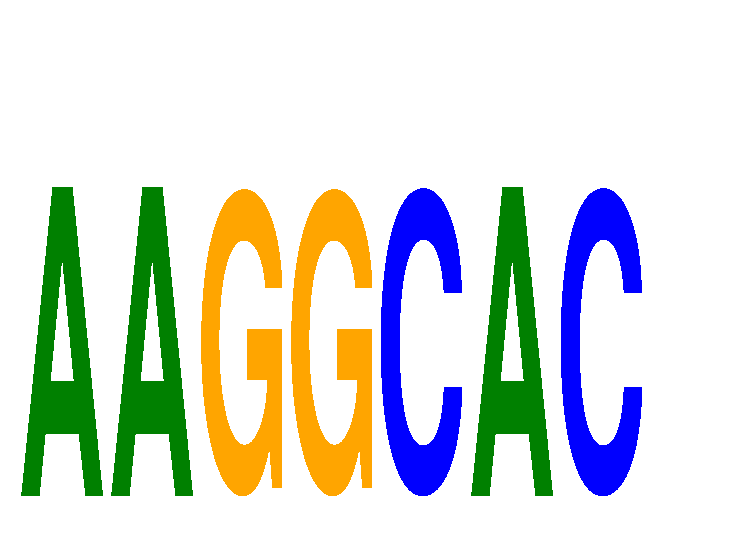Project
12D miR HR13_24
Navigation
Downloads
Results for AAGGCAC
Z-value: 1.86
miRNA associated with seed AAGGCAC
| Name | miRBASE accession |
|---|---|
|
mmu-miR-124-3p.1
|
MIMAT0000134 |
Activity profile of AAGGCAC motif
Sorted Z-values of AAGGCAC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_32619997 | 3.89 |
ENSMUST00000025833.6
|
Papss2
|
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr9_-_21312255 | 3.40 |
ENSMUST00000115433.3
ENSMUST00000003397.7 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr2_-_144331695 | 3.15 |
ENSMUST00000103171.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr11_+_61022560 | 3.03 |
ENSMUST00000089184.4
|
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr8_-_38661508 | 2.35 |
ENSMUST00000118896.1
|
Sgcz
|
sarcoglycan zeta |
| chr8_+_35375719 | 2.31 |
ENSMUST00000070481.6
|
Ppp1r3b
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B |
| chr12_-_11436607 | 2.30 |
ENSMUST00000072299.5
|
Vsnl1
|
visinin-like 1 |
| chr8_+_76899772 | 2.24 |
ENSMUST00000109913.2
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr14_-_54781886 | 2.11 |
ENSMUST00000022787.6
|
Slc7a8
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 |
| chr5_+_64970069 | 2.10 |
ENSMUST00000031080.8
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chr2_-_164443177 | 2.00 |
ENSMUST00000017153.3
|
Sdc4
|
syndecan 4 |
| chr10_-_83534130 | 2.00 |
ENSMUST00000020497.7
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr18_+_74442500 | 1.87 |
ENSMUST00000074157.6
|
Myo5b
|
myosin VB |
| chrX_+_103356464 | 1.87 |
ENSMUST00000116547.2
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr11_-_47379405 | 1.83 |
ENSMUST00000077221.5
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr5_+_66968416 | 1.82 |
ENSMUST00000038188.7
|
Limch1
|
LIM and calponin homology domains 1 |
| chr19_-_47464406 | 1.79 |
ENSMUST00000111800.2
ENSMUST00000081619.2 |
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr4_+_102254993 | 1.76 |
ENSMUST00000106908.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr14_-_110755100 | 1.73 |
ENSMUST00000078386.2
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr12_-_50649190 | 1.72 |
ENSMUST00000002765.7
|
Prkd1
|
protein kinase D1 |
| chr1_-_156204998 | 1.72 |
ENSMUST00000015628.3
|
Fam163a
|
family with sequence similarity 163, member A |
| chr18_-_61536522 | 1.71 |
ENSMUST00000171629.1
|
Arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr10_-_61147659 | 1.70 |
ENSMUST00000092498.5
ENSMUST00000137833.1 ENSMUST00000155919.1 |
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr6_+_97807014 | 1.69 |
ENSMUST00000043637.7
|
Mitf
|
microphthalmia-associated transcription factor |
| chr1_-_156674290 | 1.67 |
ENSMUST00000079625.4
|
Tor3a
|
torsin family 3, member A |
| chr12_-_4841583 | 1.65 |
ENSMUST00000020964.5
|
Fkbp1b
|
FK506 binding protein 1b |
| chr19_-_6015769 | 1.63 |
ENSMUST00000164843.1
|
Capn1
|
calpain 1 |
| chr10_+_99108135 | 1.62 |
ENSMUST00000161240.2
|
Galnt4
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 |
| chr5_+_141241490 | 1.62 |
ENSMUST00000085774.4
|
Sdk1
|
sidekick homolog 1 (chicken) |
| chr1_-_132139666 | 1.62 |
ENSMUST00000027697.5
|
Cdk18
|
cyclin-dependent kinase 18 |
| chr3_+_122729158 | 1.60 |
ENSMUST00000066728.5
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr11_+_87760533 | 1.60 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr13_+_38345716 | 1.57 |
ENSMUST00000171970.1
|
Bmp6
|
bone morphogenetic protein 6 |
| chr2_+_179442427 | 1.54 |
ENSMUST00000000314.6
|
Cdh4
|
cadherin 4 |
| chr14_+_55854115 | 1.53 |
ENSMUST00000168479.1
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr14_-_24245913 | 1.53 |
ENSMUST00000073687.6
ENSMUST00000090398.4 |
Dlg5
|
discs, large homolog 5 (Drosophila) |
| chr11_+_115187481 | 1.47 |
ENSMUST00000100235.2
ENSMUST00000061450.6 |
Tmem104
|
transmembrane protein 104 |
| chr15_+_25622525 | 1.47 |
ENSMUST00000110457.1
ENSMUST00000137601.1 |
Myo10
|
myosin X |
| chr9_-_77544870 | 1.44 |
ENSMUST00000183873.1
|
Lrrc1
|
leucine rich repeat containing 1 |
| chr12_-_104865076 | 1.44 |
ENSMUST00000109937.1
ENSMUST00000109936.1 |
Clmn
|
calmin |
| chr3_-_89387132 | 1.42 |
ENSMUST00000107433.1
|
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr2_-_152344009 | 1.42 |
ENSMUST00000040312.6
|
Trib3
|
tribbles homolog 3 (Drosophila) |
| chr6_+_108828633 | 1.42 |
ENSMUST00000089162.3
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr18_+_9212856 | 1.36 |
ENSMUST00000041080.5
|
Fzd8
|
frizzled homolog 8 (Drosophila) |
| chr3_+_89520152 | 1.36 |
ENSMUST00000000811.7
|
Kcnn3
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr3_-_58525867 | 1.36 |
ENSMUST00000029385.7
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr4_-_130275213 | 1.35 |
ENSMUST00000122374.1
|
Serinc2
|
serine incorporator 2 |
| chr11_-_109298121 | 1.31 |
ENSMUST00000020920.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr16_-_52454074 | 1.31 |
ENSMUST00000023312.7
|
Alcam
|
activated leukocyte cell adhesion molecule |
| chr18_-_10706688 | 1.29 |
ENSMUST00000002549.7
ENSMUST00000117726.1 ENSMUST00000117828.1 |
Abhd3
|
abhydrolase domain containing 3 |
| chr16_-_17132377 | 1.29 |
ENSMUST00000023453.7
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr3_-_122619663 | 1.29 |
ENSMUST00000162409.1
|
Fnbp1l
|
formin binding protein 1-like |
| chr8_-_27174623 | 1.28 |
ENSMUST00000033878.6
ENSMUST00000054212.6 |
Rab11fip1
|
RAB11 family interacting protein 1 (class I) |
| chr19_+_10688744 | 1.25 |
ENSMUST00000087951.5
|
Vps37c
|
vacuolar protein sorting 37C (yeast) |
| chr16_+_78301458 | 1.24 |
ENSMUST00000023572.7
|
Cxadr
|
coxsackie virus and adenovirus receptor |
| chrX_+_106027300 | 1.22 |
ENSMUST00000055941.6
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr6_-_137649211 | 1.22 |
ENSMUST00000134630.1
ENSMUST00000058210.6 ENSMUST00000111878.1 |
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr6_+_42245907 | 1.21 |
ENSMUST00000031897.5
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr12_-_86884808 | 1.21 |
ENSMUST00000038422.6
|
Irf2bpl
|
interferon regulatory factor 2 binding protein-like |
| chr10_-_62231208 | 1.20 |
ENSMUST00000047883.9
|
Tspan15
|
tetraspanin 15 |
| chr5_+_37050854 | 1.20 |
ENSMUST00000043794.4
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr5_+_137288273 | 1.20 |
ENSMUST00000024099.4
ENSMUST00000085934.3 |
Ache
|
acetylcholinesterase |
| chr11_+_75193783 | 1.20 |
ENSMUST00000102514.3
|
Rtn4rl1
|
reticulon 4 receptor-like 1 |
| chrX_-_106011874 | 1.20 |
ENSMUST00000033583.7
ENSMUST00000151689.1 |
Magt1
|
magnesium transporter 1 |
| chr15_-_71727815 | 1.20 |
ENSMUST00000022953.8
|
Fam135b
|
family with sequence similarity 135, member B |
| chr6_+_38381469 | 1.20 |
ENSMUST00000162554.1
ENSMUST00000161751.1 |
Ttc26
|
tetratricopeptide repeat domain 26 |
| chr17_+_17402672 | 1.19 |
ENSMUST00000115576.2
|
Lix1
|
limb expression 1 homolog (chicken) |
| chr12_+_44328882 | 1.19 |
ENSMUST00000020939.8
ENSMUST00000110748.2 |
Nrcam
|
neuron-glia-CAM-related cell adhesion molecule |
| chr10_+_41519493 | 1.18 |
ENSMUST00000019962.8
|
Cd164
|
CD164 antigen |
| chr1_+_167001417 | 1.18 |
ENSMUST00000165874.1
|
Fam78b
|
family with sequence similarity 78, member B |
| chr2_+_79635352 | 1.18 |
ENSMUST00000111785.2
|
Ssfa2
|
sperm specific antigen 2 |
| chr2_-_172043466 | 1.17 |
ENSMUST00000087950.3
|
Cbln4
|
cerebellin 4 precursor protein |
| chr12_-_4907705 | 1.17 |
ENSMUST00000020962.5
|
Ubxn2a
|
UBX domain protein 2A |
| chr4_+_140906344 | 1.16 |
ENSMUST00000030765.6
|
Padi2
|
peptidyl arginine deiminase, type II |
| chr1_-_172206775 | 1.16 |
ENSMUST00000013842.5
ENSMUST00000111247.1 |
Pea15a
|
phosphoprotein enriched in astrocytes 15A |
| chr3_-_141982224 | 1.15 |
ENSMUST00000029948.8
|
Bmpr1b
|
bone morphogenetic protein receptor, type 1B |
| chr2_+_155381808 | 1.12 |
ENSMUST00000043237.7
ENSMUST00000174685.1 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr1_-_52952834 | 1.12 |
ENSMUST00000050567.4
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr6_+_115774538 | 1.12 |
ENSMUST00000075995.5
|
Cand2
|
cullin-associated and neddylation-dissociated 2 (putative) |
| chr12_+_80790532 | 1.11 |
ENSMUST00000068519.5
|
4933426M11Rik
|
RIKEN cDNA 4933426M11 gene |
| chr15_-_67113909 | 1.11 |
ENSMUST00000092640.5
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
| chr16_+_84774123 | 1.10 |
ENSMUST00000114195.1
|
Jam2
|
junction adhesion molecule 2 |
| chr11_-_30025915 | 1.10 |
ENSMUST00000058902.5
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr3_-_85746266 | 1.08 |
ENSMUST00000118408.1
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr12_+_24831583 | 1.07 |
ENSMUST00000110942.3
ENSMUST00000078902.6 |
Mboat2
|
membrane bound O-acyltransferase domain containing 2 |
| chr1_+_74391479 | 1.06 |
ENSMUST00000027367.7
|
Ctdsp1
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
| chr9_+_77917364 | 1.06 |
ENSMUST00000034904.7
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr8_+_62951361 | 1.06 |
ENSMUST00000119068.1
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr2_-_144270852 | 1.06 |
ENSMUST00000110030.3
|
Snx5
|
sorting nexin 5 |
| chr11_-_69560186 | 1.05 |
ENSMUST00000004036.5
|
Efnb3
|
ephrin B3 |
| chr9_-_43239816 | 1.05 |
ENSMUST00000034512.5
|
Oaf
|
OAF homolog (Drosophila) |
| chr18_+_75820174 | 1.04 |
ENSMUST00000058997.7
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr5_-_69341699 | 1.03 |
ENSMUST00000054095.4
|
Kctd8
|
potassium channel tetramerisation domain containing 8 |
| chr10_-_83337845 | 1.03 |
ENSMUST00000039956.5
|
Slc41a2
|
solute carrier family 41, member 2 |
| chr3_-_41082992 | 1.02 |
ENSMUST00000058578.7
|
Pgrmc2
|
progesterone receptor membrane component 2 |
| chr3_-_131272077 | 1.01 |
ENSMUST00000029610.8
|
Hadh
|
hydroxyacyl-Coenzyme A dehydrogenase |
| chr5_-_115158169 | 1.01 |
ENSMUST00000053271.5
ENSMUST00000112121.1 |
Mlec
|
malectin |
| chr15_-_102257449 | 1.01 |
ENSMUST00000043172.8
|
Rarg
|
retinoic acid receptor, gamma |
| chr14_-_20480106 | 1.01 |
ENSMUST00000065504.9
ENSMUST00000100844.4 |
Anxa7
|
annexin A7 |
| chrX_+_41401304 | 1.00 |
ENSMUST00000076349.5
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr19_-_34879452 | 1.00 |
ENSMUST00000036584.5
|
Pank1
|
pantothenate kinase 1 |
| chr10_-_110000219 | 1.00 |
ENSMUST00000032719.7
|
Nav3
|
neuron navigator 3 |
| chr11_-_106160101 | 0.99 |
ENSMUST00000045923.3
|
Limd2
|
LIM domain containing 2 |
| chr17_+_5841307 | 0.99 |
ENSMUST00000002436.9
|
Snx9
|
sorting nexin 9 |
| chr19_+_11965817 | 0.98 |
ENSMUST00000025590.9
|
Osbp
|
oxysterol binding protein |
| chr5_+_130219706 | 0.98 |
ENSMUST00000065329.6
|
Tmem248
|
transmembrane protein 248 |
| chr1_-_69685937 | 0.97 |
ENSMUST00000027146.2
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr6_-_86397098 | 0.97 |
ENSMUST00000153723.1
ENSMUST00000032065.8 |
Pcyox1
|
prenylcysteine oxidase 1 |
| chr6_-_100287441 | 0.95 |
ENSMUST00000101118.2
|
Rybp
|
RING1 and YY1 binding protein |
| chr17_-_27204357 | 0.95 |
ENSMUST00000055117.7
|
Lemd2
|
LEM domain containing 2 |
| chr7_+_64287665 | 0.95 |
ENSMUST00000032736.4
|
Mtmr10
|
myotubularin related protein 10 |
| chr10_-_93311073 | 0.95 |
ENSMUST00000008542.5
|
Elk3
|
ELK3, member of ETS oncogene family |
| chr7_-_116443439 | 0.95 |
ENSMUST00000170430.1
|
Pik3c2a
|
phosphatidylinositol 3-kinase, C2 domain containing, alpha polypeptide |
| chr8_-_25201349 | 0.94 |
ENSMUST00000084512.4
ENSMUST00000084030.4 |
Tacc1
|
transforming, acidic coiled-coil containing protein 1 |
| chr2_-_26933781 | 0.94 |
ENSMUST00000154651.1
ENSMUST00000015011.3 |
Surf4
|
surfeit gene 4 |
| chr7_+_88430257 | 0.94 |
ENSMUST00000107256.2
|
Rab38
|
RAB38, member of RAS oncogene family |
| chr8_+_78509319 | 0.94 |
ENSMUST00000034111.8
|
Slc10a7
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 7 |
| chr10_-_77902467 | 0.94 |
ENSMUST00000057608.4
|
Lrrc3
|
leucine rich repeat containing 3 |
| chr5_-_113993873 | 0.94 |
ENSMUST00000159592.1
|
Ssh1
|
slingshot homolog 1 (Drosophila) |
| chr19_-_24477356 | 0.93 |
ENSMUST00000099556.1
|
Fam122a
|
family with sequence similarity 122, member A |
| chr11_+_57645417 | 0.93 |
ENSMUST00000066987.7
ENSMUST00000108846.1 |
Galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 |
| chr18_-_66002612 | 0.93 |
ENSMUST00000120461.1
ENSMUST00000048260.7 |
Lman1
|
lectin, mannose-binding, 1 |
| chr18_-_16809233 | 0.91 |
ENSMUST00000025166.7
|
Cdh2
|
cadherin 2 |
| chr12_-_85177288 | 0.91 |
ENSMUST00000004913.6
|
Pgf
|
placental growth factor |
| chrX_+_23693043 | 0.91 |
ENSMUST00000035766.6
ENSMUST00000101670.2 |
Wdr44
|
WD repeat domain 44 |
| chr14_-_47189406 | 0.91 |
ENSMUST00000089959.6
|
Gch1
|
GTP cyclohydrolase 1 |
| chr4_-_59549314 | 0.90 |
ENSMUST00000148331.2
ENSMUST00000030076.5 |
Ptbp3
|
polypyrimidine tract binding protein 3 |
| chr16_+_24393350 | 0.90 |
ENSMUST00000038053.6
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr2_-_93462386 | 0.90 |
ENSMUST00000123565.1
ENSMUST00000099696.1 |
Cd82
|
CD82 antigen |
| chr17_+_27057288 | 0.89 |
ENSMUST00000049308.8
|
Itpr3
|
inositol 1,4,5-triphosphate receptor 3 |
| chr1_+_166254095 | 0.89 |
ENSMUST00000111416.1
|
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr5_-_36484112 | 0.89 |
ENSMUST00000119916.1
ENSMUST00000031097.7 |
Tada2b
|
transcriptional adaptor 2B |
| chr8_+_61224162 | 0.89 |
ENSMUST00000034060.5
|
Sh3rf1
|
SH3 domain containing ring finger 1 |
| chr6_-_13839916 | 0.88 |
ENSMUST00000060442.7
|
Gpr85
|
G protein-coupled receptor 85 |
| chr18_+_84851338 | 0.88 |
ENSMUST00000160180.1
|
Cyb5
|
cytochrome b-5 |
| chr7_+_113513829 | 0.88 |
ENSMUST00000033018.8
|
Far1
|
fatty acyl CoA reductase 1 |
| chr9_+_56994932 | 0.87 |
ENSMUST00000034832.6
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr11_-_20332689 | 0.87 |
ENSMUST00000109594.1
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr13_+_96542727 | 0.87 |
ENSMUST00000077672.4
ENSMUST00000109444.2 |
Col4a3bp
|
collagen, type IV, alpha 3 (Goodpasture antigen) binding protein |
| chr1_-_161251153 | 0.86 |
ENSMUST00000051925.4
ENSMUST00000071718.5 |
Prdx6
|
peroxiredoxin 6 |
| chr17_-_84187939 | 0.86 |
ENSMUST00000060366.6
|
Zfp36l2
|
zinc finger protein 36, C3H type-like 2 |
| chr2_+_4400958 | 0.86 |
ENSMUST00000075767.7
|
Frmd4a
|
FERM domain containing 4A |
| chr10_-_7780866 | 0.85 |
ENSMUST00000124838.1
ENSMUST00000039763.7 |
Ginm1
|
glycoprotein integral membrane 1 |
| chr19_-_6840590 | 0.85 |
ENSMUST00000170516.2
ENSMUST00000025903.5 |
Rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr7_+_51879041 | 0.85 |
ENSMUST00000107591.2
|
Gas2
|
growth arrest specific 2 |
| chrX_+_159627265 | 0.85 |
ENSMUST00000112456.2
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr4_+_137862237 | 0.85 |
ENSMUST00000102518.3
|
Ece1
|
endothelin converting enzyme 1 |
| chr1_+_125561010 | 0.85 |
ENSMUST00000027580.4
|
Slc35f5
|
solute carrier family 35, member F5 |
| chr14_-_78308031 | 0.83 |
ENSMUST00000022592.7
|
Tnfsf11
|
tumor necrosis factor (ligand) superfamily, member 11 |
| chr18_-_37935429 | 0.83 |
ENSMUST00000115634.1
|
Diap1
|
diaphanous homolog 1 (Drosophila) |
| chr19_+_3323301 | 0.82 |
ENSMUST00000025835.4
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr9_-_107635330 | 0.82 |
ENSMUST00000055704.6
|
Gnai2
|
guanine nucleotide binding protein (G protein), alpha inhibiting 2 |
| chr12_+_80644212 | 0.82 |
ENSMUST00000085245.5
|
Slc39a9
|
solute carrier family 39 (zinc transporter), member 9 |
| chr19_+_7494033 | 0.80 |
ENSMUST00000170373.1
|
Atl3
|
atlastin GTPase 3 |
| chr12_-_85374696 | 0.79 |
ENSMUST00000040766.7
|
Tmed10
|
transmembrane emp24-like trafficking protein 10 (yeast) |
| chr1_-_182517447 | 0.78 |
ENSMUST00000068505.8
|
Capn2
|
calpain 2 |
| chr11_-_61855026 | 0.78 |
ENSMUST00000004920.3
|
Ulk2
|
unc-51 like kinase 2 |
| chr16_-_17928136 | 0.77 |
ENSMUST00000003622.8
|
Slc25a1
|
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
| chr14_+_25842146 | 0.77 |
ENSMUST00000022416.8
|
Anxa11
|
annexin A11 |
| chr3_+_59006978 | 0.77 |
ENSMUST00000040325.7
ENSMUST00000164225.1 ENSMUST00000040846.8 ENSMUST00000029393.8 |
Med12l
|
mediator of RNA polymerase II transcription, subunit 12 homolog (yeast)-like |
| chr2_+_35282380 | 0.77 |
ENSMUST00000028239.6
|
Gsn
|
gelsolin |
| chr18_+_38418946 | 0.77 |
ENSMUST00000025293.3
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr17_+_24488773 | 0.77 |
ENSMUST00000024958.7
|
Caskin1
|
CASK interacting protein 1 |
| chr12_+_32378692 | 0.77 |
ENSMUST00000172332.2
|
Ccdc71l
|
coiled-coil domain containing 71 like |
| chr8_+_13287887 | 0.77 |
ENSMUST00000045229.5
|
Tmco3
|
transmembrane and coiled-coil domains 3 |
| chr6_+_22875496 | 0.76 |
ENSMUST00000090568.3
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z, polypeptide 1 |
| chr11_+_49794157 | 0.76 |
ENSMUST00000020629.4
|
Gfpt2
|
glutamine fructose-6-phosphate transaminase 2 |
| chr10_-_78244602 | 0.76 |
ENSMUST00000000384.6
|
Trappc10
|
trafficking protein particle complex 10 |
| chr13_-_53377355 | 0.76 |
ENSMUST00000021920.6
|
Sptlc1
|
serine palmitoyltransferase, long chain base subunit 1 |
| chr9_+_111118070 | 0.76 |
ENSMUST00000035078.6
ENSMUST00000098340.2 |
Lrrfip2
|
leucine rich repeat (in FLII) interacting protein 2 |
| chr4_-_109476666 | 0.75 |
ENSMUST00000030284.3
|
Rnf11
|
ring finger protein 11 |
| chr9_-_14614949 | 0.74 |
ENSMUST00000013220.6
ENSMUST00000160770.1 |
Amotl1
|
angiomotin-like 1 |
| chr5_+_30588078 | 0.74 |
ENSMUST00000066295.2
|
Kcnk3
|
potassium channel, subfamily K, member 3 |
| chr12_+_32953874 | 0.74 |
ENSMUST00000076698.5
|
Sypl
|
synaptophysin-like protein |
| chr3_-_116007399 | 0.73 |
ENSMUST00000067485.3
|
Slc30a7
|
solute carrier family 30 (zinc transporter), member 7 |
| chr6_-_126645784 | 0.73 |
ENSMUST00000055168.3
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr17_-_44105774 | 0.73 |
ENSMUST00000024757.7
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr9_+_108808356 | 0.73 |
ENSMUST00000035218.7
|
Nckipsd
|
NCK interacting protein with SH3 domain |
| chr16_-_64771146 | 0.73 |
ENSMUST00000076991.6
|
4930453N24Rik
|
RIKEN cDNA 4930453N24 gene |
| chr6_-_118479237 | 0.73 |
ENSMUST00000161170.1
|
Zfp9
|
zinc finger protein 9 |
| chr5_+_124629050 | 0.72 |
ENSMUST00000037865.8
|
Atp6v0a2
|
ATPase, H+ transporting, lysosomal V0 subunit A2 |
| chr2_+_32876114 | 0.72 |
ENSMUST00000028135.8
|
Fam129b
|
family with sequence similarity 129, member B |
| chr2_+_91202885 | 0.71 |
ENSMUST00000150403.1
ENSMUST00000002172.7 ENSMUST00000155418.1 |
Acp2
|
acid phosphatase 2, lysosomal |
| chr9_+_118926453 | 0.71 |
ENSMUST00000073109.5
|
Ctdspl
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase-like |
| chr17_+_25188380 | 0.71 |
ENSMUST00000039734.5
|
Unkl
|
unkempt-like (Drosophila) |
| chr10_-_127211528 | 0.71 |
ENSMUST00000013970.7
|
Pip4k2c
|
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr18_-_80713062 | 0.71 |
ENSMUST00000170905.1
ENSMUST00000078049.4 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr2_+_180710117 | 0.71 |
ENSMUST00000029090.2
|
Gid8
|
GID complex subunit 8 homolog (S. cerevisiae) |
| chr6_-_38876163 | 0.70 |
ENSMUST00000161779.1
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr8_+_75033673 | 0.69 |
ENSMUST00000078847.5
ENSMUST00000165630.1 |
Tom1
|
target of myb1 homolog (chicken) |
| chr2_-_30415767 | 0.69 |
ENSMUST00000102855.1
ENSMUST00000028207.6 |
Crat
|
carnitine acetyltransferase |
| chr10_+_79854618 | 0.69 |
ENSMUST00000165704.1
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr12_+_108792946 | 0.68 |
ENSMUST00000021692.7
|
Yy1
|
YY1 transcription factor |
| chrX_+_99136119 | 0.68 |
ENSMUST00000052839.6
|
Efnb1
|
ephrin B1 |
| chr7_-_81454751 | 0.68 |
ENSMUST00000098331.3
ENSMUST00000178892.1 |
Cpeb1
|
cytoplasmic polyadenylation element binding protein 1 |
| chrX_+_139610612 | 0.68 |
ENSMUST00000113026.1
|
Rnf128
|
ring finger protein 128 |
| chr9_-_75611308 | 0.68 |
ENSMUST00000064433.3
|
Tmod2
|
tropomodulin 2 |
| chr2_-_132815978 | 0.67 |
ENSMUST00000039554.6
|
Trmt6
|
tRNA methyltransferase 6 |
| chr7_+_73740277 | 0.67 |
ENSMUST00000107456.2
|
Fam174b
|
family with sequence similarity 174, member B |
| chr6_+_7844806 | 0.67 |
ENSMUST00000040159.4
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGCAC
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.7 | 2.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.7 | 2.0 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.5 | 1.4 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.5 | 1.4 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.4 | 1.3 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.4 | 1.2 | GO:1904732 | elastin biosynthetic process(GO:0051542) regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.4 | 1.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.4 | 1.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.4 | 2.0 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.4 | 1.5 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.4 | 1.1 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.4 | 2.9 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.4 | 1.4 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.4 | 0.4 | GO:0038189 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) |
| 0.3 | 1.4 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.3 | 0.9 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) regulation of zinc ion transport(GO:0071579) |
| 0.3 | 0.9 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.3 | 1.8 | GO:0032439 | endosome localization(GO:0032439) |
| 0.3 | 1.2 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.3 | 1.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.3 | 0.9 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.3 | 1.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) negative regulation of lymphocyte migration(GO:2000402) |
| 0.3 | 0.9 | GO:0035627 | ER to Golgi ceramide transport(GO:0035621) ceramide transport(GO:0035627) |
| 0.3 | 0.9 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.3 | 0.8 | GO:0034959 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.3 | 1.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.3 | 1.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.3 | 0.8 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.3 | 1.6 | GO:0032349 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) |
| 0.3 | 0.8 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.3 | 3.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.3 | 0.8 | GO:1900149 | positive regulation of Schwann cell migration(GO:1900149) |
| 0.3 | 1.0 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.3 | 1.0 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.2 | 1.7 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.2 | 0.2 | GO:1901963 | canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cell proliferation involved in outflow tract morphogenesis(GO:1901963) |
| 0.2 | 1.2 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.2 | 1.0 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 0.7 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.2 | 1.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.2 | 1.6 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.2 | 0.9 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.2 | 1.3 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.2 | 0.7 | GO:0043134 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) positive regulation of relaxation of muscle(GO:1901079) |
| 0.2 | 0.7 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.2 | 0.9 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.2 | 1.5 | GO:0072205 | negative regulation of hippo signaling(GO:0035331) protein localization to adherens junction(GO:0071896) metanephric collecting duct development(GO:0072205) |
| 0.2 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.2 | 0.8 | GO:0048294 | negative regulation of isotype switching to IgE isotypes(GO:0048294) |
| 0.2 | 0.6 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.2 | 1.0 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.2 | 1.3 | GO:1904222 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.2 | 1.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.6 | GO:1903774 | positive regulation of viral budding via host ESCRT complex(GO:1903774) |
| 0.2 | 0.7 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.2 | 0.9 | GO:0060754 | positive regulation of mast cell chemotaxis(GO:0060754) |
| 0.2 | 0.5 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.2 | 0.5 | GO:0061738 | late endosomal microautophagy(GO:0061738) regulation of centriole elongation(GO:1903722) |
| 0.2 | 0.5 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.2 | 2.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.2 | 0.5 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.2 | 0.5 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.2 | 0.7 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.2 | 0.5 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.2 | 0.5 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.2 | 0.6 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.2 | 0.9 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.2 | 0.5 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.2 | 0.5 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.1 | 1.0 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.1 | 0.4 | GO:0018008 | N-terminal peptidyl-glycine N-myristoylation(GO:0018008) |
| 0.1 | 0.4 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.1 | 0.9 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) regulation of terminal button organization(GO:2000331) |
| 0.1 | 1.6 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 2.2 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.1 | 0.9 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.3 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.1 | 0.6 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.1 | 0.6 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.1 | 0.3 | GO:1902263 | apoptotic process involved in embryonic digit morphogenesis(GO:1902263) |
| 0.1 | 0.7 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.1 | 1.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.5 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.1 | 1.7 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.8 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.1 | 0.5 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.1 | 0.8 | GO:0070197 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.4 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.1 | 1.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.1 | 0.5 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.1 | 0.5 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.1 | 0.6 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.1 | 0.7 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.1 | 0.9 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.5 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.1 | 0.3 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.6 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.1 | 0.3 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 0.7 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.6 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 1.2 | GO:0031468 | nuclear envelope reassembly(GO:0031468) |
| 0.1 | 0.3 | GO:2000630 | positive regulation of miRNA metabolic process(GO:2000630) |
| 0.1 | 0.5 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.1 | 0.6 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.1 | 0.8 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.1 | 0.6 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.1 | 0.4 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.1 | 0.6 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.6 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.5 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.1 | 0.1 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.1 | 0.3 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.6 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.1 | 0.7 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 1.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.1 | 0.5 | GO:1902261 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.8 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 1.7 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.2 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) positive regulation of protein lipidation(GO:1903061) |
| 0.1 | 1.4 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.1 | 0.8 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.1 | 0.3 | GO:0009197 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.1 | 0.4 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.7 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.1 | 1.3 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.7 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.5 | GO:1903689 | regulation of wound healing, spreading of epidermal cells(GO:1903689) |
| 0.1 | 0.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.4 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.1 | 0.7 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.1 | 0.7 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 2.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 1.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 0.8 | GO:0075071 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.1 | 0.9 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 1.2 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.1 | 0.9 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.4 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 0.4 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.5 | GO:0043201 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.1 | 0.8 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.1 | 1.2 | GO:0045136 | development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.9 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 1.0 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.5 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.2 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.4 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.3 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.1 | 0.8 | GO:0032274 | gonadotropin secretion(GO:0032274) |
| 0.1 | 0.5 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.1 | 0.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 2.3 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.1 | 0.1 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.1 | 0.7 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 0.3 | GO:0009816 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 0.3 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.1 | 0.3 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 0.1 | GO:0060535 | trachea cartilage morphogenesis(GO:0060535) |
| 0.1 | 0.3 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.2 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.8 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.1 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.1 | 1.8 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.1 | 0.4 | GO:0006621 | protein retention in ER lumen(GO:0006621) maintenance of protein localization in endoplasmic reticulum(GO:0035437) |
| 0.1 | 1.2 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.1 | 0.2 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.1 | 3.2 | GO:0046676 | negative regulation of insulin secretion(GO:0046676) |
| 0.1 | 0.2 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.1 | 0.3 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.4 | GO:0039702 | viral budding via host ESCRT complex(GO:0039702) |
| 0.1 | 0.6 | GO:0015791 | polyol transport(GO:0015791) |
| 0.1 | 0.5 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.4 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.1 | 0.2 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.1 | 0.2 | GO:0048289 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.1 | 0.5 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.3 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.1 | 0.6 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.1 | 1.0 | GO:0044126 | regulation of growth of symbiont in host(GO:0044126) |
| 0.1 | 0.5 | GO:0002441 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.1 | 0.5 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 1.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.7 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0014870 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.0 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.0 | 0.3 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.3 | GO:0071397 | cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.1 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.2 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.0 | 0.5 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.3 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.3 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.5 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 0.4 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.2 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.2 | GO:0046208 | spermine catabolic process(GO:0046208) |
| 0.0 | 0.6 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) optic placode formation involved in camera-type eye formation(GO:0046619) |
| 0.0 | 1.6 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.2 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 1.3 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.9 | GO:0098869 | hydrogen peroxide catabolic process(GO:0042744) cellular oxidant detoxification(GO:0098869) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.5 | GO:0006622 | protein targeting to lysosome(GO:0006622) |
| 0.0 | 0.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 1.0 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.3 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 1.6 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.8 | GO:0006829 | zinc II ion transport(GO:0006829) |
| 0.0 | 0.7 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.2 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 1.9 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.3 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 0.8 | GO:1903077 | negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.7 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.7 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 2.9 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.5 | GO:0048490 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.2 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.0 | 0.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 1.5 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.0 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of nitric oxide mediated signal transduction(GO:0010751) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.0 | 0.9 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.4 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0015874 | norepinephrine transport(GO:0015874) |
| 0.0 | 0.6 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.1 | GO:0051127 | positive regulation of actin nucleation(GO:0051127) |
| 0.0 | 0.5 | GO:0086012 | membrane depolarization during cardiac muscle cell action potential(GO:0086012) |
| 0.0 | 0.1 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.5 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.2 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.8 | GO:0046470 | phosphatidylcholine metabolic process(GO:0046470) |
| 0.0 | 1.0 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:1902254 | negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902254) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.1 | GO:0072720 | response to sorbitol(GO:0072708) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.2 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.2 | GO:0044247 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.8 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.6 | GO:0038066 | p38MAPK cascade(GO:0038066) |
| 0.0 | 0.5 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.7 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.1 | GO:0010980 | regulation of vitamin D 24-hydroxylase activity(GO:0010979) positive regulation of vitamin D 24-hydroxylase activity(GO:0010980) bile acid signaling pathway(GO:0038183) |
| 0.0 | 0.4 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.6 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.2 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.2 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.0 | 0.2 | GO:0031507 | heterochromatin assembly(GO:0031507) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0042795 | snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.3 | GO:1901186 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) positive regulation of ERBB signaling pathway(GO:1901186) |
| 0.0 | 0.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
| 0.0 | 0.3 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.5 | GO:0030835 | negative regulation of actin filament depolymerization(GO:0030835) |
| 0.0 | 0.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.2 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.2 | GO:0030033 | microvillus assembly(GO:0030033) |
| 0.0 | 0.2 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.0 | 0.8 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.2 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:2000773 | negative regulation of cellular senescence(GO:2000773) |
| 0.0 | 0.2 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.4 | GO:0018279 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.5 | GO:0000045 | autophagosome assembly(GO:0000045) |
| 0.0 | 0.0 | GO:0045924 | regulation of female receptivity(GO:0045924) |
| 0.0 | 0.4 | GO:0006336 | DNA replication-independent nucleosome assembly(GO:0006336) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:1900086 | positive regulation of peptidyl-tyrosine autophosphorylation(GO:1900086) |
| 0.0 | 0.1 | GO:0032264 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 4.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 1.6 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.3 | 1.0 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.3 | 1.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 2.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.2 | 1.8 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.2 | 0.8 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.2 | 0.5 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.2 | 0.5 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.2 | 0.9 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 2.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 0.3 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.7 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 1.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 1.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 1.2 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.4 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.1 | 0.6 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.5 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 4.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 1.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.8 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.1 | 1.0 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.1 | 0.7 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 1.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.7 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.1 | 0.8 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.1 | 1.8 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.8 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 1.0 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.1 | 0.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.7 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 1.8 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.5 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.1 | 1.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.1 | 1.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.5 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.1 | 1.4 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 1.5 | GO:0034385 | very-low-density lipoprotein particle(GO:0034361) triglyceride-rich lipoprotein particle(GO:0034385) |
| 0.1 | 0.5 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 0.3 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.1 | 0.8 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.7 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.3 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.6 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.1 | 0.4 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.9 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.3 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.3 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.8 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.8 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 1.5 | GO:0031231 | intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 2.1 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.5 | GO:0044754 | autolysosome(GO:0044754) |
| 0.0 | 0.7 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.8 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 2.1 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.5 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 1.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.9 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.9 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.3 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 1.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 1.6 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 1.2 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.5 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.2 | GO:0036396 | MIS complex(GO:0036396) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 1.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.5 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.6 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 1.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.4 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.8 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.7 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 2.7 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.5 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.4 | GO:0032994 | protein-lipid complex(GO:0032994) plasma lipoprotein particle(GO:0034358) lipoprotein particle(GO:1990777) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 1.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.1 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.5 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.6 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.5 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 2.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.8 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.7 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 1.0 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 2.0 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.2 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.4 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.6 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 0.3 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.2 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 4.4 | GO:0000323 | lytic vacuole(GO:0000323) lysosome(GO:0005764) |
| 0.0 | 2.1 | GO:0019898 | extrinsic component of membrane(GO:0019898) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.7 | 2.0 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.4 | 1.3 | GO:0052739 | phosphatidylserine 1-acylhydrolase activity(GO:0052739) 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.3 | 1.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.3 | 2.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.3 | 0.9 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.3 | 1.3 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.3 | 0.9 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.3 | 1.2 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.3 | 1.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.3 | 1.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.3 | 0.6 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.3 | 1.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.3 | 1.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 2.4 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.3 | 1.3 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.3 | 0.8 | GO:0015142 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.3 | 0.8 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.2 | 2.5 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.2 | 0.9 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 1.8 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 0.7 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.2 | 0.9 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.2 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 0.5 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.2 | 0.7 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.2 | 2.2 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.2 | 0.5 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.2 | 0.7 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.2 | 0.5 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.2 | 0.8 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.2 | 0.7 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.2 | 1.4 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.2 | 0.5 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.2 | 0.9 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.1 | 0.4 | GO:0019107 | glycylpeptide N-tetradecanoyltransferase activity(GO:0004379) myristoyltransferase activity(GO:0019107) |
| 0.1 | 1.7 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 2.9 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 0.4 | GO:0034188 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.1 | 0.7 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 1.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.5 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 1.0 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.1 | 0.4 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 1.5 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 3.8 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 1.7 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 1.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.8 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.1 | 1.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.1 | 0.3 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.1 | 1.5 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.1 | 0.3 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.4 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.1 | 1.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.6 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 1.0 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.1 | 0.7 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.5 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.1 | 0.6 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.1 | 2.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.2 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.1 | 0.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.1 | 1.1 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 1.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.1 | 0.4 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.4 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.5 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.9 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 2.8 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.6 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.1 | 0.3 | GO:0001888 | glucuronyl-galactosyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0001888) |
| 0.1 | 0.8 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.1 | 0.5 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.1 | 0.6 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.7 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 2.0 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.2 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 0.9 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 1.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 0.4 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.1 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 1.4 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.1 | 0.4 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.1 | 0.4 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.1 | 0.7 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) |
| 0.1 | 0.3 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.1 | 0.5 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.1 | 0.7 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.1 | 0.5 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 1.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.6 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.1 | 2.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.1 | 1.2 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.1 | 0.4 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 1.6 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.1 | 0.3 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.1 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.5 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 2.1 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.1 | 0.4 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.2 | GO:0052901 | polyamine oxidase activity(GO:0046592) spermine:oxygen oxidoreductase (spermidine-forming) activity(GO:0052901) |
| 0.1 | 0.6 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.2 | GO:0086007 | voltage-gated calcium channel activity involved in cardiac muscle cell action potential(GO:0086007) |
| 0.1 | 0.7 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.1 | 1.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.8 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 3.1 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.1 | 0.7 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 0.3 | GO:0019798 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.3 | GO:0033142 | progesterone receptor binding(GO:0033142) |
| 0.1 | 0.3 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.1 | 1.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.1 | 0.3 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.8 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.1 | 0.3 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.1 | 0.9 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.2 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.4 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.0 | 1.1 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.8 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 1.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.9 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.3 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.7 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.5 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 1.9 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.6 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.6 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.8 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.3 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 1.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 1.4 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.4 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.5 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.7 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.5 | GO:0048038 | quinone binding(GO:0048038) |
| 0.0 | 2.2 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.6 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.7 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 2.2 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 1.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:1902121 | lithocholic acid binding(GO:1902121) D3 vitamins binding(GO:1902271) |
| 0.0 | 0.0 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.6 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.5 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.1 | GO:0003945 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.6 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.5 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.5 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.9 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.5 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.6 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 1.4 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.5 | GO:0030145 | manganese ion binding(GO:0030145) |
| 0.0 | 1.0 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 1.2 | GO:0043130 | ubiquitin binding(GO:0043130) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.5 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.9 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 5.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.1 | 0.8 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 0.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.1 | 1.6 | ST ADRENERGIC | Adrenergic Pathway |
| 0.1 | 1.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 2.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.6 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 1.5 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.7 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.7 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.4 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.7 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 1.3 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 2.3 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.6 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.5 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.8 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 1.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 1.8 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.6 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.5 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.6 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.6 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.7 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.9 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 3.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.2 | 2.4 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.2 | 7.8 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.2 | 1.8 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.1 | 0.6 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.1 | 0.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.1 | 1.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.9 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.1 | 1.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.1 | 1.1 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 0.4 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.1 | 0.4 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.1 | 1.9 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 1.1 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 2.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.9 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.5 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.1 | 1.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.9 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.1 | 0.5 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.1 | 1.8 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.8 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.1 | 1.7 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.2 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.0 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 1.3 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |
| 0.0 | 1.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.7 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.8 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 1.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 2.0 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.9 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 1.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.4 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.7 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.9 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.7 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 1.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 2.6 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.9 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 1.1 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.5 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 0.6 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.3 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.6 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.5 | REACTOME UNFOLDED PROTEIN RESPONSE | Genes involved in Unfolded Protein Response |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.1 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |