Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
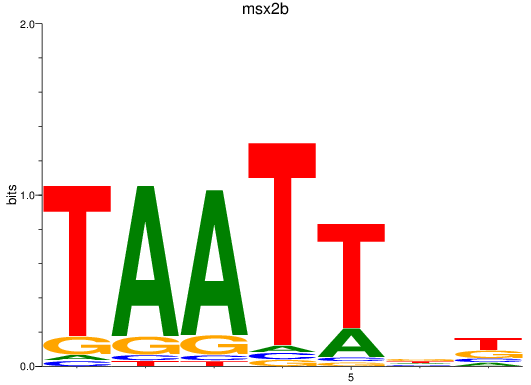
Results for msx2b
Z-value: 0.78
Transcription factors associated with msx2b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
msx2b
|
ENSDARG00000101023 | muscle segment homeobox 2b |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| msx2b | dr11_v1_chr21_-_41147818_41147818 | -0.22 | 3.9e-01 | Click! |
Activity profile of msx2b motif
Sorted Z-values of msx2b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr22_-_15957041 | 2.38 |
ENSDART00000149236
ENSDART00000187500 ENSDART00000176304 ENSDART00000080047 ENSDART00000190068 |
eps15l1a
|
epidermal growth factor receptor pathway substrate 15-like 1a |
| chr12_-_2993095 | 2.25 |
ENSDART00000152316
|
si:dkey-202c14.3
|
si:dkey-202c14.3 |
| chr14_-_49063157 | 1.83 |
ENSDART00000021260
|
sept8b
|
septin 8b |
| chr22_-_15562933 | 1.82 |
ENSDART00000141528
|
ankmy1
|
ankyrin repeat and MYND domain containing 1 |
| chr11_+_25259058 | 1.71 |
ENSDART00000154109
|
tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr17_+_50248152 | 1.60 |
ENSDART00000153998
|
zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chr5_-_61970674 | 1.60 |
ENSDART00000143161
|
znf541
|
zinc finger protein 541 |
| chr22_-_547748 | 1.40 |
ENSDART00000037455
ENSDART00000140101 |
ccnd3
|
cyclin D3 |
| chr20_-_29474859 | 1.40 |
ENSDART00000152906
ENSDART00000045249 |
scg5
|
secretogranin V |
| chr19_-_47527093 | 1.32 |
ENSDART00000171379
ENSDART00000171140 ENSDART00000114886 |
scg5
|
secretogranin V |
| chr7_-_6470431 | 1.29 |
ENSDART00000081359
|
zgc:110425
|
zgc:110425 |
| chr25_+_26895394 | 1.28 |
ENSDART00000155820
|
si:dkey-42p14.3
|
si:dkey-42p14.3 |
| chr20_-_43533096 | 1.22 |
ENSDART00000145199
|
pimr134
|
Pim proto-oncogene, serine/threonine kinase, related 134 |
| chr10_-_16065185 | 1.16 |
ENSDART00000187266
|
si:dkey-184a18.5
|
si:dkey-184a18.5 |
| chr1_-_55785722 | 1.15 |
ENSDART00000142069
ENSDART00000043933 |
ndufb7
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 7 |
| chr21_-_5205617 | 1.15 |
ENSDART00000145554
ENSDART00000045284 |
rpl37
|
ribosomal protein L37 |
| chr1_-_52659548 | 1.14 |
ENSDART00000029252
ENSDART00000150862 |
aptx
|
aprataxin |
| chr7_+_34687969 | 1.11 |
ENSDART00000182013
ENSDART00000188255 ENSDART00000182728 |
plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr14_-_41272034 | 1.11 |
ENSDART00000191709
ENSDART00000074438 |
cenpi
|
centromere protein I |
| chr14_+_23520986 | 1.07 |
ENSDART00000170473
ENSDART00000175970 |
si:ch211-221f10.2
|
si:ch211-221f10.2 |
| chr3_+_18398876 | 1.07 |
ENSDART00000141100
ENSDART00000138107 |
rps2
|
ribosomal protein S2 |
| chr16_-_40508624 | 1.07 |
ENSDART00000075718
|
ndufaf6
|
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr21_-_2299002 | 1.06 |
ENSDART00000168712
|
si:ch73-299h12.6
|
si:ch73-299h12.6 |
| chr10_-_5016997 | 1.03 |
ENSDART00000101414
|
hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr25_-_28926631 | 1.00 |
ENSDART00000112850
|
etfbkmt
|
electron transfer flavoprotein beta subunit lysine methyltransferase |
| chr20_+_36628059 | 0.99 |
ENSDART00000062898
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr24_-_26328721 | 0.96 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr12_-_48477031 | 0.94 |
ENSDART00000105176
|
ndufb8
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 8 |
| chr15_+_36309070 | 0.89 |
ENSDART00000157034
|
gmnc
|
geminin coiled-coil domain containing |
| chr21_+_38002879 | 0.89 |
ENSDART00000065183
|
cldn2
|
claudin 2 |
| chr10_+_2715548 | 0.86 |
ENSDART00000130793
|
grk5
|
G protein-coupled receptor kinase 5 |
| chr22_-_30973791 | 0.81 |
ENSDART00000104728
|
ssuh2.2
|
ssu-2 homolog, tandem duplicate 2 |
| chr21_-_44009169 | 0.81 |
ENSDART00000028960
|
ndufa2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2 |
| chr23_+_35714574 | 0.79 |
ENSDART00000164616
|
tuba1c
|
tubulin, alpha 1c |
| chr8_+_23738122 | 0.79 |
ENSDART00000062983
|
rpl10a
|
ribosomal protein L10a |
| chr2_+_33747509 | 0.79 |
ENSDART00000134187
|
si:dkey-31m5.5
|
si:dkey-31m5.5 |
| chr16_-_35394422 | 0.76 |
ENSDART00000180593
|
si:dkey-34d22.3
|
si:dkey-34d22.3 |
| chr5_+_24882633 | 0.74 |
ENSDART00000111302
|
rhbdd3
|
rhomboid domain containing 3 |
| chr1_+_34203817 | 0.69 |
ENSDART00000191432
ENSDART00000046094 |
arl6
|
ADP-ribosylation factor-like 6 |
| chr7_+_65309738 | 0.69 |
ENSDART00000169228
|
vat1l
|
vesicle amine transport 1-like |
| chr19_-_1947403 | 0.68 |
ENSDART00000113951
ENSDART00000151293 ENSDART00000134074 |
znrf2a
|
zinc and ring finger 2a |
| chr14_-_32089117 | 0.68 |
ENSDART00000158014
|
si:ch211-69b22.5
|
si:ch211-69b22.5 |
| chr11_+_6881001 | 0.66 |
ENSDART00000170331
|
klhl26
|
kelch-like family member 26 |
| chr1_+_42225060 | 0.65 |
ENSDART00000138740
ENSDART00000101306 |
ctnna2
|
catenin (cadherin-associated protein), alpha 2 |
| chr2_+_6127593 | 0.65 |
ENSDART00000184007
|
fzr1b
|
fizzy/cell division cycle 20 related 1b |
| chr3_-_31619463 | 0.64 |
ENSDART00000124559
|
moto
|
minamoto |
| chr3_-_33427803 | 0.64 |
ENSDART00000075495
|
rpl23
|
ribosomal protein L23 |
| chr3_-_13461361 | 0.62 |
ENSDART00000080807
|
fbxw9
|
F-box and WD repeat domain containing 9 |
| chr9_-_24031461 | 0.62 |
ENSDART00000021218
|
rpe
|
ribulose-5-phosphate-3-epimerase |
| chr23_-_35064785 | 0.62 |
ENSDART00000172240
|
BX294434.1
|
|
| chr11_+_11360641 | 0.61 |
ENSDART00000159765
|
si:dkey-23f9.7
|
si:dkey-23f9.7 |
| chr4_+_14900042 | 0.60 |
ENSDART00000018261
|
akr1b1
|
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr21_+_19062124 | 0.57 |
ENSDART00000134746
|
rpl17
|
ribosomal protein L17 |
| chr4_-_5455506 | 0.55 |
ENSDART00000156593
ENSDART00000154676 |
si:dkey-14d8.22
|
si:dkey-14d8.22 |
| chr20_-_2134620 | 0.55 |
ENSDART00000064375
|
tmem244
|
transmembrane protein 244 |
| chr2_+_38373272 | 0.55 |
ENSDART00000113111
|
psmb5
|
proteasome subunit beta 5 |
| chr6_+_59984772 | 0.52 |
ENSDART00000048449
|
mtrf1
|
mitochondrial translational release factor 1 |
| chr15_-_19669770 | 0.51 |
ENSDART00000152707
|
si:dkey-4p15.3
|
si:dkey-4p15.3 |
| chr15_+_17074185 | 0.50 |
ENSDART00000046648
|
clptm1
|
cleft lip and palate associated transmembrane protein 1 |
| chr6_-_30210378 | 0.50 |
ENSDART00000157359
ENSDART00000113924 |
lrrc7
|
leucine rich repeat containing 7 |
| chr5_-_2689753 | 0.50 |
ENSDART00000172699
|
gng10
|
guanine nucleotide binding protein (G protein), gamma 10 |
| chr23_+_43489182 | 0.50 |
ENSDART00000162062
|
znf341
|
zinc finger protein 341 |
| chr15_-_23784600 | 0.48 |
ENSDART00000059354
|
rad1
|
RAD1 homolog (S. pombe) |
| chr11_+_3254524 | 0.48 |
ENSDART00000159459
|
pmela
|
premelanosome protein a |
| chr8_+_17069577 | 0.47 |
ENSDART00000131847
|
si:dkey-190g11.3
|
si:dkey-190g11.3 |
| chr22_-_18164671 | 0.47 |
ENSDART00000014057
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr13_-_4223955 | 0.46 |
ENSDART00000113060
|
dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr15_+_1796313 | 0.44 |
ENSDART00000126253
|
fam124b
|
family with sequence similarity 124B |
| chr14_-_8453192 | 0.43 |
ENSDART00000136947
|
eif1ad
|
eukaryotic translation initiation factor 1A domain containing |
| chr4_+_5506952 | 0.43 |
ENSDART00000032857
ENSDART00000160222 |
mapk11
|
mitogen-activated protein kinase 11 |
| chr11_+_25539698 | 0.43 |
ENSDART00000035602
|
cxxc1b
|
CXXC finger protein 1b |
| chr23_-_24493768 | 0.42 |
ENSDART00000135559
ENSDART00000046933 |
sult1st5
|
sulfotransferase family 1, cytosolic sulfotransferase 5 |
| chr5_-_7834582 | 0.42 |
ENSDART00000162626
ENSDART00000157661 |
pdlim5a
|
PDZ and LIM domain 5a |
| chr5_-_38777852 | 0.42 |
ENSDART00000131603
|
si:dkey-58f10.4
|
si:dkey-58f10.4 |
| chr1_+_11977426 | 0.42 |
ENSDART00000103399
|
tspan5b
|
tetraspanin 5b |
| chr20_+_38031439 | 0.41 |
ENSDART00000153208
|
galnt14
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr19_-_26923957 | 0.41 |
ENSDART00000182390
|
skiv2l
|
SKI2 homolog, superkiller viralicidic activity 2-like |
| chr24_-_21923930 | 0.40 |
ENSDART00000131944
|
tagln3b
|
transgelin 3b |
| chr14_-_7137808 | 0.40 |
ENSDART00000054803
|
trpt1
|
tRNA phosphotransferase 1 |
| chr14_+_35369979 | 0.40 |
ENSDART00000144702
ENSDART00000169712 |
clint1a
|
clathrin interactor 1a |
| chr22_-_18164835 | 0.39 |
ENSDART00000143189
|
rfxank
|
regulatory factor X-associated ankyrin-containing protein |
| chr8_+_22930627 | 0.39 |
ENSDART00000187860
|
sypa
|
synaptophysin a |
| chr16_+_41570653 | 0.38 |
ENSDART00000102665
|
aste1a
|
asteroid homolog 1a |
| chr15_+_32711663 | 0.37 |
ENSDART00000157854
ENSDART00000167515 |
postnb
|
periostin, osteoblast specific factor b |
| chr20_-_53366137 | 0.37 |
ENSDART00000146001
|
wasf1
|
WAS protein family, member 1 |
| chr11_-_44484952 | 0.36 |
ENSDART00000166674
ENSDART00000188016 |
mfn1b
|
mitofusin 1b |
| chr9_-_2572790 | 0.36 |
ENSDART00000135076
ENSDART00000016710 |
scrn3
|
secernin 3 |
| chr5_+_13427826 | 0.35 |
ENSDART00000083359
|
sec14l8
|
SEC14-like lipid binding 8 |
| chr19_-_22346582 | 0.35 |
ENSDART00000045675
ENSDART00000169065 |
slc52a2
zgc:109744
|
solute carrier family 52 (riboflavin transporter), member 2 zgc:109744 |
| chr10_+_9697097 | 0.35 |
ENSDART00000167821
|
rabgap1
|
RAB GTPase activating protein 1 |
| chr23_-_31974060 | 0.35 |
ENSDART00000168087
|
BX927210.1
|
|
| chr5_+_67268110 | 0.33 |
ENSDART00000148116
|
si:ch211-110p13.9
|
si:ch211-110p13.9 |
| chr24_-_3783497 | 0.32 |
ENSDART00000158354
|
adarb2
|
adenosine deaminase, RNA-specific, B2 (non-functional) |
| chr14_-_4273396 | 0.31 |
ENSDART00000127318
|
frmpd1b
|
FERM and PDZ domain containing 1b |
| chr3_+_25195966 | 0.30 |
ENSDART00000112338
|
il2rb
|
interleukin 2 receptor, beta |
| chr7_-_24005268 | 0.30 |
ENSDART00000173608
|
si:dkey-183c6.9
|
si:dkey-183c6.9 |
| chr25_-_18953322 | 0.30 |
ENSDART00000155927
|
si:ch211-68a17.7
|
si:ch211-68a17.7 |
| chr3_-_44059902 | 0.29 |
ENSDART00000158485
ENSDART00000159088 ENSDART00000165628 |
il4r.1
|
interleukin 4 receptor, tandem duplicate 1 |
| chr24_-_31843173 | 0.29 |
ENSDART00000185782
|
steap2
|
STEAP family member 2, metalloreductase |
| chr10_-_26430168 | 0.29 |
ENSDART00000128894
|
dchs1b
|
dachsous cadherin-related 1b |
| chr20_+_34870517 | 0.29 |
ENSDART00000191797
|
ankef1a
|
ankyrin repeat and EF-hand domain containing 1a |
| chr5_+_66433287 | 0.29 |
ENSDART00000170757
|
kntc1
|
kinetochore associated 1 |
| chr12_-_5728755 | 0.28 |
ENSDART00000105887
|
dlx4b
|
distal-less homeobox 4b |
| chr8_-_44004135 | 0.28 |
ENSDART00000136269
|
rimbp2
|
RIMS binding protein 2 |
| chr6_-_27891961 | 0.28 |
ENSDART00000155116
|
im:7152348
|
im:7152348 |
| chr23_+_40460333 | 0.28 |
ENSDART00000184658
|
soga3b
|
SOGA family member 3b |
| chr7_-_13362590 | 0.27 |
ENSDART00000091616
|
sdhaf2
|
succinate dehydrogenase complex assembly factor 2 |
| chr15_-_28082310 | 0.27 |
ENSDART00000152620
|
dhrs13a.3
|
dehydrogenase/reductase (SDR family) member 13a, duplicate 3 |
| chr22_-_15717897 | 0.27 |
ENSDART00000008424
|
malt2
|
MALT paracaspase 2 |
| chr17_-_51199558 | 0.27 |
ENSDART00000154532
|
paplna
|
papilin a, proteoglycan-like sulfated glycoprotein |
| chr15_-_17813680 | 0.26 |
ENSDART00000158556
|
CT573342.2
|
|
| chr7_+_6990510 | 0.25 |
ENSDART00000138948
|
ccs
|
copper chaperone for superoxide dismutase |
| chr5_+_24179307 | 0.25 |
ENSDART00000051552
|
mpdu1a
|
mannose-P-dolichol utilization defect 1a |
| chr5_-_9216758 | 0.25 |
ENSDART00000134896
ENSDART00000147000 |
lrp13
|
low-density lipoprotein receptor related-protein 13 |
| chr15_-_4094888 | 0.24 |
ENSDART00000166307
|
TM4SF19
|
si:dkey-83h2.3 |
| chr19_-_12392819 | 0.24 |
ENSDART00000052239
|
ptpn2b
|
protein tyrosine phosphatase, non-receptor type 2, b |
| chr11_+_42765963 | 0.23 |
ENSDART00000156080
ENSDART00000179888 |
tdrd3
|
tudor domain containing 3 |
| chr18_-_33979693 | 0.23 |
ENSDART00000021215
|
si:ch211-203b20.7
|
si:ch211-203b20.7 |
| chr12_+_5081759 | 0.23 |
ENSDART00000164178
|
prrt2
|
proline-rich transmembrane protein 2 |
| chr6_-_55347616 | 0.22 |
ENSDART00000172081
|
pcif1
|
PDX1 C-terminal inhibiting factor 1 |
| chr15_-_3976035 | 0.22 |
ENSDART00000168061
|
si:ch73-309g22.1
|
si:ch73-309g22.1 |
| chr10_+_8690936 | 0.22 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr21_+_10712823 | 0.21 |
ENSDART00000123476
|
lman1
|
lectin, mannose-binding, 1 |
| chr8_-_40276035 | 0.21 |
ENSDART00000185301
ENSDART00000187610 ENSDART00000053158 ENSDART00000162020 |
kdm2ba
|
lysine (K)-specific demethylase 2Ba |
| chr17_+_12658411 | 0.20 |
ENSDART00000139918
|
gpn1
|
GPN-loop GTPase 1 |
| chr5_-_66768121 | 0.20 |
ENSDART00000141095
|
FERMT3 (1 of many)
|
im:7154036 |
| chr21_+_43402794 | 0.20 |
ENSDART00000185572
|
frmd7
|
FERM domain containing 7 |
| chr16_-_50897887 | 0.19 |
ENSDART00000156985
|
si:ch73-90p23.1
|
si:ch73-90p23.1 |
| chr13_+_23988442 | 0.18 |
ENSDART00000010918
|
agt
|
angiotensinogen |
| chr3_-_17871846 | 0.18 |
ENSDART00000074478
ENSDART00000187941 |
nkiras2
|
NFKB inhibitor interacting Ras-like 2 |
| chr15_+_19646902 | 0.17 |
ENSDART00000179822
|
lim2.3
|
lens intrinsic membrane protein 2.3 |
| chr25_-_24248000 | 0.17 |
ENSDART00000073527
|
spty2d1
|
SPT2 chromatin protein domain containing 1 |
| chr6_+_4033832 | 0.17 |
ENSDART00000159952
|
sema5bb
|
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5Bb |
| chr20_-_23426339 | 0.17 |
ENSDART00000004625
|
zar1
|
zygote arrest 1 |
| chr14_-_11456724 | 0.16 |
ENSDART00000110424
|
si:ch211-153b23.4
|
si:ch211-153b23.4 |
| chr23_-_41965557 | 0.16 |
ENSDART00000144183
|
slc1a7b
|
solute carrier family 1 (glutamate transporter), member 7b |
| chr15_+_7064819 | 0.16 |
ENSDART00000155268
|
pik3cb
|
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
| chr24_-_40860603 | 0.15 |
ENSDART00000188032
|
CU633479.7
|
|
| chr14_-_35462148 | 0.15 |
ENSDART00000045809
|
srpx2
|
sushi-repeat containing protein, X-linked 2 |
| chr24_-_14712427 | 0.15 |
ENSDART00000176316
|
jph1a
|
junctophilin 1a |
| chr24_-_24849091 | 0.15 |
ENSDART00000133649
ENSDART00000038290 |
crhb
|
corticotropin releasing hormone b |
| chr11_+_37178271 | 0.14 |
ENSDART00000161771
|
itih3b
|
inter-alpha-trypsin inhibitor heavy chain 3b |
| chr10_-_11353101 | 0.13 |
ENSDART00000092110
|
lin54
|
lin-54 DREAM MuvB core complex component |
| chr25_+_30442389 | 0.13 |
ENSDART00000003346
|
pdcd2l
|
programmed cell death 2-like |
| chr6_+_6828167 | 0.13 |
ENSDART00000181284
|
si:ch211-85n16.4
|
si:ch211-85n16.4 |
| chr18_+_10884996 | 0.13 |
ENSDART00000147613
|
mical3a
|
microtubule associated monooxygenase, calponin and LIM domain containing 3a |
| chr19_-_3255725 | 0.12 |
ENSDART00000105165
|
illr1
|
immune-related, lectin-like receptor 1 |
| chr7_+_69356661 | 0.12 |
ENSDART00000177049
|
CABZ01083862.1
|
|
| chr13_+_8958438 | 0.12 |
ENSDART00000165721
ENSDART00000169398 |
haao
|
3-hydroxyanthranilate 3,4-dioxygenase |
| chr10_+_39952995 | 0.12 |
ENSDART00000183077
|
BX927333.1
|
|
| chr5_+_9246458 | 0.12 |
ENSDART00000081772
|
susd1
|
sushi domain containing 1 |
| chr11_+_29856032 | 0.12 |
ENSDART00000079150
|
grpr
|
gastrin-releasing peptide receptor |
| chr7_-_41812355 | 0.12 |
ENSDART00000016105
|
vps35
|
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr8_-_16788626 | 0.11 |
ENSDART00000191652
|
CR759968.2
|
|
| chr8_+_3426522 | 0.11 |
ENSDART00000159466
|
CU929199.1
|
|
| chr3_+_24571163 | 0.11 |
ENSDART00000123017
|
sp100.3
|
SP110 nuclear body protein, tandem duplicate 3 |
| chr14_+_23874062 | 0.11 |
ENSDART00000172149
|
sh3rf2
|
SH3 domain containing ring finger 2 |
| chr1_+_12068800 | 0.10 |
ENSDART00000055566
|
xpa
|
xeroderma pigmentosum, complementation group A |
| chr10_-_41157135 | 0.10 |
ENSDART00000134851
|
aak1b
|
AP2 associated kinase 1b |
| chr2_-_24398324 | 0.10 |
ENSDART00000165226
|
zgc:154006
|
zgc:154006 |
| chr12_+_28574863 | 0.10 |
ENSDART00000153284
|
tbkbp1
|
TBK1 binding protein 1 |
| chr23_-_44903048 | 0.10 |
ENSDART00000149103
|
fhdc5
|
FH2 domain containing 5 |
| chr17_+_8542203 | 0.10 |
ENSDART00000158873
|
CU462878.2
|
|
| chr17_+_31185276 | 0.10 |
ENSDART00000062887
|
disp2
|
dispatched homolog 2 (Drosophila) |
| chr2_+_37176814 | 0.10 |
ENSDART00000048277
ENSDART00000176827 |
copa
|
coatomer protein complex, subunit alpha |
| chr11_+_17984167 | 0.09 |
ENSDART00000020283
ENSDART00000188329 |
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr15_+_17031111 | 0.09 |
ENSDART00000175378
|
plin2
|
perilipin 2 |
| chr9_+_29548195 | 0.09 |
ENSDART00000176057
|
rnf17
|
ring finger protein 17 |
| chr13_+_50151407 | 0.08 |
ENSDART00000031858
|
gpr137ba
|
G protein-coupled receptor 137Ba |
| chr17_+_45607580 | 0.08 |
ENSDART00000122583
|
si:ch211-202f3.4
|
si:ch211-202f3.4 |
| chr4_-_6373735 | 0.08 |
ENSDART00000140100
|
MDFIC
|
si:ch73-156e19.1 |
| chr21_-_31013817 | 0.08 |
ENSDART00000065504
|
ncbp3
|
nuclear cap binding subunit 3 |
| chr6_-_15641686 | 0.07 |
ENSDART00000135583
|
mlpha
|
melanophilin a |
| chr22_-_31672566 | 0.07 |
ENSDART00000128247
|
lsm3
|
LSM3 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr11_+_17984354 | 0.07 |
ENSDART00000179986
|
rpusd4
|
RNA pseudouridylate synthase domain containing 4 |
| chr14_-_12307522 | 0.06 |
ENSDART00000163900
|
myot
|
myotilin |
| chr18_-_7539469 | 0.06 |
ENSDART00000101296
|
si:dkey-30c15.2
|
si:dkey-30c15.2 |
| chr19_+_12406583 | 0.06 |
ENSDART00000013865
ENSDART00000151535 |
seh1l
|
SEH1-like (S. cerevisiae) |
| chr6_-_29515709 | 0.06 |
ENSDART00000180205
|
pex5la
|
peroxisomal biogenesis factor 5-like a |
| chr3_+_22935183 | 0.06 |
ENSDART00000157378
|
hdac5
|
histone deacetylase 5 |
| chr4_+_78058635 | 0.06 |
ENSDART00000162631
ENSDART00000191619 |
CABZ01075274.1
|
|
| chr19_+_20540843 | 0.05 |
ENSDART00000192500
|
kcnh8
|
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr17_-_6730247 | 0.05 |
ENSDART00000031091
|
vsnl1b
|
visinin-like 1b |
| chr2_-_10600950 | 0.05 |
ENSDART00000160216
|
BX323564.1
|
|
| chr20_-_20355577 | 0.04 |
ENSDART00000018500
|
hif1ab
|
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) b |
| chr14_+_1007169 | 0.04 |
ENSDART00000172676
|
f8
|
coagulation factor VIII, procoagulant component |
| chr10_+_25726694 | 0.03 |
ENSDART00000140308
|
ugt5d1
|
UDP glucuronosyltransferase 5 family, polypeptide D1 |
| chr25_+_25085349 | 0.03 |
ENSDART00000192166
|
si:ch73-182e20.4
|
si:ch73-182e20.4 |
| chr2_-_23778180 | 0.03 |
ENSDART00000136782
|
si:dkey-24c2.7
|
si:dkey-24c2.7 |
| chr22_+_31295791 | 0.03 |
ENSDART00000092447
|
grip2b
|
glutamate receptor interacting protein 2b |
| chr17_-_10122204 | 0.03 |
ENSDART00000160751
|
BX088587.1
|
|
| chr18_+_7073130 | 0.03 |
ENSDART00000101216
ENSDART00000148947 |
si:dkey-88e18.2
|
si:dkey-88e18.2 |
| chr13_-_40238813 | 0.02 |
ENSDART00000044963
|
loxl4
|
lysyl oxidase-like 4 |
| chr18_+_33264609 | 0.02 |
ENSDART00000050639
|
v2ra20
|
vomeronasal 2 receptor, a20 |
| chr15_+_17030941 | 0.02 |
ENSDART00000062069
|
plin2
|
perilipin 2 |
| chr6_-_46861676 | 0.02 |
ENSDART00000188712
ENSDART00000190148 |
igfn1.3
|
immunoglobulin-like and fibronectin type III domain containing 1, tandem duplicate 3 |
| chr7_+_29512673 | 0.02 |
ENSDART00000173895
|
si:dkey-182o15.5
|
si:dkey-182o15.5 |
| chr7_+_15313443 | 0.02 |
ENSDART00000045385
|
mespba
|
mesoderm posterior ba |
| chr16_+_14707960 | 0.02 |
ENSDART00000137912
|
col14a1a
|
collagen, type XIV, alpha 1a |
| chr19_+_22727940 | 0.02 |
ENSDART00000052509
|
trhrb
|
thyrotropin-releasing hormone receptor b |
Network of associatons between targets according to the STRING database.
First level regulatory network of msx2b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0050995 | negative regulation of lipid catabolic process(GO:0050995) |
| 0.2 | 1.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.2 | 0.6 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.2 | 0.9 | GO:1903251 | multi-ciliated epithelial cell differentiation(GO:1903251) |
| 0.2 | 0.5 | GO:0070126 | mitochondrial translational termination(GO:0070126) |
| 0.2 | 0.7 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.2 | 0.5 | GO:0097435 | fibril organization(GO:0097435) |
| 0.2 | 0.5 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.4 | GO:1904088 | regulation of convergent extension involved in axis elongation(GO:1901232) positive regulation of epiboly involved in gastrulation with mouth forming second(GO:1904088) |
| 0.1 | 0.7 | GO:1904668 | positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.1 | 2.2 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.1 | 0.3 | GO:1903673 | mitotic cytokinetic process(GO:1902410) mitotic cleavage furrow formation(GO:1903673) |
| 0.1 | 1.4 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 0.3 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.1 | 0.6 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.1 | 0.4 | GO:0032776 | DNA methylation on cytosine(GO:0032776) |
| 0.1 | 0.3 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.1 | 0.3 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 1.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.1 | 0.9 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 1.7 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.1 | 0.3 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.1 | 0.4 | GO:0032218 | riboflavin transport(GO:0032218) |
| 0.1 | 2.4 | GO:0046883 | regulation of hormone secretion(GO:0046883) |
| 0.1 | 0.2 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.3 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 1.1 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 1.0 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 1.2 | GO:0000302 | response to reactive oxygen species(GO:0000302) |
| 0.0 | 0.4 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.5 | GO:0032088 | negative regulation of NF-kappaB transcription factor activity(GO:0032088) |
| 0.0 | 0.4 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0070914 | UV-damage excision repair(GO:0070914) nucleotide-excision repair involved in interstrand cross-link repair(GO:1901255) |
| 0.0 | 0.7 | GO:0042541 | hemoglobin biosynthetic process(GO:0042541) |
| 0.0 | 0.1 | GO:0043420 | anthranilate metabolic process(GO:0043420) |
| 0.0 | 0.1 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.2 | GO:0000455 | enzyme-directed rRNA pseudouridine synthesis(GO:0000455) |
| 0.0 | 0.3 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.0 | 0.4 | GO:0045682 | regulation of epidermis development(GO:0045682) |
| 0.0 | 0.5 | GO:0018023 | peptidyl-lysine trimethylation(GO:0018023) |
| 0.0 | 0.4 | GO:2000601 | positive regulation of actin nucleation(GO:0051127) positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.4 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.4 | GO:0016203 | muscle attachment(GO:0016203) |
| 0.0 | 1.0 | GO:0043297 | apical junction assembly(GO:0043297) bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.1 | GO:0051876 | pigment granule dispersal(GO:0051876) |
| 0.0 | 0.7 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 1.8 | GO:0061640 | cytoskeleton-dependent cytokinesis(GO:0061640) |
| 0.0 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.2 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:1902262 | apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.4 | GO:0051923 | sulfation(GO:0051923) |
| 0.0 | 0.3 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:2000369 | regulation of clathrin-mediated endocytosis(GO:2000369) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.2 | GO:0001990 | regulation of systemic arterial blood pressure by hormone(GO:0001990) endocrine process(GO:0050886) |
| 0.0 | 2.5 | GO:0016197 | endosomal transport(GO:0016197) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.1 | 0.5 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.1 | 0.4 | GO:0055087 | Ski complex(GO:0055087) |
| 0.1 | 2.9 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.8 | GO:0031105 | septin ring(GO:0005940) septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 1.7 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.7 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 2.7 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 2.0 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 1.4 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.5 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.1 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 1.1 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.5 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.4 | GO:0030125 | clathrin vesicle coat(GO:0030125) |
| 0.0 | 1.2 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.6 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 1.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.1 | 0.7 | GO:1990757 | anaphase-promoting complex binding(GO:0010997) ubiquitin ligase activator activity(GO:1990757) |
| 0.1 | 0.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 2.1 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 0.4 | GO:0070004 | cysteine-type exopeptidase activity(GO:0070004) |
| 0.1 | 0.4 | GO:0032217 | riboflavin transporter activity(GO:0032217) |
| 0.1 | 1.3 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 0.2 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.1 | 0.9 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.5 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.3 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.6 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.3 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.5 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.3 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 1.1 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 1.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.4 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) alpha-actinin binding(GO:0051393) |
| 0.0 | 0.5 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.2 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 2.4 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol-3,4-bisphosphate 5-kinase activity(GO:0052812) |
| 0.0 | 0.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.4 | GO:0004707 | MAP kinase activity(GO:0004707) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.9 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.4 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.1 | 2.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.8 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 1.1 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 4.2 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 1.4 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 1.6 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.5 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.5 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.2 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.5 | REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | Genes involved in Activation of ATR in response to replication stress |