000142941.8
ENSMUST00000150624.9 ENSMUST00000100429.11 ENSMUST00000182879.8 |
Celf2
|
CUGBP, Elav-like family member 2 |
| chr14_+_119375753 | 3.60 |
ENSMUST00000065904.5
|
Hs6st3
|
heparan sulfate 6-O-sulfotransferase 3 |
| chr2_-_144174066 | 3.58 |
ENSMUST00000037423.4
|
Ovol2
|
ovo like zinc finger 2 |
| chr6_+_99669640 | 3.46 |
ENSMUST00000101122.3
|
Gpr27
|
G protein-coupled receptor 27 |
| chr11_-_116226175 | 3.44 |
ENSMUST00000036215.8
|
Foxj1
|
forkhead box J1 |
| chr2_-_152793607 | 3.44 |
ENSMUST00000109811.4
|
Dusp15
|
dual specificity phosphatase-like 15 |
| chr5_-_142495408 | 3.43 |
ENSMUST00000110784.8
|
Radil
|
Ras association and DIL domains |
| chr19_-_6285827 | 3.39 |
ENSMUST00000025695.10
|
Ppp2r5b
|
protein phosphatase 2, regulatory subunit B', beta |
| chr1_+_153541339 | 3.39 |
ENSMUST00000147700.8
ENSMUST00000147482.8 |
Rgs8
|
regulator of G-protein signaling 8 |
| chr7_+_143838191 | 3.30 |
ENSMUST00000097929.4
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr9_-_108338111 | 3.27 |
ENSMUST00000193895.6
|
Klhdc8b
|
kelch domain containing 8B |
| chr13_-_95359543 | 3.20 |
ENSMUST00000162292.8
|
Pde8b
|
phosphodiesterase 8B |
| chr8_-_91544021 | 3.16 |
ENSMUST00000209208.2
|
Gm19935
|
predicted gene, 19935 |
| chr4_-_108690805 | 3.07 |
ENSMUST00000102742.11
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr17_+_86475205 | 3.07 |
ENSMUST00000097275.9
|
Prkce
|
protein kinase C, epsilon |
| chr11_-_119910986 | 3.04 |
ENSMUST00000134319.2
|
Aatk
|
apoptosis-associated tyrosine kinase |
| chr19_+_4905158 | 3.00 |
ENSMUST00000119694.3
ENSMUST00000237504.2 ENSMUST00000237011.2 |
Ctsf
|
cathepsin F |
| chr19_+_47167444 | 2.97 |
ENSMUST00000235326.2
|
Neurl1a
|
neuralized E3 ubiquitin protein ligase 1A |
| chr18_-_24736848 | 2.93 |
ENSMUST00000070726.10
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr19_+_5497575 | 2.88 |
ENSMUST00000025850.7
ENSMUST00000236774.2 |
Fosl1
|
fos-like antigen 1 |
| chr16_+_20551853 | 2.80 |
ENSMUST00000115423.8
ENSMUST00000007171.13 ENSMUST00000232646.2 |
Chrd
|
chordin |
| chr12_+_102094977 | 2.80 |
ENSMUST00000159329.8
|
Slc24a4
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr7_-_16790594 | 2.77 |
ENSMUST00000037762.11
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr7_+_28466160 | 2.71 |
ENSMUST00000122915.8
ENSMUST00000072965.5 ENSMUST00000170068.9 |
Sirt2
|
sirtuin 2 |
| chr11_-_6425877 | 2.69 |
ENSMUST00000179343.3
|
Purb
|
purine rich element binding protein B |
| chr7_-_100581314 | 2.61 |
ENSMUST00000107032.3
|
Arhgef17
|
Rho guanine nucleotide exchange factor (GEF) 17 |
| chr15_-_64794139 | 2.59 |
ENSMUST00000023007.7
ENSMUST00000228014.2 |
Adcy8
|
adenylate cyclase 8 |
| chr18_-_24736521 | 2.51 |
ENSMUST00000154205.2
|
Slc39a6
|
solute carrier family 39 (metal ion transporter), member 6 |
| chr17_-_51486196 | 2.44 |
ENSMUST00000024717.10
ENSMUST00000224528.2 |
Tbc1d5
|
TBC1 domain family, member 5 |
| chrX_-_48886577 | 2.43 |
ENSMUST00000033442.14
ENSMUST00000114891.2 |
Igsf1
|
immunoglobulin superfamily, member 1 |
| chr5_-_3943907 | 2.43 |
ENSMUST00000117463.2
ENSMUST00000044746.5 |
Mterf1a
|
mitochondrial transcription termination factor 1a |
| chr12_+_79075924 | 2.34 |
ENSMUST00000039928.7
|
Plekhh1
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr12_-_75224099 | 2.31 |
ENSMUST00000042299.4
|
Kcnh5
|
potassium voltage-gated channel, subfamily H (eag-related), member 5 |
| chr17_+_37357451 | 2.24 |
ENSMUST00000172789.2
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chr19_+_45352173 | 2.21 |
ENSMUST00000223764.2
ENSMUST00000065601.13 ENSMUST00000224102.2 ENSMUST00000111936.4 |
Btrc
|
beta-transducin repeat containing protein |
| chrX_-_7606445 | 2.11 |
ENSMUST00000128289.8
|
Ccdc120
|
coiled-coil domain containing 120 |
| chr7_-_16020668 | 2.02 |
ENSMUST00000150528.9
ENSMUST00000118976.9 ENSMUST00000146609.3 |
Ccdc9
|
coiled-coil domain containing 9 |
| chr11_+_61847589 | 2.00 |
ENSMUST00000201671.4
ENSMUST00000202178.4 |
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr11_+_101358990 | 2.00 |
ENSMUST00000001347.7
|
Rnd2
|
Rho family GTPase 2 |
| chr4_+_141095415 | 1.99 |
ENSMUST00000006380.5
|
Fam131c
|
family with sequence similarity 131, member C |
| chr8_-_45835234 | 1.88 |
ENSMUST00000093526.13
|
Fam149a
|
family with sequence similarity 149, member A |
| chr4_-_124744454 | 1.85 |
ENSMUST00000125776.8
ENSMUST00000163946.2 ENSMUST00000106190.10 |
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr17_+_79359617 | 1.84 |
ENSMUST00000233916.2
|
Qpct
|
glutaminyl-peptide cyclotransferase (glutaminyl cyclase) |
| chr19_+_45352514 | 1.82 |
ENSMUST00000224318.2
ENSMUST00000223684.2 |
Btrc
|
beta-transducin repeat containing protein |
| chr5_+_115573055 | 1.80 |
ENSMUST00000136586.6
|
Msi1
|
musashi RNA-binding protein 1 |
| chr17_-_34250474 | 1.77 |
ENSMUST00000171872.3
ENSMUST00000025186.16 |
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr7_+_6442835 | 1.74 |
ENSMUST00000168341.2
|
Olfr1344
|
olfactory receptor 1344 |
| chr5_+_43390513 | 1.73 |
ENSMUST00000166713.9
ENSMUST00000169035.8 ENSMUST00000114065.9 |
Cpeb2
|
cytoplasmic polyadenylation element binding protein 2 |
| chrX_-_106446928 | 1.72 |
ENSMUST00000033591.6
|
Itm2a
|
integral membrane protein 2A |
| chr1_-_186437760 | 1.67 |
ENSMUST00000195201.2
|
Tgfb2
|
transforming growth factor, beta 2 |
| chr1_+_91107725 | 1.65 |
ENSMUST00000188475.7
ENSMUST00000097648.6 |
Ramp1
|
receptor (calcitonin) activity modifying protein 1 |
| chr5_-_44259293 | 1.58 |
ENSMUST00000074113.13
|
Prom1
|
prominin 1 |
| chr10_-_81201642 | 1.53 |
ENSMUST00000020456.5
|
4930404N11Rik
|
RIKEN cDNA 4930404N11 gene |
| chr9_+_50662625 | 1.51 |
ENSMUST00000217475.2
|
Cryab
|
crystallin, alpha B |
| chr12_-_98541293 | 1.50 |
ENSMUST00000110113.3
|
Kcnk10
|
potassium channel, subfamily K, member 10 |
| chr2_+_120807498 | 1.49 |
ENSMUST00000067582.14
|
Tmem62
|
transmembrane protein 62 |
| chr14_-_24054273 | 1.49 |
ENSMUST00000188285.7
ENSMUST00000190044.7 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_-_71198091 | 1.47 |
ENSMUST00000151937.8
|
Slc25a12
|
solute carrier family 25 (mitochondrial carrier, Aralar), member 12 |
| chr15_+_32244947 | 1.42 |
ENSMUST00000067458.7
|
Sema5a
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
| chr5_+_4242367 | 1.37 |
ENSMUST00000177258.2
|
Mterf1b
|
mitochondrial transcription termination factor 1b |
| chr7_+_28466658 | 1.36 |
ENSMUST00000155327.8
|
Sirt2
|
sirtuin 2 |
| chr18_-_36648850 | 1.34 |
ENSMUST00000025363.7
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr11_+_61847622 | 1.32 |
ENSMUST00000202389.4
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr8_-_37081091 | 1.31 |
ENSMUST00000033923.14
|
Dlc1
|
deleted in liver cancer 1 |
| chr9_+_108337726 | 1.30 |
ENSMUST00000061209.7
ENSMUST00000193269.2 |
Ccdc71
|
coiled-coil domain containing 71 |
| chr17_-_34250616 | 1.26 |
ENSMUST00000169397.9
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr7_-_12514534 | 1.24 |
ENSMUST00000211392.2
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr11_+_75623695 | 1.23 |
ENSMUST00000067664.10
|
Ywhae
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide |
| chr12_-_5425682 | 1.17 |
ENSMUST00000020958.9
|
Klhl29
|
kelch-like 29 |
| chr3_-_8732316 | 1.14 |
ENSMUST00000042412.5
|
Hey1
|
hairy/enhancer-of-split related with YRPW motif 1 |
| chr2_+_130137068 | 1.09 |
ENSMUST00000110288.9
|
Ebf4
|
early B cell factor 4 |
| chr5_-_44259010 | 1.09 |
ENSMUST00000087441.11
|
Prom1
|
prominin 1 |
| chr4_-_124744266 | 1.06 |
ENSMUST00000137769.3
|
1110065P20Rik
|
RIKEN cDNA 1110065P20 gene |
| chr5_+_105563605 | 1.05 |
ENSMUST00000112707.3
|
Lrrc8b
|
leucine rich repeat containing 8 family, member B |
| chr18_+_10725532 | 1.04 |
ENSMUST00000052838.11
|
Mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chr11_+_78234300 | 1.00 |
ENSMUST00000002127.14
ENSMUST00000108295.8 |
Unc119
|
unc-119 lipid binding chaperone |
| chr3_-_63836796 | 0.92 |
ENSMUST00000061706.7
|
E130311K13Rik
|
RIKEN cDNA E130311K13 gene |
| chr7_-_67409432 | 0.91 |
ENSMUST00000208815.2
ENSMUST00000074233.12 ENSMUST00000208231.2 ENSMUST00000051389.10 |
Synm
|
synemin, intermediate filament protein |
| chr9_-_58462720 | 0.90 |
ENSMUST00000165365.3
|
Cd276
|
CD276 antigen |
| chr2_-_153371861 | 0.90 |
ENSMUST00000035346.14
|
Nol4l
|
nucleolar protein 4-like |
| chr17_-_83939316 | 0.86 |
ENSMUST00000234613.2
ENSMUST00000051482.2 |
Kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr2_-_166904902 | 0.85 |
ENSMUST00000048988.14
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr14_+_25980463 | 0.81 |
ENSMUST00000173155.8
|
Duxbl1
|
double homeobox B-like 1 |
| chr1_-_189654535 | 0.80 |
ENSMUST00000027897.8
|
Smyd2
|
SET and MYND domain containing 2 |
| chr1_-_166066298 | 0.69 |
ENSMUST00000038782.4
ENSMUST00000194057.6 |
Mael
|
maelstrom spermatogenic transposon silencer |
| chr4_-_72119070 | 0.61 |
ENSMUST00000102848.9
ENSMUST00000072695.13 ENSMUST00000107337.8 ENSMUST00000074216.14 |
Tle1
|
transducin-like enhancer of split 1 |
| chr11_+_93886906 | 0.61 |
ENSMUST00000041956.14
|
Spag9
|
sperm associated antigen 9 |
| chr2_-_166904625 | 0.57 |
ENSMUST00000128676.2
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr14_-_54878804 | 0.52 |
ENSMUST00000067784.8
|
Cdh24
|
cadherin-like 24 |
| chr17_-_32607859 | 0.51 |
ENSMUST00000087703.12
ENSMUST00000170603.3 |
Wiz
|
widely-interspaced zinc finger motifs |
| chr18_+_10725650 | 0.50 |
ENSMUST00000165555.8
|
Mib1
|
mindbomb E3 ubiquitin protein ligase 1 |
| chrX_-_73129195 | 0.50 |
ENSMUST00000140399.3
ENSMUST00000123362.8 ENSMUST00000100750.10 ENSMUST00000033770.13 |
Mecp2
|
methyl CpG binding protein 2 |
| chr13_+_15638466 | 0.48 |
ENSMUST00000110510.4
|
Gli3
|
GLI-Kruppel family member GLI3 |
| chr6_-_143045493 | 0.46 |
ENSMUST00000204655.3
ENSMUST00000111758.9 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr8_-_88686188 | 0.46 |
ENSMUST00000109655.9
|
Zfp423
|
zinc finger protein 423 |
| chr9_-_105372330 | 0.41 |
ENSMUST00000038118.15
|
Atp2c1
|
ATPase, Ca++-sequestering |
| chr14_+_121148625 | 0.41 |
ENSMUST00000032898.9
|
Ipo5
|
importin 5 |
| chr16_+_14179421 | 0.38 |
ENSMUST00000100167.10
ENSMUST00000154748.8 ENSMUST00000134776.8 |
Abcc1
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
| chr4_-_156281935 | 0.36 |
ENSMUST00000180572.2
|
Agrn
|
agrin |
| chr11_-_59678462 | 0.36 |
ENSMUST00000125307.2
|
Pld6
|
phospholipase D family, member 6 |
| chr7_-_44320244 | 0.34 |
ENSMUST00000048102.15
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr9_-_105372235 | 0.28 |
ENSMUST00000176190.8
ENSMUST00000163879.9 ENSMUST00000112558.10 ENSMUST00000176363.9 |
Atp2c1
|
ATPase, Ca++-sequestering |
| chr12_-_91712783 | 0.26 |
ENSMUST00000166967.2
|
Ston2
|
stonin 2 |
| chr6_+_61157279 | 0.26 |
ENSMUST00000126214.8
|
Ccser1
|
coiled-coil serine rich 1 |
| chr15_-_97902515 | 0.23 |
ENSMUST00000088355.12
|
Col2a1
|
collagen, type II, alpha 1 |
| chr4_-_108690741 | 0.20 |
ENSMUST00000102740.8
ENSMUST00000102741.8 |
Btf3l4
|
basic transcription factor 3-like 4 |
| chr15_-_97902576 | 0.20 |
ENSMUST00000023123.15
|
Col2a1
|
collagen, type II, alpha 1 |
| chr1_+_85992341 | 0.20 |
ENSMUST00000027432.9
|
Psmd1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr19_-_60456742 | 0.15 |
ENSMUST00000051277.4
|
Prlhr
|
prolactin releasing hormone receptor |
| chr4_-_62278761 | 0.13 |
ENSMUST00000107461.2
ENSMUST00000084528.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr7_+_97229065 | 0.09 |
ENSMUST00000107153.3
|
Rsf1
|
remodeling and spacing factor 1 |
| chr14_-_24054186 | 0.05 |
ENSMUST00000188991.7
ENSMUST00000224468.2 |
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr7_+_139900771 | 0.02 |
ENSMUST00000214594.2
|
Olfr525
|
olfactory receptor 525 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 11.2 | 33.7 | GO:0031283 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 6.0 | 18.0 | GO:0040040 | thermosensory behavior(GO:0040040) |
| 3.2 | 9.6 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 2.8 | 17.0 | GO:0060082 | response to carbon monoxide(GO:0034465) eye blink reflex(GO:0060082) |
| 2.8 | 8.3 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 2.6 | 10.3 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 2.3 | 11.6 | GO:0050976 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 2.3 | 18.5 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 2.1 | 4.2 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 2.0 | 10.2 | GO:0099558 | maintenance of synapse structure(GO:0099558) |
| 1.9 | 5.6 | GO:0051385 | response to mineralocorticoid(GO:0051385) |
| 1.8 | 12.9 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 1.8 | 10.6 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 1.7 | 11.7 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 1.6 | 6.2 | GO:0017055 | negative regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0017055) |
| 1.4 | 4.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 1.4 | 20.2 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 1.4 | 4.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 1.3 | 5.3 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 1.3 | 3.8 | GO:0006393 | termination of mitochondrial transcription(GO:0006393) |
| 1.2 | 9.9 | GO:0035106 | operant conditioning(GO:0035106) |
| 1.1 | 3.4 | GO:0002514 | B cell tolerance induction(GO:0002514) regulation of B cell tolerance induction(GO:0002661) positive regulation of B cell tolerance induction(GO:0002663) |
| 1.0 | 12.2 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 1.0 | 14.9 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.8 | 7.3 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.7 | 3.5 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.7 | 5.9 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.6 | 1.8 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.6 | 3.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.6 | 3.6 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.6 | 1.7 | GO:1905006 | endocardial cushion fusion(GO:0003274) positive regulation of activation-induced cell death of T cells(GO:0070237) negative regulation of epithelial to mesenchymal transition involved in endocardial cushion formation(GO:1905006) |
| 0.5 | 1.6 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.5 | 2.7 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.5 | 17.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.5 | 6.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.5 | 5.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.5 | 2.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.5 | 2.8 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.4 | 1.3 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.4 | 3.4 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.4 | 8.4 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.3 | 8.5 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 1.2 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.3 | 23.1 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.3 | 1.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.3 | 8.3 | GO:1900452 | regulation of long term synaptic depression(GO:1900452) |
| 0.3 | 5.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.3 | 3.8 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.2 | 1.0 | GO:2001287 | negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.2 | 1.7 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.2 | 1.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.2 | 7.0 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.2 | 3.0 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.2 | 7.9 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.2 | 1.7 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 7.1 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.2 | 9.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.2 | 2.0 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.2 | 0.9 | GO:0031443 | fast-twitch skeletal muscle fiber contraction(GO:0031443) |
| 0.2 | 1.4 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.1 | 9.3 | GO:0048713 | regulation of oligodendrocyte differentiation(GO:0048713) |
| 0.1 | 1.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.1 | 0.9 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.1 | 0.4 | GO:0034635 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 5.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.1 | 3.3 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.1 | 0.4 | GO:1903406 | regulation of sodium:potassium-exchanging ATPase activity(GO:1903406) |
| 0.1 | 0.4 | GO:0008358 | oocyte construction(GO:0007308) oocyte axis specification(GO:0007309) oocyte anterior/posterior axis specification(GO:0007314) pole plasm assembly(GO:0007315) maternal determination of anterior/posterior axis, embryo(GO:0008358) P granule organization(GO:0030719) |
| 0.1 | 4.0 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.1 | 2.8 | GO:0097186 | amelogenesis(GO:0097186) |
| 0.1 | 10.1 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 4.8 | GO:0097009 | energy homeostasis(GO:0097009) |
| 0.1 | 0.6 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 3.6 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 6.5 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.1 | 0.5 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.1 | 2.6 | GO:0007616 | long-term memory(GO:0007616) |
| 0.1 | 12.4 | GO:0046546 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.1 | 0.8 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 7.4 | GO:1901379 | regulation of potassium ion transmembrane transport(GO:1901379) |
| 0.1 | 0.6 | GO:2000811 | negative regulation of anoikis(GO:2000811) |
| 0.1 | 0.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 1.3 | GO:0051895 | negative regulation of focal adhesion assembly(GO:0051895) |
| 0.0 | 0.4 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.5 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.4 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.7 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 6.5 | GO:0009749 | response to glucose(GO:0009749) |
| 0.0 | 6.1 | GO:0045807 | positive regulation of endocytosis(GO:0045807) |
| 0.0 | 5.6 | GO:0001764 | neuron migration(GO:0001764) |
| 0.0 | 18.5 | GO:0007264 | small GTPase mediated signal transduction(GO:0007264) |
| 0.0 | 4.7 | GO:0010977 | negative regulation of neuron projection development(GO:0010977) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 2.0 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 2.9 | GO:0017157 | regulation of exocytosis(GO:0017157) |
| 0.0 | 0.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.9 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 1.2 | GO:0000187 | activation of MAPK activity(GO:0000187) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.6 | 12.9 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 2.2 | 33.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 2.0 | 10.2 | GO:0044301 | climbing fiber(GO:0044301) |
| 1.7 | 7.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 1.4 | 4.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 1.2 | 16.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 1.0 | 17.0 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.7 | 9.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.5 | 18.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.5 | 15.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.5 | 13.0 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.4 | 20.2 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.4 | 2.7 | GO:0071914 | prominosome(GO:0071914) |
| 0.4 | 17.2 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.3 | 5.4 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.3 | 21.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.3 | 1.7 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 2.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.2 | 3.3 | GO:0005883 | neurofilament(GO:0005883) |
| 0.2 | 2.7 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.2 | 3.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.7 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 9.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 2.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.1 | 0.9 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 20.1 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 1.7 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 2.9 | GO:0042734 | presynaptic membrane(GO:0042734) |
| 0.1 | 3.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 3.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.1 | 14.3 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.1 | 5.1 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.1 | 4.8 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 19.3 | GO:0014069 | postsynaptic density(GO:0014069) |
| 0.0 | 0.3 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 4.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 1.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 7.9 | GO:0030659 | cytoplasmic vesicle membrane(GO:0030659) |
| 0.0 | 0.4 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 1.0 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.5 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 6.0 | GO:0030017 | sarcomere(GO:0030017) |
| 0.0 | 1.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 1.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0031597 | proteasome regulatory particle, base subcomplex(GO:0008540) cytosolic proteasome complex(GO:0031597) |
| 0.0 | 3.8 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 6.3 | GO:0043025 | neuronal cell body(GO:0043025) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.0 | 20.2 | GO:0045183 | translation factor activity, non-nucleic acid binding(GO:0045183) |
| 4.6 | 18.5 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 2.2 | 17.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 2.2 | 12.9 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 2.1 | 17.0 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 1.9 | 5.6 | GO:0019002 | GMP binding(GO:0019002) |
| 1.8 | 10.6 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 1.6 | 18.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 1.6 | 4.8 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 1.6 | 6.2 | GO:0002153 | steroid receptor RNA activator RNA binding(GO:0002153) |
| 1.1 | 4.3 | GO:0038100 | nodal binding(GO:0038100) |
| 1.0 | 4.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.9 | 3.6 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.8 | 4.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.8 | 23.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.8 | 3.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) ethanol binding(GO:0035276) |
| 0.7 | 2.7 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.7 | 5.3 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.6 | 3.8 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.6 | 1.7 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.5 | 2.8 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.4 | 7.9 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.4 | 1.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.4 | 2.6 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.4 | 1.4 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.3 | 5.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.3 | 1.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.3 | 14.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.3 | 1.7 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.3 | 6.0 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.3 | 2.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.2 | 6.3 | GO:0070402 | aldo-keto reductase (NADP) activity(GO:0004033) NADPH binding(GO:0070402) |
| 0.2 | 9.9 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 10.8 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.2 | 8.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.2 | 12.1 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.2 | 0.7 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.2 | 8.7 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.1 | 4.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 2.4 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.1 | 15.0 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.1 | 1.7 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 9.4 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.1 | 0.8 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.1 | 0.4 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 7.3 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.1 | 0.5 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.1 | 1.1 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.1 | 0.4 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 14.9 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.1 | 1.8 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.1 | 4.9 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.1 | 48.5 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 1.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 5.3 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.9 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 5.9 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.6 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 1.3 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 2.7 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 3.0 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 1.7 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 13.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 2.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 12.9 | GO:0004930 | G-protein coupled receptor activity(GO:0004930) |
| 0.0 | 3.0 | GO:0004713 | protein tyrosine kinase activity(GO:0004713) |
| 0.0 | 1.4 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 2.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.7 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.2 | 9.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.1 | 1.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 5.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 2.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 6.2 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.1 | 5.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 5.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.1 | 1.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 4.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 2.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 9.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 3.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.4 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.7 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 6.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 17.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.8 | 23.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.6 | 12.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.4 | 5.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.4 | 5.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.3 | 21.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.3 | 8.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.2 | 4.8 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.2 | 12.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 4.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.2 | 2.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.1 | 4.0 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.8 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.1 | 15.6 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.1 | 3.4 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.1 | 3.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.1 | 3.1 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.1 | 11.5 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.1 | 6.2 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.1 | 0.9 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 7.7 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.1 | 1.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 7.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.9 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 5.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.7 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |