Project
12D miR HR13_24
Navigation
Downloads
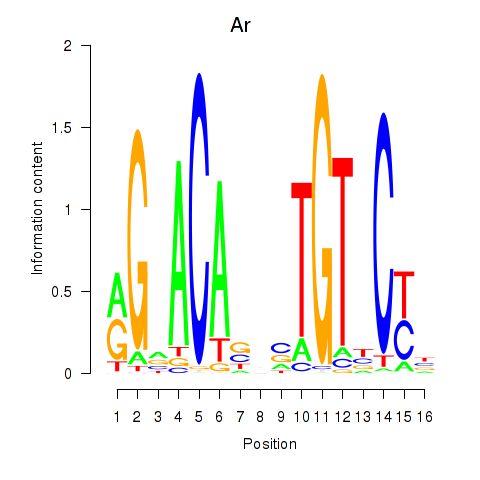
Results for Ar
Z-value: 1.05
Transcription factors associated with Ar
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ar
|
ENSMUSG00000046532.7 | androgen receptor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ar | mm10_v2_chrX_+_98149666_98149721 | 0.43 | 1.9e-01 | Click! |
Activity profile of Ar motif
Sorted Z-values of Ar motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_5455467 | 1.20 |
ENSMUST00000180867.1
|
Gm3194
|
predicted gene 3194 |
| chr14_-_7090812 | 1.17 |
ENSMUST00000179898.1
|
Gm3696
|
predicted gene 3696 |
| chr2_+_119167758 | 1.05 |
ENSMUST00000057454.3
|
Gchfr
|
GTP cyclohydrolase I feedback regulator |
| chr14_+_65806066 | 1.04 |
ENSMUST00000139644.1
|
Pbk
|
PDZ binding kinase |
| chr7_-_97417730 | 1.02 |
ENSMUST00000043077.7
|
Thrsp
|
thyroid hormone responsive |
| chr14_-_6742332 | 0.92 |
ENSMUST00000163850.1
|
Gm3636
|
predicted gene 3636 |
| chr14_+_4023941 | 0.91 |
ENSMUST00000096184.4
|
Gm5796
|
predicted gene 5796 |
| chr14_+_3688783 | 0.88 |
ENSMUST00000170639.1
|
Gm3043
|
predicted gene 3043 |
| chr14_-_6875155 | 0.86 |
ENSMUST00000170465.2
|
Gm3636
|
predicted gene 3636 |
| chr8_-_13254154 | 0.83 |
ENSMUST00000033825.4
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr18_+_33794882 | 0.83 |
ENSMUST00000146010.2
|
2410004N09Rik
|
RIKEN cDNA 2410004N09 gene |
| chr5_+_130257029 | 0.81 |
ENSMUST00000100662.3
ENSMUST00000040213.6 |
Tyw1
|
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chr16_-_55895279 | 0.80 |
ENSMUST00000099705.3
|
Nxpe3
|
neurexophilin and PC-esterase domain family, member 3 |
| chr10_+_26822560 | 0.78 |
ENSMUST00000135866.1
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr14_+_4339563 | 0.76 |
ENSMUST00000112778.3
|
2610042L04Rik
|
RIKEN cDNA 2610042L04 gene |
| chr13_+_99100698 | 0.76 |
ENSMUST00000181742.1
|
Gm807
|
predicted gene 807 |
| chr14_-_6287250 | 0.75 |
ENSMUST00000170104.2
|
Gm3411
|
predicted gene 3411 |
| chr14_-_5389049 | 0.74 |
ENSMUST00000177986.1
|
Gm3500
|
predicted gene 3500 |
| chr11_-_61453992 | 0.73 |
ENSMUST00000060255.7
ENSMUST00000054927.7 ENSMUST00000102661.3 |
Rnf112
|
ring finger protein 112 |
| chr14_-_5807958 | 0.73 |
ENSMUST00000112758.3
ENSMUST00000096171.5 |
Gm3383
|
predicted gene 3383 |
| chr7_-_44974781 | 0.71 |
ENSMUST00000063761.7
|
Cpt1c
|
carnitine palmitoyltransferase 1c |
| chr7_+_102702331 | 0.67 |
ENSMUST00000094124.3
|
Olfr558
|
olfactory receptor 558 |
| chr14_-_7100621 | 0.67 |
ENSMUST00000167923.1
|
Gm3696
|
predicted gene 3696 |
| chr14_+_55618023 | 0.66 |
ENSMUST00000002395.7
|
Rec8
|
REC8 homolog (yeast) |
| chr14_-_7022599 | 0.64 |
ENSMUST00000100895.3
|
Gm10406
|
predicted gene 10406 |
| chr3_-_113574242 | 0.63 |
ENSMUST00000142505.2
|
Amy1
|
amylase 1, salivary |
| chr14_-_7332395 | 0.62 |
ENSMUST00000100886.3
|
Gm5797
|
predicted gene 5797 |
| chr1_+_131797381 | 0.61 |
ENSMUST00000112393.2
ENSMUST00000048660.5 |
Pm20d1
|
peptidase M20 domain containing 1 |
| chr14_+_5050629 | 0.60 |
ENSMUST00000112770.2
|
4930555G01Rik
|
RIKEN cDNA 4930555G01 gene |
| chr2_-_129297205 | 0.60 |
ENSMUST00000052708.6
|
Ckap2l
|
cytoskeleton associated protein 2-like |
| chr3_-_113574758 | 0.58 |
ENSMUST00000106540.1
|
Amy1
|
amylase 1, salivary |
| chr10_+_75566257 | 0.58 |
ENSMUST00000129232.1
ENSMUST00000143792.1 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr14_+_4415448 | 0.58 |
ENSMUST00000168866.1
|
Gm3164
|
predicted gene 3164 |
| chr14_-_5741577 | 0.58 |
ENSMUST00000177556.1
|
Gm3373
|
predicted gene 3373 |
| chr14_-_5373401 | 0.58 |
ENSMUST00000178058.1
|
Gm3500
|
predicted gene 3500 |
| chr18_+_33794915 | 0.57 |
ENSMUST00000179138.1
|
2410004N09Rik
|
RIKEN cDNA 2410004N09 gene |
| chr2_+_152962485 | 0.57 |
ENSMUST00000099197.2
ENSMUST00000103155.3 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr3_-_113577743 | 0.56 |
ENSMUST00000067980.5
|
Amy1
|
amylase 1, salivary |
| chr6_+_91156772 | 0.55 |
ENSMUST00000143621.1
|
Hdac11
|
histone deacetylase 11 |
| chr15_-_85821733 | 0.55 |
ENSMUST00000064370.4
|
Pkdrej
|
polycystic kidney disease (polycystin) and REJ (sperm receptor for egg jelly homolog, sea urchin) |
| chr17_+_33810515 | 0.53 |
ENSMUST00000048560.4
ENSMUST00000172649.1 ENSMUST00000173789.1 |
Kank3
|
KN motif and ankyrin repeat domains 3 |
| chr7_+_28982832 | 0.53 |
ENSMUST00000085835.6
|
Map4k1
|
mitogen-activated protein kinase kinase kinase kinase 1 |
| chr1_+_170644523 | 0.53 |
ENSMUST00000046792.8
|
Olfml2b
|
olfactomedin-like 2B |
| chr17_-_36642661 | 0.52 |
ENSMUST00000087165.4
ENSMUST00000087167.4 |
H2-M9
|
histocompatibility 2, M region locus 9 |
| chrX_+_159459125 | 0.52 |
ENSMUST00000043151.5
ENSMUST00000112470.1 ENSMUST00000156172.1 |
Map7d2
|
MAP7 domain containing 2 |
| chr11_-_87108656 | 0.52 |
ENSMUST00000051395.8
|
Prr11
|
proline rich 11 |
| chr14_+_3224440 | 0.51 |
ENSMUST00000112797.4
|
D830030K20Rik
|
RIKEN cDNA D830030K20 gene |
| chr2_+_55435918 | 0.51 |
ENSMUST00000067101.3
|
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr13_-_67306412 | 0.49 |
ENSMUST00000049705.7
|
Zfp457
|
zinc finger protein 457 |
| chr2_-_91963507 | 0.49 |
ENSMUST00000028667.3
|
Dgkz
|
diacylglycerol kinase zeta |
| chr10_+_41810528 | 0.48 |
ENSMUST00000099931.3
|
Sesn1
|
sestrin 1 |
| chr14_-_118923070 | 0.48 |
ENSMUST00000047208.5
|
Dzip1
|
DAZ interacting protein 1 |
| chr14_+_3395093 | 0.47 |
ENSMUST00000112728.2
ENSMUST00000177913.1 |
Gm10413
|
predicted gene 10413 |
| chr14_+_3634512 | 0.47 |
ENSMUST00000112802.2
ENSMUST00000179486.1 |
Gm3012
|
predicted gene 3012 |
| chr9_+_73101836 | 0.46 |
ENSMUST00000172578.1
|
Khdc3
|
KH domain containing 3, subcortical maternal complex member |
| chr6_+_91157373 | 0.46 |
ENSMUST00000155007.1
|
Hdac11
|
histone deacetylase 11 |
| chr14_-_5725943 | 0.46 |
ENSMUST00000169087.2
|
Gm3373
|
predicted gene 3373 |
| chr7_+_12922290 | 0.46 |
ENSMUST00000108539.1
ENSMUST00000004554.7 ENSMUST00000147435.1 ENSMUST00000137329.1 |
Rps5
|
ribosomal protein S5 |
| chr6_-_116461024 | 0.46 |
ENSMUST00000164547.1
ENSMUST00000170186.1 |
Alox5
|
arachidonate 5-lipoxygenase |
| chr2_+_3704787 | 0.45 |
ENSMUST00000115054.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr6_-_69243445 | 0.45 |
ENSMUST00000101325.3
|
Igkv4-71
|
immunoglobulin kappa chain variable 4-71 |
| chr13_+_108316332 | 0.44 |
ENSMUST00000051594.5
|
Depdc1b
|
DEP domain containing 1B |
| chrX_+_134308084 | 0.41 |
ENSMUST00000081064.5
ENSMUST00000101251.1 ENSMUST00000129782.1 |
Cenpi
|
centromere protein I |
| chr7_-_126676357 | 0.41 |
ENSMUST00000106371.1
ENSMUST00000106372.3 ENSMUST00000155419.1 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr1_-_180193653 | 0.41 |
ENSMUST00000159914.1
|
Adck3
|
aarF domain containing kinase 3 |
| chr8_-_13254096 | 0.41 |
ENSMUST00000171619.1
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr4_+_104913456 | 0.40 |
ENSMUST00000106803.2
ENSMUST00000106804.1 |
1700024P16Rik
|
RIKEN cDNA 1700024P16 gene |
| chr2_-_28563362 | 0.40 |
ENSMUST00000028161.5
|
Cel
|
carboxyl ester lipase |
| chr6_+_15196949 | 0.40 |
ENSMUST00000151301.1
ENSMUST00000131414.1 ENSMUST00000140557.1 ENSMUST00000115469.1 |
Foxp2
|
forkhead box P2 |
| chr4_+_9269285 | 0.39 |
ENSMUST00000038841.7
|
Clvs1
|
clavesin 1 |
| chr6_+_91156665 | 0.39 |
ENSMUST00000041736.4
|
Hdac11
|
histone deacetylase 11 |
| chr1_+_172698046 | 0.37 |
ENSMUST00000038495.3
|
Crp
|
C-reactive protein, pentraxin-related |
| chr10_-_7212222 | 0.37 |
ENSMUST00000015346.5
|
Cnksr3
|
Cnksr family member 3 |
| chr11_+_69125896 | 0.37 |
ENSMUST00000021268.2
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr8_+_123186235 | 0.37 |
ENSMUST00000019422.4
|
Dpep1
|
dipeptidase 1 (renal) |
| chr16_+_31428745 | 0.36 |
ENSMUST00000115227.3
|
Bdh1
|
3-hydroxybutyrate dehydrogenase, type 1 |
| chr1_+_87796467 | 0.36 |
ENSMUST00000097661.3
|
5830472F04Rik
|
RIKEN cDNA 5830472F04 gene |
| chr2_-_11502067 | 0.36 |
ENSMUST00000028114.6
ENSMUST00000049849.6 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr8_+_71406003 | 0.36 |
ENSMUST00000119976.1
ENSMUST00000120725.1 |
Ankle1
|
ankyrin repeat and LEM domain containing 1 |
| chr10_-_78464853 | 0.35 |
ENSMUST00000105385.1
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr5_+_7960445 | 0.34 |
ENSMUST00000115421.1
|
Steap4
|
STEAP family member 4 |
| chr2_-_11502025 | 0.33 |
ENSMUST00000114846.2
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr19_+_4099998 | 0.33 |
ENSMUST00000049658.7
|
Pitpnm1
|
phosphatidylinositol transfer protein, membrane-associated 1 |
| chr6_-_116461151 | 0.33 |
ENSMUST00000026795.6
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr14_-_32685246 | 0.32 |
ENSMUST00000096038.3
|
3425401B19Rik
|
RIKEN cDNA 3425401B19 gene |
| chr2_-_11502090 | 0.32 |
ENSMUST00000179584.1
ENSMUST00000170196.2 ENSMUST00000171188.2 ENSMUST00000114845.3 ENSMUST00000114844.1 ENSMUST00000100411.2 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr2_-_65364000 | 0.32 |
ENSMUST00000155962.1
ENSMUST00000112420.1 ENSMUST00000152324.1 |
Slc38a11
|
solute carrier family 38, member 11 |
| chr6_+_147042755 | 0.32 |
ENSMUST00000036111.7
|
Mrps35
|
mitochondrial ribosomal protein S35 |
| chr5_+_25246775 | 0.32 |
ENSMUST00000144971.1
|
Galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 |
| chr7_+_28169744 | 0.31 |
ENSMUST00000042405.6
|
Fbl
|
fibrillarin |
| chr8_-_119605199 | 0.31 |
ENSMUST00000093099.6
|
Taf1c
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, C |
| chr3_-_63899437 | 0.30 |
ENSMUST00000159188.1
ENSMUST00000177143.1 |
Plch1
|
phospholipase C, eta 1 |
| chr7_+_101378183 | 0.30 |
ENSMUST00000084895.5
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr4_+_11123950 | 0.30 |
ENSMUST00000142297.1
|
Gm11827
|
predicted gene 11827 |
| chr7_+_82648595 | 0.30 |
ENSMUST00000141726.1
ENSMUST00000179489.1 ENSMUST00000039881.3 |
Eftud1
|
elongation factor Tu GTP binding domain containing 1 |
| chr17_-_23677432 | 0.29 |
ENSMUST00000167059.1
ENSMUST00000024698.8 |
Tnfrsf12a
|
tumor necrosis factor receptor superfamily, member 12a |
| chr2_+_109280738 | 0.29 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chr1_-_180193475 | 0.29 |
ENSMUST00000160482.1
ENSMUST00000170472.1 |
Adck3
|
aarF domain containing kinase 3 |
| chr1_+_74588289 | 0.29 |
ENSMUST00000113733.3
ENSMUST00000027358.4 |
Bcs1l
|
BCS1-like (yeast) |
| chr10_+_127078886 | 0.28 |
ENSMUST00000039259.6
|
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr10_-_78464969 | 0.28 |
ENSMUST00000041616.8
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr7_-_126676428 | 0.27 |
ENSMUST00000106373.1
|
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr17_+_32036098 | 0.27 |
ENSMUST00000081339.6
|
Rrp1b
|
ribosomal RNA processing 1 homolog B (S. cerevisiae) |
| chr4_-_44066960 | 0.27 |
ENSMUST00000173234.1
ENSMUST00000173274.1 |
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr2_-_131063508 | 0.27 |
ENSMUST00000147333.1
|
Adam33
|
a disintegrin and metallopeptidase domain 33 |
| chr14_+_3188255 | 0.27 |
ENSMUST00000100920.3
ENSMUST00000171150.1 ENSMUST00000166275.1 |
Gm5795
|
predicted gene 5795 |
| chr9_+_110419750 | 0.26 |
ENSMUST00000035061.6
|
Ngp
|
neutrophilic granule protein |
| chr4_-_137409777 | 0.26 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr3_-_144760841 | 0.26 |
ENSMUST00000059091.5
|
Clca1
|
chloride channel calcium activated 1 |
| chr2_-_105399286 | 0.25 |
ENSMUST00000006128.6
|
Rcn1
|
reticulocalbin 1 |
| chr10_+_69534208 | 0.25 |
ENSMUST00000182439.1
ENSMUST00000092434.5 ENSMUST00000092432.5 ENSMUST00000092431.5 ENSMUST00000054167.8 ENSMUST00000047061.6 |
Ank3
|
ankyrin 3, epithelial |
| chr10_-_14718191 | 0.25 |
ENSMUST00000020016.4
|
Gje1
|
gap junction protein, epsilon 1 |
| chr16_-_97763712 | 0.25 |
ENSMUST00000019386.8
|
Ripk4
|
receptor-interacting serine-threonine kinase 4 |
| chr2_-_132029845 | 0.25 |
ENSMUST00000028814.8
|
Rassf2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr8_+_95703037 | 0.24 |
ENSMUST00000073139.7
ENSMUST00000080666.7 |
Ndrg4
|
N-myc downstream regulated gene 4 |
| chr11_-_69948145 | 0.24 |
ENSMUST00000179298.1
ENSMUST00000018710.6 ENSMUST00000135437.1 ENSMUST00000141837.2 ENSMUST00000142500.1 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr18_+_61220467 | 0.24 |
ENSMUST00000025468.7
|
Pde6a
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr17_+_33524170 | 0.24 |
ENSMUST00000087623.6
|
Adamts10
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 10 |
| chr11_-_120453473 | 0.24 |
ENSMUST00000026452.2
|
Pde6g
|
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr2_-_150255591 | 0.24 |
ENSMUST00000063463.5
|
Gm21994
|
predicted gene 21994 |
| chr8_+_22411340 | 0.24 |
ENSMUST00000033934.3
|
Mrps31
|
mitochondrial ribosomal protein S31 |
| chr13_-_59675754 | 0.23 |
ENSMUST00000022039.5
ENSMUST00000095739.2 |
Golm1
|
golgi membrane protein 1 |
| chr3_+_10366903 | 0.23 |
ENSMUST00000029049.5
|
Chmp4c
|
charged multivesicular body protein 4C |
| chr11_-_48946148 | 0.23 |
ENSMUST00000104958.1
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr6_-_129451906 | 0.23 |
ENSMUST00000037481.7
|
Clec1a
|
C-type lectin domain family 1, member a |
| chr15_-_102524615 | 0.23 |
ENSMUST00000023814.7
|
Npff
|
neuropeptide FF-amide peptide precursor |
| chr4_-_154899077 | 0.23 |
ENSMUST00000030935.3
ENSMUST00000132281.1 |
Fam213b
|
family with sequence similarity 213, member B |
| chr1_+_166130467 | 0.22 |
ENSMUST00000166860.1
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr10_-_99759658 | 0.22 |
ENSMUST00000056085.4
|
Csl
|
citrate synthase like |
| chr6_-_136922169 | 0.21 |
ENSMUST00000032343.6
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr5_+_112288734 | 0.21 |
ENSMUST00000151947.1
|
Tpst2
|
protein-tyrosine sulfotransferase 2 |
| chr7_-_89633124 | 0.21 |
ENSMUST00000172178.1
|
A230065N10Rik
|
RIKEN cDNA A230065N10 gene |
| chr1_+_166130238 | 0.21 |
ENSMUST00000060833.7
ENSMUST00000166159.1 |
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr15_-_97721111 | 0.21 |
ENSMUST00000023105.3
|
Endou
|
endonuclease, polyU-specific |
| chr14_+_3846245 | 0.20 |
ENSMUST00000180934.1
|
Gm3033
|
predicted gene 3033 |
| chr4_+_88094599 | 0.20 |
ENSMUST00000097992.3
|
Focad
|
focadhesin |
| chr13_-_47043116 | 0.20 |
ENSMUST00000110118.1
ENSMUST00000124948.1 ENSMUST00000021806.3 ENSMUST00000136864.1 |
Tpmt
|
thiopurine methyltransferase |
| chr14_-_19602581 | 0.20 |
ENSMUST00000096121.5
ENSMUST00000163800.1 |
Gm5458
|
predicted gene 5458 |
| chr11_+_33963013 | 0.20 |
ENSMUST00000020362.2
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr7_-_139683797 | 0.20 |
ENSMUST00000129990.1
ENSMUST00000130453.1 |
9330101J02Rik
|
RIKEN cDNA 9330101J02 gene |
| chr16_+_13358375 | 0.20 |
ENSMUST00000149359.1
|
Mkl2
|
MKL/myocardin-like 2 |
| chr2_-_26380600 | 0.20 |
ENSMUST00000114115.2
ENSMUST00000035427.4 |
Snapc4
|
small nuclear RNA activating complex, polypeptide 4 |
| chr11_-_120796369 | 0.20 |
ENSMUST00000143139.1
ENSMUST00000129955.1 ENSMUST00000026151.4 ENSMUST00000167023.1 ENSMUST00000106133.1 ENSMUST00000106135.1 |
Dus1l
|
dihydrouridine synthase 1-like (S. cerevisiae) |
| chr1_-_78488846 | 0.19 |
ENSMUST00000068333.7
ENSMUST00000170217.1 |
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chr1_+_172555932 | 0.19 |
ENSMUST00000061835.3
|
Vsig8
|
V-set and immunoglobulin domain containing 8 |
| chr8_-_115707778 | 0.19 |
ENSMUST00000109104.1
|
Maf
|
avian musculoaponeurotic fibrosarcoma (v-maf) AS42 oncogene homolog |
| chr17_+_34187545 | 0.18 |
ENSMUST00000170086.1
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr9_+_65630552 | 0.18 |
ENSMUST00000055844.8
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr1_-_173942445 | 0.18 |
ENSMUST00000042228.8
ENSMUST00000081216.5 ENSMUST00000129829.1 ENSMUST00000123708.1 ENSMUST00000111210.2 |
Ifi203
Mndal
|
interferon activated gene 203 myeloid nuclear differentiation antigen like |
| chr9_-_22085391 | 0.18 |
ENSMUST00000179422.1
ENSMUST00000098937.3 ENSMUST00000177967.1 ENSMUST00000180180.1 |
Ecsit
|
ECSIT homolog (Drosophila) |
| chr5_+_121204477 | 0.18 |
ENSMUST00000031617.9
|
Rpl6
|
ribosomal protein L6 |
| chr11_+_6389061 | 0.17 |
ENSMUST00000109787.1
|
Zmiz2
|
zinc finger, MIZ-type containing 2 |
| chr8_+_72646679 | 0.17 |
ENSMUST00000161386.1
ENSMUST00000093427.4 |
Nwd1
|
NACHT and WD repeat domain containing 1 |
| chr13_-_54688246 | 0.17 |
ENSMUST00000122935.1
ENSMUST00000128257.1 |
Rnf44
|
ring finger protein 44 |
| chr18_-_61211380 | 0.17 |
ENSMUST00000148829.1
|
Slc26a2
|
solute carrier family 26 (sulfate transporter), member 2 |
| chr1_-_163289214 | 0.17 |
ENSMUST00000183691.1
|
Prrx1
|
paired related homeobox 1 |
| chr6_-_126645784 | 0.17 |
ENSMUST00000055168.3
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr19_-_5560570 | 0.16 |
ENSMUST00000025861.1
|
Ovol1
|
OVO homolog-like 1 (Drosophila) |
| chr4_-_9643638 | 0.16 |
ENSMUST00000108333.1
ENSMUST00000108334.1 ENSMUST00000108335.1 ENSMUST00000152526.1 ENSMUST00000103004.3 |
Asph
|
aspartate-beta-hydroxylase |
| chr15_+_52040107 | 0.16 |
ENSMUST00000090025.4
|
Aard
|
alanine and arginine rich domain containing protein |
| chr2_+_136057927 | 0.16 |
ENSMUST00000057503.6
|
Lamp5
|
lysosomal-associated membrane protein family, member 5 |
| chr13_-_106936907 | 0.16 |
ENSMUST00000080856.7
|
Ipo11
|
importin 11 |
| chr1_-_133801031 | 0.16 |
ENSMUST00000143567.1
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr8_+_4243264 | 0.16 |
ENSMUST00000110996.1
|
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr17_+_34187789 | 0.16 |
ENSMUST00000041633.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_-_146963742 | 0.15 |
ENSMUST00000125217.1
ENSMUST00000110564.1 ENSMUST00000066675.3 ENSMUST00000016654.2 ENSMUST00000110566.1 ENSMUST00000140526.1 |
Mtif3
|
mitochondrial translational initiation factor 3 |
| chr7_+_49759100 | 0.15 |
ENSMUST00000085272.5
|
Htatip2
|
HIV-1 tat interactive protein 2, homolog (human) |
| chr13_+_108316395 | 0.15 |
ENSMUST00000171178.1
|
Depdc1b
|
DEP domain containing 1B |
| chr8_-_40308331 | 0.15 |
ENSMUST00000118639.1
|
Fgf20
|
fibroblast growth factor 20 |
| chr4_+_48124903 | 0.15 |
ENSMUST00000107721.1
ENSMUST00000153502.1 ENSMUST00000107720.2 ENSMUST00000064765.4 |
Stx17
|
syntaxin 17 |
| chr7_-_144939823 | 0.15 |
ENSMUST00000093962.4
|
Ccnd1
|
cyclin D1 |
| chr1_+_132008285 | 0.15 |
ENSMUST00000146432.1
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr2_+_30266513 | 0.15 |
ENSMUST00000091132.6
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr11_-_75439551 | 0.15 |
ENSMUST00000128330.1
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr8_+_110919916 | 0.15 |
ENSMUST00000117534.1
ENSMUST00000034197.4 |
St3gal2
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 2 |
| chr18_+_62657285 | 0.15 |
ENSMUST00000162511.1
|
Spink10
|
serine peptidase inhibitor, Kazal type 10 |
| chr6_-_136835426 | 0.15 |
ENSMUST00000068293.3
ENSMUST00000111894.1 |
Smco3
|
single-pass membrane protein with coiled-coil domains 3 |
| chr15_-_79742518 | 0.15 |
ENSMUST00000089311.4
ENSMUST00000046259.7 |
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr17_-_35969724 | 0.15 |
ENSMUST00000043757.8
|
Abcf1
|
ATP-binding cassette, sub-family F (GCN20), member 1 |
| chr2_-_59948155 | 0.14 |
ENSMUST00000153136.1
|
Baz2b
|
bromodomain adjacent to zinc finger domain, 2B |
| chr16_-_95459245 | 0.14 |
ENSMUST00000176345.1
ENSMUST00000121809.2 ENSMUST00000118113.1 ENSMUST00000122199.1 |
Erg
|
avian erythroblastosis virus E-26 (v-ets) oncogene related |
| chr4_+_138395198 | 0.14 |
ENSMUST00000062902.6
|
AB041806
|
hypothetical protein, MNCb-2457 |
| chr17_-_45549655 | 0.14 |
ENSMUST00000180252.1
|
Tmem151b
|
transmembrane protein 151B |
| chr17_-_12940317 | 0.14 |
ENSMUST00000160378.1
ENSMUST00000043923.5 |
Acat3
|
acetyl-Coenzyme A acetyltransferase 3 |
| chr1_+_180568913 | 0.14 |
ENSMUST00000027777.6
|
Parp1
|
poly (ADP-ribose) polymerase family, member 1 |
| chr6_+_37870786 | 0.14 |
ENSMUST00000120428.1
ENSMUST00000031859.7 |
Trim24
|
tripartite motif-containing 24 |
| chr8_-_107065632 | 0.14 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr15_-_58214882 | 0.14 |
ENSMUST00000022986.6
|
Fbxo32
|
F-box protein 32 |
| chr16_-_20316750 | 0.14 |
ENSMUST00000182741.1
|
Cyp2ab1
|
cytochrome P450, family 2, subfamily ab, polypeptide 1 |
| chr2_-_73485733 | 0.14 |
ENSMUST00000102680.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr12_+_104406704 | 0.14 |
ENSMUST00000021506.5
|
Serpina3n
|
serine (or cysteine) peptidase inhibitor, clade A, member 3N |
| chr8_+_72646728 | 0.13 |
ENSMUST00000161254.1
|
Nwd1
|
NACHT and WD repeat domain containing 1 |
| chr2_+_131234043 | 0.13 |
ENSMUST00000041362.5
ENSMUST00000110199.2 |
Mavs
|
mitochondrial antiviral signaling protein |
| chr15_+_100469034 | 0.13 |
ENSMUST00000037001.8
|
Letmd1
|
LETM1 domain containing 1 |
| chr11_-_78422217 | 0.13 |
ENSMUST00000001122.5
|
Slc13a2
|
solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 |
| chr15_-_98607611 | 0.13 |
ENSMUST00000096224.4
|
Adcy6
|
adenylate cyclase 6 |
| chr10_+_69533803 | 0.13 |
ENSMUST00000182155.1
ENSMUST00000183169.1 ENSMUST00000183148.1 |
Ank3
|
ankyrin 3, epithelial |
| chr1_-_160792908 | 0.13 |
ENSMUST00000028049.7
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr11_+_4135233 | 0.13 |
ENSMUST00000124670.1
|
Rnf215
|
ring finger protein 215 |
| chr11_-_106750628 | 0.13 |
ENSMUST00000068021.2
|
Pecam1
|
platelet/endothelial cell adhesion molecule 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ar
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0042823 | pyridoxal phosphate biosynthetic process(GO:0042823) |
| 0.2 | 0.8 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.2 | 0.8 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.2 | 1.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 1.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.7 | GO:2000275 | regulation of oxidative phosphorylation uncoupler activity(GO:2000275) |
| 0.1 | 0.3 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.1 | 0.5 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.4 | GO:0045950 | negative regulation of mitotic recombination(GO:0045950) |
| 0.1 | 0.2 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.1 | 0.2 | GO:0044878 | mitotic cytokinesis checkpoint(GO:0044878) |
| 0.1 | 0.6 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.1 | 0.5 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.1 | 0.3 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.3 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.3 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.1 | 0.2 | GO:0071286 | cellular response to magnesium ion(GO:0071286) |
| 0.1 | 0.2 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.3 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 0.2 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.1 | 0.6 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) |
| 0.1 | 0.4 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.2 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.2 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.7 | GO:0009437 | carnitine metabolic process(GO:0009437) |
| 0.0 | 0.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.5 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.2 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:1902572 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.0 | 0.3 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.4 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0071651 | positive regulation of chemokine (C-C motif) ligand 5 production(GO:0071651) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.2 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 0.5 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.5 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 1.0 | GO:0010866 | regulation of triglyceride biosynthetic process(GO:0010866) |
| 0.0 | 0.1 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.0 | 0.7 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.0 | 0.1 | GO:0006548 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) |
| 0.0 | 0.7 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.4 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.3 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) regulation of thymocyte migration(GO:2000410) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.0 | 0.1 | GO:1903094 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.0 | 1.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.4 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) negative regulation of gastric acid secretion(GO:0060455) |
| 0.0 | 0.1 | GO:0070562 | regulation of vitamin D receptor signaling pathway(GO:0070562) |
| 0.0 | 0.4 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:1990743 | protein sialylation(GO:1990743) |
| 0.0 | 0.2 | GO:2001135 | regulation of endocytic recycling(GO:2001135) |
| 0.0 | 0.2 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 0.1 | GO:0090309 | DNA methylation on cytosine within a CG sequence(GO:0010424) positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.8 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.8 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.0 | 1.5 | GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032435) |
| 0.0 | 0.2 | GO:2000580 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.2 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.2 | GO:0060046 | regulation of acrosome reaction(GO:0060046) |
| 0.0 | 0.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:2000482 | regulation of interleukin-8 secretion(GO:2000482) |
| 0.0 | 0.1 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0046010 | positive regulation of circadian sleep/wake cycle, non-REM sleep(GO:0046010) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0010536 | positive regulation of activation of Janus kinase activity(GO:0010536) |
| 0.0 | 0.1 | GO:1901907 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.0 | 0.4 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 0.1 | GO:0045650 | negative regulation of macrophage differentiation(GO:0045650) |
| 0.0 | 0.1 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) cellular response to laminar fluid shear stress(GO:0071499) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.1 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.0 | 0.1 | GO:0060666 | pulmonary myocardium development(GO:0003350) dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.1 | 0.2 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 0.2 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 0.3 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 0.2 | GO:0071920 | cleavage body(GO:0071920) |
| 0.1 | 0.3 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.5 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.5 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 1.1 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.7 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.4 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.2 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.3 | 0.8 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.3 | 1.8 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.2 | 0.7 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 0.6 | GO:0031403 | lithium ion binding(GO:0031403) |
| 0.1 | 0.7 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 0.4 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 0.5 | GO:0070573 | metallodipeptidase activity(GO:0070573) |
| 0.1 | 1.0 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.3 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.1 | 0.3 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.1 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.1 | 0.5 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.3 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 0.6 | GO:0036374 | glutathione hydrolase activity(GO:0036374) |
| 0.1 | 0.3 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.1 | 0.8 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 0.2 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.1 | 1.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.4 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.3 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.5 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.5 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.0 | 1.0 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 0.3 | GO:0035014 | phosphatidylinositol 3-kinase regulator activity(GO:0035014) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.3 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.5 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0033677 | DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.4 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0000298 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 1.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.5 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 1.0 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.7 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 1.2 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.0 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |