Project
PRJNA438478: RNAseq of wild type zebrafish germline
Navigation
Downloads
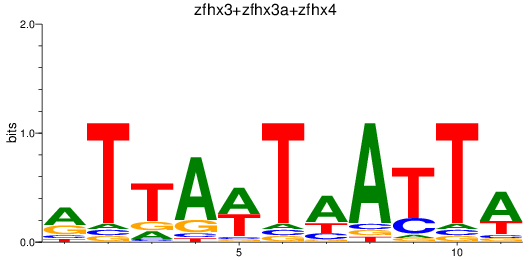
Results for zfhx3+zfhx3a+zfhx4
Z-value: 0.58
Transcription factors associated with zfhx3+zfhx3a+zfhx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
zfhx3a
|
ENSDARG00000073944 | si_ch73-386h18.1 |
|
zfhx4
|
ENSDARG00000075542 | zinc finger homeobox 4 |
|
zfhx3
|
ENSDARG00000103057 | zinc finger homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| zfhx4 | dr11_v1_chr24_-_23320223_23320223 | 0.93 | 1.6e-08 | Click! |
| si:ch73-386h18.1 | dr11_v1_chr18_+_6054816_6054816 | 0.85 | 8.4e-06 | Click! |
| zfhx3 | dr11_v1_chr7_-_68373495_68373495 | 0.83 | 2.3e-05 | Click! |
Activity profile of zfhx3+zfhx3a+zfhx4 motif
Sorted Z-values of zfhx3+zfhx3a+zfhx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_31075972 | 1.74 |
ENSDART00000122927
|
si:ch211-198b3.4
|
si:ch211-198b3.4 |
| chr7_+_20587506 | 1.67 |
ENSDART00000172416
ENSDART00000170633 |
si:dkey-19b23.7
|
si:dkey-19b23.7 |
| chr15_+_34988148 | 1.58 |
ENSDART00000076269
|
ccdc105
|
coiled-coil domain containing 105 |
| chr9_+_52613820 | 1.53 |
ENSDART00000168753
ENSDART00000165580 ENSDART00000164769 |
si:ch211-241j8.2
|
si:ch211-241j8.2 |
| chr18_+_7611298 | 1.51 |
ENSDART00000062156
|
odf3b
|
outer dense fiber of sperm tails 3B |
| chr1_+_44003654 | 1.07 |
ENSDART00000131337
|
si:dkey-22i16.10
|
si:dkey-22i16.10 |
| chr25_+_34915576 | 1.03 |
ENSDART00000073441
|
sntb2
|
syntrophin, beta 2 |
| chr11_-_2838699 | 0.99 |
ENSDART00000066189
|
lhfpl5a
|
LHFPL tetraspan subfamily member 5a |
| chr16_-_42894628 | 0.95 |
ENSDART00000045600
|
hfe2
|
hemochromatosis type 2 |
| chr22_+_20720808 | 0.93 |
ENSDART00000171321
|
si:dkey-211f22.5
|
si:dkey-211f22.5 |
| chr24_-_26328721 | 0.91 |
ENSDART00000125468
|
apodb
|
apolipoprotein Db |
| chr5_-_30715225 | 0.86 |
ENSDART00000016758
|
ftr82
|
finTRIM family, member 82 |
| chr18_+_36914221 | 0.85 |
ENSDART00000147656
|
si:ch211-238g23.1
|
si:ch211-238g23.1 |
| chr5_+_6954162 | 0.84 |
ENSDART00000086666
|
stpg2
|
sperm-tail PG-rich repeat containing 2 |
| chr25_+_34915762 | 0.83 |
ENSDART00000191776
|
sntb2
|
syntrophin, beta 2 |
| chr23_+_20689255 | 0.82 |
ENSDART00000182420
|
usp21
|
ubiquitin specific peptidase 21 |
| chr16_-_27566552 | 0.63 |
ENSDART00000142102
|
zgc:153215
|
zgc:153215 |
| chr13_+_27314795 | 0.58 |
ENSDART00000128726
|
eef1a1a
|
eukaryotic translation elongation factor 1 alpha 1a |
| chr20_+_36629173 | 0.58 |
ENSDART00000161241
|
ephx1
|
epoxide hydrolase 1, microsomal (xenobiotic) |
| chr20_+_27298783 | 0.58 |
ENSDART00000013861
|
ppp4r4
|
protein phosphatase 4, regulatory subunit 4 |
| chr17_+_8212477 | 0.57 |
ENSDART00000064665
|
slc18b1
|
solute carrier family 18, subfamily B, member 1 |
| chr8_-_18042089 | 0.55 |
ENSDART00000132596
|
dio1
|
iodothyronine deiodinase 1 |
| chr5_+_13385837 | 0.53 |
ENSDART00000191190
|
ccl19a.1
|
chemokine (C-C motif) ligand 19a, tandem duplicate 1 |
| chr15_-_17869115 | 0.51 |
ENSDART00000112838
|
atf5b
|
activating transcription factor 5b |
| chr1_+_54013457 | 0.49 |
ENSDART00000012104
ENSDART00000126339 |
dla
|
deltaA |
| chr12_-_35885349 | 0.47 |
ENSDART00000085662
|
cep131
|
centrosomal protein 131 |
| chr21_-_20765338 | 0.46 |
ENSDART00000135940
|
ghrb
|
growth hormone receptor b |
| chr4_+_9669717 | 0.45 |
ENSDART00000004604
|
si:dkey-153k10.9
|
si:dkey-153k10.9 |
| chr8_-_25074319 | 0.45 |
ENSDART00000134190
|
sort1b
|
sortilin 1b |
| chr21_-_13123176 | 0.44 |
ENSDART00000144866
ENSDART00000024616 |
fam219aa
|
family with sequence similarity 219, member Aa |
| chr22_-_34551568 | 0.43 |
ENSDART00000148147
|
rnf123
|
ring finger protein 123 |
| chr4_+_57881965 | 0.42 |
ENSDART00000162234
|
si:dkeyp-44b5.4
|
si:dkeyp-44b5.4 |
| chr22_-_15602593 | 0.42 |
ENSDART00000036075
|
tpm4a
|
tropomyosin 4a |
| chr4_+_7698862 | 0.39 |
ENSDART00000009909
|
elk3
|
ELK3, ETS-domain protein |
| chr12_-_19151708 | 0.35 |
ENSDART00000057124
|
tefa
|
thyrotrophic embryonic factor a |
| chr15_+_28685892 | 0.34 |
ENSDART00000155815
ENSDART00000060244 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr20_-_9095105 | 0.33 |
ENSDART00000140792
|
oma1
|
OMA1 zinc metallopeptidase |
| chr7_+_56577906 | 0.32 |
ENSDART00000184023
|
hp
|
haptoglobin |
| chr4_-_22338906 | 0.32 |
ENSDART00000131402
|
golgb1
|
golgin B1 |
| chr22_+_13886821 | 0.31 |
ENSDART00000130585
ENSDART00000105711 |
sh3bp4a
|
SH3-domain binding protein 4a |
| chr20_+_41756996 | 0.29 |
ENSDART00000186393
|
fam184a
|
family with sequence similarity 184, member A |
| chr10_-_42131408 | 0.29 |
ENSDART00000076693
|
stambpa
|
STAM binding protein a |
| chr25_-_16818195 | 0.29 |
ENSDART00000185215
|
dyrk4
|
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr5_-_65158203 | 0.28 |
ENSDART00000171656
|
sh2d3cb
|
SH2 domain containing 3Cb |
| chr18_+_29950233 | 0.27 |
ENSDART00000146431
|
atmin
|
ATM interactor |
| chr10_-_11261565 | 0.25 |
ENSDART00000146727
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr25_+_28679672 | 0.24 |
ENSDART00000139965
ENSDART00000134072 |
cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr19_+_14573998 | 0.24 |
ENSDART00000022076
|
fam46bb
|
family with sequence similarity 46, member Bb |
| chr7_-_38658411 | 0.24 |
ENSDART00000109463
ENSDART00000017155 |
npsn
|
nephrosin |
| chr22_-_38480186 | 0.24 |
ENSDART00000171704
|
soul4
|
heme-binding protein soul4 |
| chr3_-_34337969 | 0.23 |
ENSDART00000151634
|
tnrc6c1
|
trinucleotide repeat containing 6C1 |
| chr17_-_51893123 | 0.23 |
ENSDART00000103350
ENSDART00000017329 |
numb
|
numb homolog (Drosophila) |
| chr18_+_2228737 | 0.23 |
ENSDART00000165301
|
rab27a
|
RAB27A, member RAS oncogene family |
| chr10_-_17179415 | 0.23 |
ENSDART00000131751
|
depdc5
|
DEP domain containing 5 |
| chr3_-_16055432 | 0.22 |
ENSDART00000123621
ENSDART00000023859 |
atp6v0ca
|
ATPase H+ transporting V0 subunit ca |
| chr25_+_24291156 | 0.22 |
ENSDART00000083407
|
b4galnt4a
|
beta-1,4-N-acetyl-galactosaminyl transferase 4a |
| chr10_-_11261386 | 0.22 |
ENSDART00000189946
|
ptbp3
|
polypyrimidine tract binding protein 3 |
| chr7_+_42460302 | 0.22 |
ENSDART00000004120
|
adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr16_-_41787421 | 0.21 |
ENSDART00000147210
|
si:dkey-199f5.8
|
si:dkey-199f5.8 |
| chr21_+_9689103 | 0.21 |
ENSDART00000165132
|
mapk10
|
mitogen-activated protein kinase 10 |
| chr1_+_44838706 | 0.21 |
ENSDART00000162779
ENSDART00000147357 |
kdm2aa
|
lysine (K)-specific demethylase 2Aa |
| chr14_-_25111496 | 0.21 |
ENSDART00000158108
|
si:rp71-1d10.8
|
si:rp71-1d10.8 |
| chr21_+_13150937 | 0.21 |
ENSDART00000102251
|
sptan1
|
spectrin alpha, non-erythrocytic 1 |
| chr5_+_32345187 | 0.20 |
ENSDART00000147132
|
c9
|
complement component 9 |
| chr8_+_15254564 | 0.20 |
ENSDART00000024433
|
slc5a9
|
solute carrier family 5 (sodium/sugar cotransporter), member 9 |
| chr23_+_24589633 | 0.20 |
ENSDART00000143694
|
mlh3
|
mutL homolog 3 (E. coli) |
| chr18_-_17399291 | 0.20 |
ENSDART00000192075
ENSDART00000060949 ENSDART00000188506 |
zfpm1
|
zinc finger protein, FOG family member 1 |
| chr23_+_45200481 | 0.20 |
ENSDART00000004357
ENSDART00000111126 ENSDART00000193560 ENSDART00000190476 |
psip1b
|
PC4 and SFRS1 interacting protein 1b |
| chr8_+_18830759 | 0.20 |
ENSDART00000089079
|
mpnd
|
MPN domain containing |
| chr4_+_18804317 | 0.20 |
ENSDART00000101043
|
slc26a3.2
|
solute carrier family 26 (anion exchanger), member 3, tandem duplicate 2 |
| chr8_-_25569920 | 0.20 |
ENSDART00000136869
|
prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chr3_+_40170216 | 0.19 |
ENSDART00000011568
|
syngr3a
|
synaptogyrin 3a |
| chr18_-_16181952 | 0.19 |
ENSDART00000157824
|
slc6a15
|
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr10_+_42423318 | 0.18 |
ENSDART00000134282
|
npy8ar
|
neuropeptide Y receptor Y8a |
| chr13_+_45524475 | 0.16 |
ENSDART00000074567
ENSDART00000019113 |
maco1b
|
macoilin 1b |
| chr20_-_40755614 | 0.16 |
ENSDART00000061247
|
cx32.3
|
connexin 32.3 |
| chr12_-_28848015 | 0.14 |
ENSDART00000153200
|
si:ch211-194k22.8
|
si:ch211-194k22.8 |
| chr5_+_28830643 | 0.13 |
ENSDART00000051448
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr25_-_35599887 | 0.13 |
ENSDART00000153827
|
clpxb
|
caseinolytic mitochondrial matrix peptidase chaperone subunit b |
| chr12_+_27061720 | 0.13 |
ENSDART00000153426
|
srcap
|
Snf2-related CREBBP activator protein |
| chr16_-_44127307 | 0.13 |
ENSDART00000104583
ENSDART00000151936 ENSDART00000058685 ENSDART00000190830 |
zfpm2a
|
zinc finger protein, FOG family member 2a |
| chr15_+_29472065 | 0.13 |
ENSDART00000154343
|
gdpd5b
|
glycerophosphodiester phosphodiesterase domain containing 5b |
| chr6_-_43283122 | 0.12 |
ENSDART00000186022
|
frmd4ba
|
FERM domain containing 4Ba |
| chr19_+_3056450 | 0.12 |
ENSDART00000141324
ENSDART00000082353 |
hsf1
|
heat shock transcription factor 1 |
| chr15_-_17868870 | 0.11 |
ENSDART00000170950
|
atf5b
|
activating transcription factor 5b |
| chr5_+_28830388 | 0.11 |
ENSDART00000149150
|
serpina7
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr17_-_30975707 | 0.10 |
ENSDART00000138346
|
evla
|
Enah/Vasp-like a |
| chr10_+_8690936 | 0.09 |
ENSDART00000144218
|
si:dkey-27b3.4
|
si:dkey-27b3.4 |
| chr15_+_28685625 | 0.08 |
ENSDART00000188797
ENSDART00000166036 |
nova2
|
neuro-oncological ventral antigen 2 |
| chr23_+_13375359 | 0.08 |
ENSDART00000151947
|
ZNF512B
|
si:dkey-256k13.2 |
| chr4_-_5302866 | 0.08 |
ENSDART00000138590
|
SNAP91 (1 of many)
|
si:ch211-214j24.9 |
| chr6_+_20647155 | 0.08 |
ENSDART00000193477
|
slc19a1
|
solute carrier family 19 (folate transporter), member 1 |
| chr24_+_14713776 | 0.07 |
ENSDART00000134475
|
gdap1
|
ganglioside induced differentiation associated protein 1 |
| chr25_-_3230187 | 0.07 |
ENSDART00000160545
|
cib1
|
calcium and integrin binding 1 (calmyrin) |
| chr25_+_18965430 | 0.06 |
ENSDART00000169742
|
tdg.1
|
thymine DNA glycosylase, tandem duplicate 1 |
| chr22_-_24818066 | 0.06 |
ENSDART00000143443
|
vtg6
|
vitellogenin 6 |
| chr17_+_2162916 | 0.06 |
ENSDART00000103775
|
pak6a
|
p21 protein (Cdc42/Rac)-activated kinase 6a |
| chr16_+_28383758 | 0.06 |
ENSDART00000059038
ENSDART00000141061 |
itga8
|
integrin, alpha 8 |
| chr2_-_27609189 | 0.05 |
ENSDART00000040555
|
lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr6_+_30456788 | 0.04 |
ENSDART00000121492
|
FP236735.1
|
|
| chr24_-_21470766 | 0.04 |
ENSDART00000178121
|
atp8a2
|
ATPase phospholipid transporting 8A2 |
| chr7_-_20865005 | 0.03 |
ENSDART00000190752
|
fis1
|
fission, mitochondrial 1 |
| chr20_-_40750953 | 0.02 |
ENSDART00000061256
|
cx28.9
|
connexin 28.9 |
| chr1_+_22851261 | 0.02 |
ENSDART00000193925
|
gtf2e2
|
general transcription factor IIE, polypeptide 2, beta |
| chr5_+_9382301 | 0.02 |
ENSDART00000124017
|
ugt2a7
|
UDP glucuronosyltransferase 2 family, polypeptide A7 |
| chr7_+_4194306 | 0.02 |
ENSDART00000190454
|
si:dkey-28d5.5
|
si:dkey-28d5.5 |
| chr4_-_20118468 | 0.02 |
ENSDART00000078587
|
si:dkey-159a18.1
|
si:dkey-159a18.1 |
| chr17_-_7371564 | 0.02 |
ENSDART00000060336
|
rab32b
|
RAB32b, member RAS oncogene family |
| chr16_-_46645396 | 0.02 |
ENSDART00000131485
|
tmem176l.2
|
transmembrane protein 176l.2 |
| chr4_-_32642681 | 0.02 |
ENSDART00000188695
ENSDART00000193974 |
si:dkey-265e7.1
|
si:dkey-265e7.1 |
| chr7_-_17412559 | 0.01 |
ENSDART00000163020
|
nitr3a
|
novel immune-type receptor 3a |
| chr11_+_7432533 | 0.01 |
ENSDART00000180977
|
adgrl2a
|
adhesion G protein-coupled receptor L2a |
| chr22_+_19220459 | 0.00 |
ENSDART00000163070
|
si:dkey-21e2.7
|
si:dkey-21e2.7 |
Network of associatons between targets according to the STRING database.
First level regulatory network of zfhx3+zfhx3a+zfhx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.1 | 0.5 | GO:0055016 | hypochord development(GO:0055016) |
| 0.1 | 0.5 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
| 0.1 | 0.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.1 | 0.6 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.1 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.2 | GO:0098908 | regulation of neuronal action potential(GO:0098908) |
| 0.1 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.3 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.5 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) response to growth hormone(GO:0060416) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.8 | GO:0021551 | central nervous system morphogenesis(GO:0021551) |
| 0.0 | 0.2 | GO:0023041 | neuronal signal transduction(GO:0023041) |
| 0.0 | 0.9 | GO:0007568 | aging(GO:0007568) |
| 0.0 | 1.0 | GO:0060122 | detection of mechanical stimulus involved in sensory perception(GO:0050974) inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.1 | GO:0098838 | folic acid transport(GO:0015884) drug transport(GO:0015893) methotrexate transport(GO:0051958) reduced folate transmembrane transport(GO:0098838) |
| 0.0 | 0.2 | GO:0060036 | notochord cell vacuolation(GO:0060036) |
| 0.0 | 0.2 | GO:2000036 | regulation of stem cell population maintenance(GO:2000036) |
| 0.0 | 0.6 | GO:0009636 | response to toxic substance(GO:0009636) |
| 0.0 | 0.2 | GO:0007530 | sex determination(GO:0007530) |
| 0.0 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.2 | GO:0045974 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.4 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.5 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) |
| 0.0 | 0.3 | GO:0033138 | positive regulation of peptidyl-serine phosphorylation(GO:0033138) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 0.5 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.1 | 1.9 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.1 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.1 | 0.2 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.5 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.2 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.1 | GO:0000812 | Swr1 complex(GO:0000812) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0033961 | cis-stilbene-oxide hydrolase activity(GO:0033961) |
| 0.1 | 0.5 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.2 | GO:0033842 | N-acetyl-beta-glucosaminyl-glycoprotein 4-beta-N-acetylgalactosaminyltransferase activity(GO:0033842) |
| 0.0 | 0.2 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.5 | GO:0070122 | isopeptidase activity(GO:0070122) |
| 0.0 | 0.1 | GO:0015350 | reduced folate carrier activity(GO:0008518) methotrexate transporter activity(GO:0015350) |
| 0.0 | 0.2 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.5 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.2 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID BMP PATHWAY | BMP receptor signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 1.0 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |