Project
PRJNA375882: Comprehensive Mouse Transcriptomic BodyMap
Navigation
Downloads
Results for Zscan4c
Z-value: 1.26
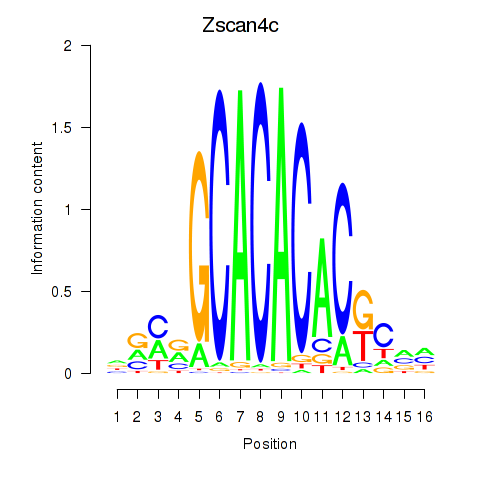
Transcription factors associated with Zscan4c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zscan4c
|
ENSMUSG00000054272.7 | Zscan4c |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zscan4c | mm39_v1_chr7_+_10739672_10739672 | -0.17 | 1.6e-01 | Click! |
Activity profile of Zscan4c motif
Sorted Z-values of Zscan4c motif
Network of associatons between targets according to the STRING database.
First level regulatory network of Zscan4c
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_98705791 | 24.41 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr2_+_102488985 | 20.37 |
ENSMUST00000080210.10
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr2_+_102489558 | 16.17 |
ENSMUST00000111213.8
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr6_-_55658242 | 14.11 |
ENSMUST00000044767.10
|
Neurod6
|
neurogenic differentiation 6 |
| chr4_-_46991842 | 13.27 |
ENSMUST00000107749.4
|
Gabbr2
|
gamma-aminobutyric acid (GABA) B receptor, 2 |
| chr15_-_37458768 | 11.65 |
ENSMUST00000116445.9
|
Ncald
|
neurocalcin delta |
| chr4_+_123077286 | 11.35 |
ENSMUST00000126995.2
|
Hpcal4
|
hippocalcin-like 4 |
| chr13_+_58955506 | 10.97 |
ENSMUST00000079828.7
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr9_-_44710480 | 10.73 |
ENSMUST00000214833.2
ENSMUST00000213972.2 ENSMUST00000214431.2 ENSMUST00000213363.2 ENSMUST00000114705.9 ENSMUST00000002100.8 |
Tmem25
|
transmembrane protein 25 |
| chr9_+_26645024 | 10.33 |
ENSMUST00000160899.8
ENSMUST00000161431.3 ENSMUST00000159799.8 |
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr9_+_89791943 | 9.89 |
ENSMUST00000189545.2
ENSMUST00000034909.11 ENSMUST00000034912.6 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr9_+_21848282 | 9.70 |
ENSMUST00000046371.13
|
Plppr2
|
phospholipid phosphatase related 2 |
| chr3_-_158267771 | 9.44 |
ENSMUST00000199890.5
ENSMUST00000238317.3 ENSMUST00000200137.5 ENSMUST00000106044.6 |
Lrrc7
|
leucine rich repeat containing 7 |
| chrX_+_47608122 | 9.01 |
ENSMUST00000033430.3
|
Rab33a
|
RAB33A, member RAS oncogene family |
| chr1_+_182591425 | 8.50 |
ENSMUST00000155229.7
ENSMUST00000153348.8 |
Susd4
|
sushi domain containing 4 |
| chr8_+_63404228 | 8.38 |
ENSMUST00000118003.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr10_-_114638202 | 8.26 |
ENSMUST00000239411.2
|
Trhde
|
TRH-degrading enzyme |
| chr9_-_49710190 | 7.97 |
ENSMUST00000114476.8
ENSMUST00000193547.6 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr9_-_49710058 | 7.91 |
ENSMUST00000192584.2
ENSMUST00000166811.9 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr3_+_13538266 | 7.69 |
ENSMUST00000211860.2
|
Ralyl
|
RALY RNA binding protein-like |
| chr8_+_63404395 | 7.68 |
ENSMUST00000119068.8
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr13_+_58955675 | 7.51 |
ENSMUST00000224402.2
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2 |
| chr18_-_23174698 | 7.14 |
ENSMUST00000097651.10
|
Nol4
|
nucleolar protein 4 |
| chr9_+_36744016 | 7.12 |
ENSMUST00000214772.2
|
Fez1
|
fasciculation and elongation protein zeta 1 (zygin I) |
| chr16_-_9812410 | 7.00 |
ENSMUST00000115835.8
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr16_-_9812787 | 6.95 |
ENSMUST00000199708.5
|
Grin2a
|
glutamate receptor, ionotropic, NMDA2A (epsilon 1) |
| chr5_-_129030111 | 6.00 |
ENSMUST00000196085.5
|
Rimbp2
|
RIMS binding protein 2 |
| chr9_+_26645141 | 5.80 |
ENSMUST00000115269.9
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr5_-_129030367 | 5.66 |
ENSMUST00000111346.6
ENSMUST00000200470.5 |
Rimbp2
|
RIMS binding protein 2 |
| chr19_+_8641369 | 5.65 |
ENSMUST00000035444.10
ENSMUST00000163785.2 |
Chrm1
|
cholinergic receptor, muscarinic 1, CNS |
| chrX_-_63320543 | 5.62 |
ENSMUST00000114679.2
ENSMUST00000069926.14 |
Slitrk4
|
SLIT and NTRK-like family, member 4 |
| chr19_-_21449916 | 5.57 |
ENSMUST00000087600.10
|
Gda
|
guanine deaminase |
| chr19_-_41732104 | 5.52 |
ENSMUST00000025993.10
|
Slit1
|
slit guidance ligand 1 |
| chr15_-_85462271 | 5.49 |
ENSMUST00000229191.2
|
Wnt7b
|
wingless-type MMTV integration site family, member 7B |
| chr11_-_101676076 | 5.01 |
ENSMUST00000164750.8
ENSMUST00000107176.8 ENSMUST00000017868.7 |
Etv4
|
ets variant 4 |
| chr11_-_37126709 | 4.75 |
ENSMUST00000102801.8
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr15_-_77037972 | 4.69 |
ENSMUST00000111581.4
ENSMUST00000166610.8 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr15_-_77037756 | 4.68 |
ENSMUST00000227314.2
ENSMUST00000227930.2 ENSMUST00000227533.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr10_+_74802996 | 4.63 |
ENSMUST00000037813.5
|
Gnaz
|
guanine nucleotide binding protein, alpha z subunit |
| chr19_+_16933471 | 4.46 |
ENSMUST00000087689.5
|
Prune2
|
prune homolog 2 |
| chr1_+_132119169 | 4.46 |
ENSMUST00000188169.7
ENSMUST00000112357.9 ENSMUST00000188175.2 |
Lemd1
Gm29695
|
LEM domain containing 1 predicted gene, 29695 |
| chr9_-_50650663 | 4.32 |
ENSMUST00000117093.2
ENSMUST00000121634.8 |
Dixdc1
|
DIX domain containing 1 |
| chr2_+_57128309 | 4.22 |
ENSMUST00000112618.9
ENSMUST00000028167.3 |
Gpd2
|
glycerol phosphate dehydrogenase 2, mitochondrial |
| chr2_+_169475436 | 4.15 |
ENSMUST00000109157.2
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr14_+_57762191 | 3.97 |
ENSMUST00000089494.6
|
Il17d
|
interleukin 17D |
| chr16_-_38533597 | 3.96 |
ENSMUST00000023487.5
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr3_-_63758672 | 3.96 |
ENSMUST00000162269.9
ENSMUST00000159676.9 ENSMUST00000175947.8 |
Plch1
|
phospholipase C, eta 1 |
| chr2_+_179684288 | 3.82 |
ENSMUST00000041126.9
|
Ss18l1
|
SS18, nBAF chromatin remodeling complex subunit like 1 |
| chr2_-_115895528 | 3.73 |
ENSMUST00000028639.13
ENSMUST00000102538.11 |
Meis2
|
Meis homeobox 2 |
| chr10_+_81128795 | 3.68 |
ENSMUST00000163075.8
ENSMUST00000105327.10 ENSMUST00000045469.15 |
Pip5k1c
|
phosphatidylinositol-4-phosphate 5-kinase, type 1 gamma |
| chr5_+_19431976 | 3.56 |
ENSMUST00000197354.5
ENSMUST00000088516.10 ENSMUST00000197443.5 |
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr15_-_77037926 | 3.36 |
ENSMUST00000228087.2
|
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr4_+_17853452 | 3.28 |
ENSMUST00000029881.10
|
Mmp16
|
matrix metallopeptidase 16 |
| chr14_+_122712809 | 3.24 |
ENSMUST00000075888.6
|
Zic2
|
zinc finger protein of the cerebellum 2 |
| chr2_-_35995283 | 3.01 |
ENSMUST00000112960.8
ENSMUST00000112967.12 ENSMUST00000112963.8 |
Lhx6
|
LIM homeobox protein 6 |
| chr5_-_44956932 | 2.96 |
ENSMUST00000199261.2
ENSMUST00000199534.5 |
Ldb2
|
LIM domain binding 2 |
| chr3_+_133942244 | 2.94 |
ENSMUST00000181904.3
|
Cxxc4
|
CXXC finger 4 |
| chr6_+_128376729 | 2.84 |
ENSMUST00000001561.12
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr2_-_91013193 | 2.72 |
ENSMUST00000111370.9
ENSMUST00000111376.8 ENSMUST00000099723.9 |
Madd
|
MAP-kinase activating death domain |
| chr3_-_107667499 | 2.70 |
ENSMUST00000153114.2
ENSMUST00000118593.8 ENSMUST00000120243.8 |
Csf1
|
colony stimulating factor 1 (macrophage) |
| chr2_-_115894993 | 2.59 |
ENSMUST00000074285.8
|
Meis2
|
Meis homeobox 2 |
| chr7_-_45519853 | 2.58 |
ENSMUST00000211713.2
|
Grin2d
|
glutamate receptor, ionotropic, NMDA2D (epsilon 4) |
| chr19_-_6117815 | 2.52 |
ENSMUST00000162575.8
ENSMUST00000159084.8 ENSMUST00000161718.8 ENSMUST00000162810.8 ENSMUST00000025713.12 ENSMUST00000113543.9 ENSMUST00000160417.8 ENSMUST00000161528.2 |
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr11_+_61967821 | 2.50 |
ENSMUST00000092415.9
ENSMUST00000201015.4 ENSMUST00000202744.4 ENSMUST00000201723.4 ENSMUST00000202179.2 |
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr2_-_115895202 | 2.49 |
ENSMUST00000110906.9
|
Meis2
|
Meis homeobox 2 |
| chr5_-_44957016 | 2.46 |
ENSMUST00000199256.5
|
Ldb2
|
LIM domain binding 2 |
| chr9_-_50639367 | 2.34 |
ENSMUST00000117646.8
|
Dixdc1
|
DIX domain containing 1 |
| chr6_+_128376844 | 2.16 |
ENSMUST00000120405.4
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr19_+_28941292 | 2.11 |
ENSMUST00000045674.4
|
Plpp6
|
phospholipid phosphatase 6 |
| chr5_-_44956981 | 2.08 |
ENSMUST00000070748.10
|
Ldb2
|
LIM domain binding 2 |
| chr2_+_30254239 | 2.08 |
ENSMUST00000077977.14
ENSMUST00000140075.9 ENSMUST00000142801.8 ENSMUST00000100214.10 |
Miga2
|
mitoguardin 2 |
| chr1_+_182591771 | 1.99 |
ENSMUST00000193660.6
|
Susd4
|
sushi domain containing 4 |
| chr10_+_18345706 | 1.87 |
ENSMUST00000162891.8
ENSMUST00000100054.4 |
Nhsl1
|
NHS-like 1 |
| chr2_+_19914573 | 1.87 |
ENSMUST00000114610.8
|
Etl4
|
enhancer trap locus 4 |
| chr13_-_76205115 | 1.80 |
ENSMUST00000056130.8
|
Gpr150
|
G protein-coupled receptor 150 |
| chr9_-_31824758 | 1.66 |
ENSMUST00000116615.5
|
Barx2
|
BarH-like homeobox 2 |
| chr2_-_165997551 | 1.62 |
ENSMUST00000109249.9
ENSMUST00000146497.9 |
Sulf2
|
sulfatase 2 |
| chr6_-_122317484 | 1.58 |
ENSMUST00000112600.9
|
Phc1
|
polyhomeotic 1 |
| chr19_-_38807600 | 1.54 |
ENSMUST00000025963.8
|
Noc3l
|
NOC3 like DNA replication regulator |
| chr3_+_125197722 | 1.17 |
ENSMUST00000173932.8
|
Ndst4
|
N-deacetylase/N-sulfotransferase (heparin glucosaminyl) 4 |
| chr5_+_99002293 | 1.16 |
ENSMUST00000031278.6
ENSMUST00000200388.2 |
Bmp3
|
bone morphogenetic protein 3 |
| chr13_+_55517545 | 1.12 |
ENSMUST00000063771.14
|
Rgs14
|
regulator of G-protein signaling 14 |
| chr17_-_63188139 | 1.12 |
ENSMUST00000078839.5
ENSMUST00000076840.12 |
Efna5
|
ephrin A5 |
| chr13_-_9815276 | 1.08 |
ENSMUST00000130151.8
|
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr8_-_49008881 | 0.91 |
ENSMUST00000110345.8
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr13_-_9815350 | 0.87 |
ENSMUST00000110636.9
ENSMUST00000152725.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr7_+_46490899 | 0.83 |
ENSMUST00000147535.8
|
Ldha
|
lactate dehydrogenase A |
| chr12_-_69939931 | 0.80 |
ENSMUST00000049239.8
ENSMUST00000110570.8 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr9_+_21279299 | 0.79 |
ENSMUST00000214852.2
ENSMUST00000115414.3 |
Ilf3
|
interleukin enhancer binding factor 3 |
| chr17_-_28779678 | 0.77 |
ENSMUST00000114785.3
ENSMUST00000025062.5 |
Clps
|
colipase, pancreatic |
| chr6_-_122317156 | 0.75 |
ENSMUST00000159384.8
|
Phc1
|
polyhomeotic 1 |
| chr19_-_28941264 | 0.74 |
ENSMUST00000161813.2
|
Spata6l
|
spermatogenesis associated 6 like |
| chr4_-_136620376 | 0.61 |
ENSMUST00000046332.6
|
C1qc
|
complement component 1, q subcomponent, C chain |
| chr9_+_37466989 | 0.60 |
ENSMUST00000213126.2
|
Siae
|
sialic acid acetylesterase |
| chr18_+_37108994 | 0.49 |
ENSMUST00000193984.2
|
Gm37388
|
predicted gene, 37388 |
| chr2_-_165997179 | 0.42 |
ENSMUST00000088086.4
|
Sulf2
|
sulfatase 2 |
| chr9_+_43978369 | 0.41 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr2_-_35994819 | 0.37 |
ENSMUST00000148852.4
|
Lhx6
|
LIM homeobox protein 6 |
| chr18_-_79152504 | 0.33 |
ENSMUST00000025430.11
|
Setbp1
|
SET binding protein 1 |
| chr5_-_68004743 | 0.25 |
ENSMUST00000072971.13
ENSMUST00000113652.8 ENSMUST00000113651.8 ENSMUST00000037380.15 |
Atp8a1
|
ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 |
| chr15_+_44482545 | 0.18 |
ENSMUST00000227691.2
|
Ebag9
|
estrogen receptor-binding fragment-associated gene 9 |
| chr13_-_9815173 | 0.16 |
ENSMUST00000062658.15
ENSMUST00000222358.2 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr12_-_69939762 | 0.12 |
ENSMUST00000110567.8
ENSMUST00000171211.8 |
Map4k5
|
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr12_+_52550775 | 0.11 |
ENSMUST00000219443.2
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr2_+_85804239 | 0.11 |
ENSMUST00000217244.2
|
Olfr1029
|
olfactory receptor 1029 |
| chr2_+_169474916 | 0.08 |
ENSMUST00000109159.3
|
Tshz2
|
teashirt zinc finger family member 2 |
| chr17_+_78815493 | 0.06 |
ENSMUST00000024880.11
ENSMUST00000232859.2 |
Vit
|
vitrin |
| chr13_-_9814901 | 0.02 |
ENSMUST00000223421.2
ENSMUST00000128658.8 |
Zmynd11
|
zinc finger, MYND domain containing 11 |
| chr9_-_57168777 | 0.02 |
ENSMUST00000217657.2
|
1700017B05Rik
|
RIKEN cDNA 1700017B05 gene |
| chr1_+_87983189 | 0.01 |
ENSMUST00000173325.2
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.2 | 36.5 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 4.0 | 15.9 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 3.1 | 18.5 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 2.6 | 10.5 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 1.7 | 13.9 | GO:0033058 | directional locomotion(GO:0033058) |
| 1.4 | 5.6 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 1.4 | 5.5 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 1.2 | 14.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 1.1 | 5.6 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 1.0 | 12.7 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.9 | 5.5 | GO:0033563 | dorsal/ventral axon guidance(GO:0033563) |
| 0.9 | 2.7 | GO:1902277 | negative regulation of pancreatic amylase secretion(GO:1902277) |
| 0.9 | 2.7 | GO:1902226 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.9 | 16.1 | GO:0030204 | chondroitin sulfate metabolic process(GO:0030204) |
| 0.8 | 3.2 | GO:1900224 | positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.6 | 3.4 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.5 | 14.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.5 | 5.0 | GO:0060762 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.5 | 6.7 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.4 | 7.1 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.4 | 4.2 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.3 | 2.0 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) glial cell-derived neurotrophic factor receptor signaling pathway(GO:0035860) |
| 0.3 | 9.0 | GO:2000785 | regulation of autophagosome assembly(GO:2000785) |
| 0.3 | 3.6 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.3 | 5.7 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.3 | 13.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.3 | 3.7 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.2 | 9.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.2 | 1.9 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.2 | 9.7 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.2 | 3.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.2 | 6.8 | GO:0043171 | peptide catabolic process(GO:0043171) |
| 0.1 | 1.7 | GO:0042637 | catagen(GO:0042637) |
| 0.1 | 11.7 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.1 | 9.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.1 | 1.1 | GO:0071372 | cellular response to follicle-stimulating hormone stimulus(GO:0071372) |
| 0.1 | 0.8 | GO:0019660 | lactate biosynthetic process from pyruvate(GO:0019244) glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.1 | 11.7 | GO:0003073 | regulation of systemic arterial blood pressure(GO:0003073) |
| 0.1 | 4.6 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.1 | 2.6 | GO:0050966 | detection of mechanical stimulus involved in sensory perception of pain(GO:0050966) |
| 0.1 | 0.6 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.1 | 3.8 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.1 | 2.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 5.6 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 1.0 | GO:0010669 | epithelial structure maintenance(GO:0010669) |
| 0.0 | 2.5 | GO:0006695 | cholesterol biosynthetic process(GO:0006695) |
| 0.0 | 1.3 | GO:0034243 | regulation of transcription elongation from RNA polymerase II promoter(GO:0034243) |
| 0.0 | 2.2 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.2 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 9.4 | GO:0010976 | positive regulation of neuron projection development(GO:0010976) |
| 0.0 | 5.0 | GO:0007219 | Notch signaling pathway(GO:0007219) |
| 0.0 | 0.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.0 | 0.8 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 2.7 | GO:0048015 | phosphatidylinositol-mediated signaling(GO:0048015) |
| 0.0 | 1.9 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.7 | 13.3 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.8 | 4.2 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.7 | 2.7 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.7 | 16.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.7 | 36.5 | GO:0030673 | axolemma(GO:0030673) |
| 0.4 | 5.6 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.3 | 18.5 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.3 | 9.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.2 | 9.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.2 | 3.6 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 2.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 3.8 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 16.4 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.1 | 4.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 6.7 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 37.4 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 12.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 0.8 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 2.7 | GO:0008021 | synaptic vesicle(GO:0008021) |
| 0.0 | 2.5 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 19.5 | GO:0005730 | nucleolus(GO:0005730) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 6.1 | 36.5 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 3.1 | 18.5 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 2.7 | 16.1 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 2.2 | 13.3 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 1.4 | 4.2 | GO:0004368 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 1.4 | 5.5 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 1.2 | 16.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 1.1 | 5.6 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.9 | 5.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.9 | 2.7 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.7 | 16.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.6 | 9.7 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.6 | 15.9 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.6 | 4.6 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.4 | 3.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.4 | 11.5 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.4 | 3.7 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.4 | 1.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.3 | 2.0 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.2 | 11.7 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.2 | 3.7 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.2 | 13.8 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.2 | 11.3 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.2 | 4.0 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.1 | 5.0 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.1 | 1.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 5.6 | GO:0016814 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amidines(GO:0016814) |
| 0.1 | 9.9 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.1 | 0.8 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 2.5 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.1 | 4.2 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.1 | 0.9 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 37.0 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 2.7 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.1 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 11.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 9.4 | GO:0008022 | protein C-terminus binding(GO:0008022) |
| 0.0 | 8.1 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 4.0 | GO:0017124 | SH3 domain binding(GO:0017124) |
| 0.0 | 2.9 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 3.8 | GO:0003713 | transcription coactivator activity(GO:0003713) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 16.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.3 | 24.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.3 | 4.6 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.2 | 13.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.2 | 15.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.2 | 2.7 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.2 | 3.7 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.2 | 5.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.1 | 9.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.1 | 5.0 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 2.9 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 4.3 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.1 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 2.7 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID TNF PATHWAY | TNF receptor signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 26.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.7 | 11.7 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.6 | 13.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.5 | 15.6 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.3 | 27.0 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.3 | 5.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.3 | 15.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.2 | 3.7 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 5.6 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.1 | 3.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 4.6 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.1 | 3.6 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 2.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.1 | 18.5 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.1 | 0.6 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 5.5 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |
| 0.1 | 3.1 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 1.2 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.8 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.8 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |