Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
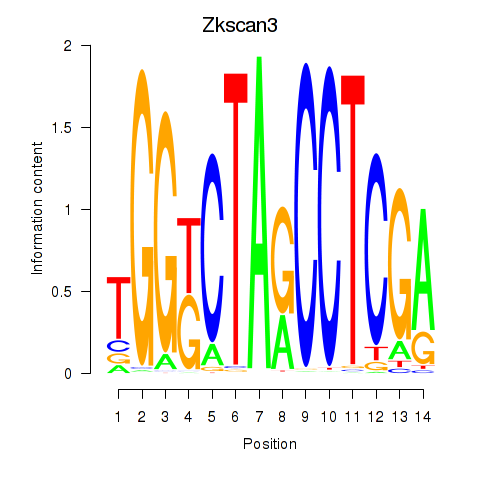
Results for Zkscan3
Z-value: 0.68
Transcription factors associated with Zkscan3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zkscan3
|
ENSMUSG00000021327.20 | Zkscan3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zkscan3 | mm39_v1_chr13_-_21586858_21586925 | 0.01 | 9.4e-01 | Click! |
Activity profile of Zkscan3 motif
Sorted Z-values of Zkscan3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zkscan3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_100720063 | 7.12 |
ENSMUST00000031264.12
|
Plac8
|
placenta-specific 8 |
| chr2_+_118603247 | 6.72 |
ENSMUST00000061360.4
ENSMUST00000130293.8 |
Phgr1
|
proline/histidine/glycine-rich 1 |
| chr1_+_160806241 | 4.99 |
ENSMUST00000195760.2
|
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr17_-_34694911 | 4.93 |
ENSMUST00000065841.5
|
Btnl4
|
butyrophilin-like 4 |
| chr11_+_115802828 | 4.65 |
ENSMUST00000132961.2
|
Smim6
|
small integral membrane protein 6 |
| chr6_+_71176811 | 4.56 |
ENSMUST00000067492.8
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr17_-_34736326 | 4.09 |
ENSMUST00000075483.5
|
Btnl6
|
butyrophilin-like 6 |
| chr2_-_30084124 | 3.69 |
ENSMUST00000113659.8
ENSMUST00000113660.2 |
Kyat1
|
kynurenine aminotransferase 1 |
| chr1_+_160806194 | 3.31 |
ENSMUST00000064725.11
ENSMUST00000191936.2 |
Serpinc1
|
serine (or cysteine) peptidase inhibitor, clade C (antithrombin), member 1 |
| chr9_+_53212871 | 3.26 |
ENSMUST00000051014.2
|
Exph5
|
exophilin 5 |
| chr16_+_32555015 | 3.22 |
ENSMUST00000239554.1
|
Muc4
|
mucin 4 |
| chr2_+_154042291 | 3.10 |
ENSMUST00000028987.7
|
Bpifb1
|
BPI fold containing family B, member 1 |
| chr6_+_125048230 | 2.93 |
ENSMUST00000140346.9
ENSMUST00000171989.3 |
Lpar5
|
lysophosphatidic acid receptor 5 |
| chr6_-_113696809 | 2.89 |
ENSMUST00000203770.3
ENSMUST00000064993.8 |
Ghrl
|
ghrelin |
| chr7_+_3339059 | 2.81 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr1_+_58834621 | 2.80 |
ENSMUST00000191201.7
|
Casp8
|
caspase 8 |
| chr6_-_43643093 | 2.78 |
ENSMUST00000114644.8
ENSMUST00000067888.14 |
Tpk1
|
thiamine pyrophosphokinase |
| chr7_+_3339077 | 2.72 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr1_+_58834532 | 2.66 |
ENSMUST00000027189.15
|
Casp8
|
caspase 8 |
| chr19_+_10611574 | 2.58 |
ENSMUST00000055115.9
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr1_+_180770016 | 2.29 |
ENSMUST00000027800.15
|
Tmem63a
|
transmembrane protein 63a |
| chr6_+_149041289 | 2.18 |
ENSMUST00000126406.8
|
Etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr1_+_180770064 | 2.15 |
ENSMUST00000159436.8
|
Tmem63a
|
transmembrane protein 63a |
| chr2_+_103926140 | 2.10 |
ENSMUST00000168176.8
ENSMUST00000040423.12 |
Cd59a
|
CD59a antigen |
| chr8_-_112417633 | 2.09 |
ENSMUST00000034435.7
|
Ctrb1
|
chymotrypsinogen B1 |
| chr3_-_129518723 | 1.96 |
ENSMUST00000199615.5
ENSMUST00000197079.5 |
Egf
|
epidermal growth factor |
| chr6_-_142418801 | 1.90 |
ENSMUST00000032371.8
|
Gys2
|
glycogen synthase 2 |
| chr16_+_17149235 | 1.80 |
ENSMUST00000023450.15
ENSMUST00000231884.2 |
Serpind1
|
serine (or cysteine) peptidase inhibitor, clade D, member 1 |
| chr11_+_35660288 | 1.78 |
ENSMUST00000018990.8
|
Pank3
|
pantothenate kinase 3 |
| chr3_+_146205864 | 1.77 |
ENSMUST00000119130.2
|
Gng5
|
guanine nucleotide binding protein (G protein), gamma 5 |
| chr11_-_75313350 | 1.77 |
ENSMUST00000125982.2
ENSMUST00000137103.8 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr4_+_144340236 | 1.72 |
ENSMUST00000094510.4
|
Aadacl4
|
arylacetamide deacetylase like 4 |
| chr16_-_56891711 | 1.71 |
ENSMUST00000067173.8
ENSMUST00000227043.2 |
Tmem45a2
|
transmembrane protein 45A2 |
| chr7_-_97827461 | 1.66 |
ENSMUST00000040971.14
|
Capn5
|
calpain 5 |
| chr11_-_75313412 | 1.61 |
ENSMUST00000138661.8
ENSMUST00000000769.14 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr10_+_75404482 | 1.59 |
ENSMUST00000134503.8
ENSMUST00000125770.8 ENSMUST00000128886.8 ENSMUST00000151212.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr10_+_75404464 | 1.50 |
ENSMUST00000145928.8
ENSMUST00000131565.8 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr6_-_97436223 | 1.43 |
ENSMUST00000113359.8
|
Frmd4b
|
FERM domain containing 4B |
| chr12_+_33004178 | 1.41 |
ENSMUST00000020885.13
|
Sypl
|
synaptophysin-like protein |
| chr3_-_95595157 | 1.35 |
ENSMUST00000015994.4
ENSMUST00000148854.2 ENSMUST00000117782.8 |
Adamtsl4
|
ADAMTS-like 4 |
| chr6_-_122833109 | 1.34 |
ENSMUST00000042081.9
|
C3ar1
|
complement component 3a receptor 1 |
| chr7_+_126358785 | 1.34 |
ENSMUST00000205657.2
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr6_+_116327853 | 1.29 |
ENSMUST00000140884.8
ENSMUST00000129170.8 |
Marchf8
|
membrane associated ring-CH-type finger 8 |
| chr13_-_104246084 | 1.25 |
ENSMUST00000224945.2
ENSMUST00000109315.5 |
Nln
|
neurolysin (metallopeptidase M3 family) |
| chrX_-_74918122 | 1.25 |
ENSMUST00000033547.14
|
Pls3
|
plastin 3 (T-isoform) |
| chr15_-_4008913 | 1.14 |
ENSMUST00000022791.9
|
Fbxo4
|
F-box protein 4 |
| chr13_-_65260716 | 1.13 |
ENSMUST00000054730.10
ENSMUST00000155487.8 |
Mfsd14b
|
major facilitator superfamily domain containing 14B |
| chr7_+_126358771 | 1.12 |
ENSMUST00000050201.11
ENSMUST00000057669.16 |
Mapk3
|
mitogen-activated protein kinase 3 |
| chr13_-_64514830 | 1.11 |
ENSMUST00000222971.2
|
Ctsl
|
cathepsin L |
| chr1_-_128256048 | 1.08 |
ENSMUST00000073490.7
|
Lct
|
lactase |
| chr8_-_71213535 | 1.04 |
ENSMUST00000038626.10
|
Mpv17l2
|
MPV17 mitochondrial membrane protein-like 2 |
| chr19_+_6952580 | 1.01 |
ENSMUST00000237084.2
ENSMUST00000236218.2 ENSMUST00000237235.2 |
Ppp1r14b
|
protein phosphatase 1, regulatory inhibitor subunit 14B |
| chr11_-_33528933 | 1.00 |
ENSMUST00000020366.8
ENSMUST00000135350.2 |
Gabrp
|
gamma-aminobutyric acid (GABA) A receptor, pi |
| chr1_-_84262274 | 1.00 |
ENSMUST00000177458.2
ENSMUST00000168574.9 |
Pid1
|
phosphotyrosine interaction domain containing 1 |
| chr1_-_40829801 | 0.95 |
ENSMUST00000039672.6
|
Mfsd9
|
major facilitator superfamily domain containing 9 |
| chr7_-_80994933 | 0.95 |
ENSMUST00000080813.5
|
Rps17
|
ribosomal protein S17 |
| chr4_-_62438122 | 0.91 |
ENSMUST00000107444.8
ENSMUST00000030090.4 |
Alad
|
aminolevulinate, delta-, dehydratase |
| chr7_-_141936355 | 0.89 |
ENSMUST00000133843.8
|
Gm49369
|
predicted gene, 49369 |
| chrX_-_17438470 | 0.88 |
ENSMUST00000176638.8
|
Fundc1
|
FUN14 domain containing 1 |
| chr8_+_84262409 | 0.84 |
ENSMUST00000214156.2
ENSMUST00000209408.4 |
Olfr370
|
olfactory receptor 370 |
| chr2_+_32536594 | 0.80 |
ENSMUST00000113272.8
ENSMUST00000009705.14 ENSMUST00000167841.8 |
Eng
|
endoglin |
| chr2_-_121001577 | 0.76 |
ENSMUST00000163766.8
ENSMUST00000146243.2 |
Zscan29
|
zinc finger SCAN domains 29 |
| chr10_+_81164552 | 0.74 |
ENSMUST00000105325.4
ENSMUST00000220312.2 |
Tbxa2r
|
thromboxane A2 receptor |
| chrX_+_162923474 | 0.65 |
ENSMUST00000073973.11
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr6_-_135145129 | 0.63 |
ENSMUST00000045855.9
|
Hebp1
|
heme binding protein 1 |
| chr11_-_102109748 | 0.60 |
ENSMUST00000131254.2
|
Hdac5
|
histone deacetylase 5 |
| chr5_-_88823472 | 0.59 |
ENSMUST00000113234.8
ENSMUST00000153565.8 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr8_-_13250535 | 0.58 |
ENSMUST00000165605.4
ENSMUST00000209691.2 ENSMUST00000211128.2 ENSMUST00000210317.2 |
Grtp1
|
GH regulated TBC protein 1 |
| chr8_-_107620147 | 0.57 |
ENSMUST00000176090.2
|
Chtf8
|
CTF8, chromosome transmission fidelity factor 8 |
| chr4_+_15957923 | 0.56 |
ENSMUST00000029879.15
ENSMUST00000149069.2 |
Nbn
|
nibrin |
| chr5_+_114141894 | 0.54 |
ENSMUST00000086599.11
|
Dao
|
D-amino acid oxidase |
| chr14_+_54413230 | 0.53 |
ENSMUST00000103700.2
|
Traj42
|
T cell receptor alpha joining 42 |
| chr12_-_112640378 | 0.47 |
ENSMUST00000130342.2
|
Akt1
|
thymoma viral proto-oncogene 1 |
| chr5_-_136003294 | 0.44 |
ENSMUST00000154181.2
ENSMUST00000111152.8 ENSMUST00000111153.8 |
Ssc4d
|
scavenger receptor cysteine rich family, 4 domains |
| chr5_+_24791796 | 0.41 |
ENSMUST00000197318.2
|
Chpf2
|
chondroitin polymerizing factor 2 |
| chr1_-_185061525 | 0.37 |
ENSMUST00000027921.11
ENSMUST00000110975.8 ENSMUST00000110974.4 |
Iars2
|
isoleucine-tRNA synthetase 2, mitochondrial |
| chr2_+_31560725 | 0.36 |
ENSMUST00000038474.14
ENSMUST00000137156.2 |
Exosc2
|
exosome component 2 |
| chr3_+_87458556 | 0.35 |
ENSMUST00000238853.2
|
Etv3l
|
ets variant 3-like |
| chr14_+_54412058 | 0.32 |
ENSMUST00000103699.2
|
Traj43
|
T cell receptor alpha joining 43 |
| chrX_-_17437801 | 0.31 |
ENSMUST00000177213.8
|
Fundc1
|
FUN14 domain containing 1 |
| chr14_+_54413727 | 0.30 |
ENSMUST00000103701.2
|
Traj41
|
T cell receptor alpha joining 41 |
| chr2_-_103609703 | 0.29 |
ENSMUST00000143188.2
|
Caprin1
|
cell cycle associated protein 1 |
| chr14_-_62718581 | 0.28 |
ENSMUST00000186010.8
|
Gm4131
|
predicted gene 4131 |
| chr15_-_37004302 | 0.26 |
ENSMUST00000228275.2
|
Zfp706
|
zinc finger protein 706 |
| chr4_-_155947819 | 0.24 |
ENSMUST00000030949.4
|
Tas1r3
|
taste receptor, type 1, member 3 |
| chr1_+_38037086 | 0.24 |
ENSMUST00000027252.8
|
Eif5b
|
eukaryotic translation initiation factor 5B |
| chr3_-_97318495 | 0.23 |
ENSMUST00000060912.4
|
Olfr1402
|
olfactory receptor 1402 |
| chr8_+_95537074 | 0.22 |
ENSMUST00000034232.3
|
Ccl17
|
chemokine (C-C motif) ligand 17 |
| chr11_-_88742285 | 0.22 |
ENSMUST00000107903.8
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chrX_-_17438520 | 0.20 |
ENSMUST00000026016.13
|
Fundc1
|
FUN14 domain containing 1 |
| chr6_-_43286488 | 0.19 |
ENSMUST00000031749.16
|
Nobox
|
NOBOX oogenesis homeobox |
| chr13_-_19579961 | 0.16 |
ENSMUST00000039694.13
|
Stard3nl
|
STARD3 N-terminal like |
| chr9_+_114517812 | 0.14 |
ENSMUST00000047404.7
|
Dync1li1
|
dynein cytoplasmic 1 light intermediate chain 1 |
| chr8_-_107620210 | 0.12 |
ENSMUST00000177068.8
ENSMUST00000176515.2 ENSMUST00000169312.2 |
Chtf8
Derpc
|
CTF8, chromosome transmission fidelity factor 8 DERPC proline and glycine rich nuclear protein |
| chr3_-_63836796 | 0.09 |
ENSMUST00000061706.7
|
E130311K13Rik
|
RIKEN cDNA E130311K13 gene |
| chr5_+_53424471 | 0.09 |
ENSMUST00000147148.5
|
Smim20
|
small integral membrane protein 20 |
| chr13_-_19579898 | 0.07 |
ENSMUST00000197565.3
ENSMUST00000221380.2 ENSMUST00000200323.3 ENSMUST00000199924.2 ENSMUST00000222869.2 |
Stard3nl
|
STARD3 N-terminal like |
| chr17_-_35984409 | 0.05 |
ENSMUST00000162266.8
ENSMUST00000160734.8 ENSMUST00000159852.2 ENSMUST00000160039.8 |
Gtf2h4
|
general transcription factor II H, polypeptide 4 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0097052 | L-kynurenine metabolic process(GO:0097052) |
| 1.0 | 8.3 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.7 | 2.2 | GO:1904736 | negative regulation of electron carrier activity(GO:1904733) regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904735) negative regulation of fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:1904736) |
| 0.7 | 2.9 | GO:2000506 | regulation of gastric mucosal blood circulation(GO:1904344) positive regulation of gastric mucosal blood circulation(GO:1904346) gastric mucosal blood circulation(GO:1990768) negative regulation of energy homeostasis(GO:2000506) |
| 0.7 | 2.8 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine-containing compound metabolic process(GO:0042723) |
| 0.7 | 5.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.7 | 4.1 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.6 | 3.2 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.6 | 2.5 | GO:2000657 | regulation of apolipoprotein binding(GO:2000656) negative regulation of apolipoprotein binding(GO:2000657) |
| 0.5 | 3.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.4 | 5.5 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.4 | 3.1 | GO:0031179 | peptide modification(GO:0031179) leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.3 | 4.6 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.3 | 3.4 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.3 | 1.6 | GO:0001905 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.3 | 1.3 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.3 | 0.8 | GO:0003032 | detection of oxygen(GO:0003032) cardiac jelly development(GO:1905072) |
| 0.3 | 3.1 | GO:0034144 | negative regulation of toll-like receptor 4 signaling pathway(GO:0034144) |
| 0.2 | 2.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.2 | 1.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.2 | 1.0 | GO:2001274 | positive regulation of fat cell proliferation(GO:0070346) negative regulation of ATP biosynthetic process(GO:2001170) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 | 1.8 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.2 | 7.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.2 | 0.5 | GO:0055130 | D-serine catabolic process(GO:0036088) D-alanine family amino acid metabolic process(GO:0046144) D-alanine metabolic process(GO:0046436) D-alanine catabolic process(GO:0055130) |
| 0.2 | 1.3 | GO:1902809 | regulation of skeletal muscle fiber differentiation(GO:1902809) |
| 0.2 | 1.0 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.2 | 0.7 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.1 | 2.9 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.1 | 0.6 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.1 | 1.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.4 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.5 | GO:0060709 | glycogen cell differentiation involved in embryonic placenta development(GO:0060709) |
| 0.1 | 1.8 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 0.4 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.1 | 1.1 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 0.7 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 1.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.6 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.9 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 1.5 | GO:0042168 | heme metabolic process(GO:0042168) |
| 0.0 | 0.2 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 1.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 1.9 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.3 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.7 | GO:1900048 | positive regulation of blood coagulation(GO:0030194) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.2 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 1.6 | GO:0007586 | digestion(GO:0007586) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.5 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.4 | 1.8 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.4 | 4.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.2 | 3.4 | GO:0043203 | axon hillock(GO:0043203) |
| 0.1 | 2.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 3.2 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.6 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.1 | 0.7 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 7.4 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.1 | 0.6 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 1.4 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 1.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 1.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.0 | 0.9 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 6.9 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 6.6 | GO:0005765 | lysosomal membrane(GO:0005765) lytic vacuole membrane(GO:0098852) |
| 0.0 | 1.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.0 | GO:0000315 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 1.3 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 1.1 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 1.1 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.6 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 5.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 1.2 | 3.7 | GO:0047316 | L-phenylalanine:pyruvate aminotransferase activity(GO:0047312) glutamine-phenylpyruvate transaminase activity(GO:0047316) L-glutamine:pyruvate aminotransferase activity(GO:0047945) |
| 1.0 | 2.9 | GO:0031768 | ghrelin receptor binding(GO:0031768) |
| 0.6 | 1.9 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.6 | 2.8 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.5 | 4.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.3 | 1.8 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 3.1 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.3 | 1.3 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 3.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 1.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.2 | 1.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.1 | 1.3 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.5 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.4 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 0.7 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.1 | 2.5 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 0.8 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.6 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.1 | 2.0 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 13.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.1 | 1.6 | GO:0001848 | complement binding(GO:0001848) |
| 0.1 | 0.7 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.1 | 1.7 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.7 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 1.0 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 2.2 | GO:0016279 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.5 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 3.1 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.8 | GO:0030165 | PDZ domain binding(GO:0030165) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.5 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.2 | 8.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.5 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 2.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 7.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.5 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 3.2 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.6 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 8.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.3 | 5.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.3 | 1.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.2 | 2.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.1 | 2.9 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.8 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 2.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.1 | 0.7 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.9 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 3.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.1 | 1.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 2.8 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.8 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.8 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.9 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 4.6 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 1.9 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.4 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 2.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |