Project
12D miR HR13_24
Navigation
Downloads
Results for Atf4
Z-value: 1.35
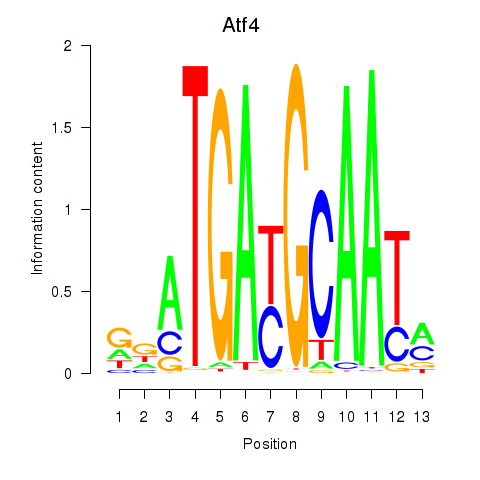
Transcription factors associated with Atf4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Atf4
|
ENSMUSG00000042406.7 | activating transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Atf4 | mm10_v2_chr15_+_80255184_80255263 | 0.58 | 6.1e-02 | Click! |
Activity profile of Atf4 motif
Sorted Z-values of Atf4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_119351222 | 4.07 |
ENSMUST00000028780.3
|
Chac1
|
ChaC, cation transport regulator 1 |
| chr10_-_83533383 | 2.80 |
ENSMUST00000146640.1
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chrX_+_164438039 | 2.65 |
ENSMUST00000033755.5
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr7_-_45615484 | 2.27 |
ENSMUST00000033099.4
|
Fgf21
|
fibroblast growth factor 21 |
| chr11_-_58534825 | 2.13 |
ENSMUST00000170009.1
|
Olfr330
|
olfactory receptor 330 |
| chr7_-_126625676 | 1.76 |
ENSMUST00000032961.3
|
Nupr1
|
nuclear protein transcription regulator 1 |
| chr7_+_78913401 | 1.50 |
ENSMUST00000118867.1
|
Isg20
|
interferon-stimulated protein |
| chrX_+_103422010 | 1.46 |
ENSMUST00000182089.1
|
Gm26992
|
predicted gene, 26992 |
| chr7_+_78913436 | 1.33 |
ENSMUST00000121645.1
|
Isg20
|
interferon-stimulated protein |
| chr7_-_143600049 | 1.27 |
ENSMUST00000105909.3
ENSMUST00000010899.7 |
Cars
|
cysteinyl-tRNA synthetase |
| chr4_-_118543210 | 1.21 |
ENSMUST00000156191.1
|
Tmem125
|
transmembrane protein 125 |
| chr11_-_20332689 | 1.18 |
ENSMUST00000109594.1
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr7_+_78913765 | 1.14 |
ENSMUST00000038142.8
|
Isg20
|
interferon-stimulated protein |
| chr11_-_20332654 | 1.08 |
ENSMUST00000004634.6
|
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr12_-_104865076 | 1.06 |
ENSMUST00000109937.1
ENSMUST00000109936.1 |
Clmn
|
calmin |
| chr1_-_71653162 | 1.03 |
ENSMUST00000055226.6
|
Fn1
|
fibronectin 1 |
| chr3_-_59130610 | 1.03 |
ENSMUST00000065220.6
ENSMUST00000091112.4 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr10_+_127290774 | 1.02 |
ENSMUST00000026475.8
ENSMUST00000139091.1 |
Ddit3
|
DNA-damage inducible transcript 3 |
| chr9_+_21835506 | 0.99 |
ENSMUST00000058777.6
|
Gm6484
|
predicted gene 6484 |
| chr15_+_89322969 | 0.97 |
ENSMUST00000066991.5
|
Adm2
|
adrenomedullin 2 |
| chr11_-_120643643 | 0.92 |
ENSMUST00000141254.1
ENSMUST00000170556.1 ENSMUST00000151876.1 ENSMUST00000026133.8 ENSMUST00000139706.1 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr15_+_99393219 | 0.85 |
ENSMUST00000159209.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr4_-_56224520 | 0.83 |
ENSMUST00000128591.1
|
2310081O03Rik
|
RIKEN cDNA 2310081O03 gene |
| chr7_+_78914216 | 0.80 |
ENSMUST00000120331.2
|
Isg20
|
interferon-stimulated protein |
| chr7_-_35056467 | 0.79 |
ENSMUST00000130491.1
|
Cebpg
|
CCAAT/enhancer binding protein (C/EBP), gamma |
| chrX_-_162565514 | 0.79 |
ENSMUST00000154424.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr7_-_109493627 | 0.77 |
ENSMUST00000106739.1
|
Trim66
|
tripartite motif-containing 66 |
| chr11_-_98400393 | 0.76 |
ENSMUST00000128897.1
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr9_+_109931863 | 0.76 |
ENSMUST00000165876.1
|
Map4
|
microtubule-associated protein 4 |
| chr11_-_75422586 | 0.73 |
ENSMUST00000138661.1
ENSMUST00000000769.7 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr15_+_99393574 | 0.73 |
ENSMUST00000162624.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr11_-_98400453 | 0.69 |
ENSMUST00000090827.5
|
Pgap3
|
post-GPI attachment to proteins 3 |
| chr15_+_99393610 | 0.69 |
ENSMUST00000159531.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr2_-_18048784 | 0.68 |
ENSMUST00000142856.1
|
Skida1
|
SKI/DACH domain containing 1 |
| chr9_+_109931774 | 0.66 |
ENSMUST00000169851.2
|
Map4
|
microtubule-associated protein 4 |
| chr19_+_44203265 | 0.66 |
ENSMUST00000026220.5
|
Scd3
|
stearoyl-coenzyme A desaturase 3 |
| chr9_+_109931458 | 0.65 |
ENSMUST00000072772.5
ENSMUST00000035055.8 |
Map4
|
microtubule-associated protein 4 |
| chr8_+_111033890 | 0.64 |
ENSMUST00000034441.7
|
Aars
|
alanyl-tRNA synthetase |
| chr10_-_127311740 | 0.59 |
ENSMUST00000037290.5
ENSMUST00000171564.1 |
Mars
|
methionine-tRNA synthetase |
| chr14_+_63436394 | 0.58 |
ENSMUST00000121288.1
|
Fam167a
|
family with sequence similarity 167, member A |
| chr3_-_105932664 | 0.58 |
ENSMUST00000098758.2
|
I830077J02Rik
|
RIKEN cDNA I830077J02 gene |
| chr13_-_101692624 | 0.58 |
ENSMUST00000035532.6
|
Pik3r1
|
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha) |
| chr18_-_64516547 | 0.54 |
ENSMUST00000025483.9
|
Nars
|
asparaginyl-tRNA synthetase |
| chr3_-_89998656 | 0.53 |
ENSMUST00000079724.4
|
Hax1
|
HCLS1 associated X-1 |
| chr11_+_100320596 | 0.53 |
ENSMUST00000152521.1
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr14_+_55540266 | 0.51 |
ENSMUST00000048781.3
|
Pck2
|
phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| chr6_+_28981490 | 0.49 |
ENSMUST00000164104.1
|
Gm3294
|
predicted gene 3294 |
| chr17_-_34187219 | 0.49 |
ENSMUST00000173831.1
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr2_-_18048347 | 0.48 |
ENSMUST00000066885.5
|
Skida1
|
SKI/DACH domain containing 1 |
| chr6_-_83317589 | 0.47 |
ENSMUST00000005810.6
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr4_+_124741844 | 0.44 |
ENSMUST00000094782.3
ENSMUST00000153837.1 ENSMUST00000154229.1 |
Inpp5b
|
inositol polyphosphate-5-phosphatase B |
| chr14_-_37135126 | 0.43 |
ENSMUST00000042564.9
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr1_+_133131143 | 0.42 |
ENSMUST00000052529.3
|
Ppp1r15b
|
protein phosphatase 1, regulatory (inhibitor) subunit 15b |
| chr7_-_3502465 | 0.39 |
ENSMUST00000065703.7
|
Tarm1
|
T cell-interacting, activating receptor on myeloid cells 1 |
| chr3_-_96263311 | 0.39 |
ENSMUST00000171473.1
|
Hist2h4
|
histone cluster 2, H4 |
| chr18_+_40256960 | 0.38 |
ENSMUST00000096572.1
|
2900055J20Rik
|
RIKEN cDNA 2900055J20 gene |
| chr7_+_44468051 | 0.38 |
ENSMUST00000118493.1
|
Josd2
|
Josephin domain containing 2 |
| chr5_+_93093428 | 0.37 |
ENSMUST00000074733.7
|
Sept11
|
septin 11 |
| chr12_-_108893197 | 0.36 |
ENSMUST00000161154.1
ENSMUST00000161410.1 |
Wars
|
tryptophanyl-tRNA synthetase |
| chr5_-_36695969 | 0.35 |
ENSMUST00000031091.9
ENSMUST00000140063.1 |
D5Ertd579e
|
DNA segment, Chr 5, ERATO Doi 579, expressed |
| chr9_-_79793378 | 0.33 |
ENSMUST00000034878.5
|
Tmem30a
|
transmembrane protein 30A |
| chr19_+_53140430 | 0.32 |
ENSMUST00000111741.2
|
Add3
|
adducin 3 (gamma) |
| chr8_+_94386486 | 0.31 |
ENSMUST00000034220.7
|
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr10_+_121739915 | 0.31 |
ENSMUST00000065600.7
ENSMUST00000136432.1 |
BC048403
|
cDNA sequence BC048403 |
| chr4_+_138879360 | 0.30 |
ENSMUST00000105804.1
|
Pla2g2e
|
phospholipase A2, group IIE |
| chrX_+_94234594 | 0.30 |
ENSMUST00000153900.1
|
Klhl15
|
kelch-like 15 |
| chr17_-_29078953 | 0.30 |
ENSMUST00000133221.1
|
Trp53cor1
|
tumor protein p53 pathway corepressor 1 |
| chr8_-_122476036 | 0.30 |
ENSMUST00000014614.3
|
Rnf166
|
ring finger protein 166 |
| chr14_-_54870913 | 0.30 |
ENSMUST00000146642.1
|
Homez
|
homeodomain leucine zipper-encoding gene |
| chr7_+_44468020 | 0.30 |
ENSMUST00000117324.1
ENSMUST00000120852.1 ENSMUST00000118628.1 |
Josd2
|
Josephin domain containing 2 |
| chrX_-_75578188 | 0.27 |
ENSMUST00000033545.5
|
Rab39b
|
RAB39B, member RAS oncogene family |
| chr3_+_89715016 | 0.27 |
ENSMUST00000098924.2
|
Adar
|
adenosine deaminase, RNA-specific |
| chr10_+_96616998 | 0.26 |
ENSMUST00000038377.7
|
Btg1
|
B cell translocation gene 1, anti-proliferative |
| chr11_-_101171302 | 0.26 |
ENSMUST00000164474.1
ENSMUST00000043397.7 |
Plekhh3
|
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr8_+_75093591 | 0.26 |
ENSMUST00000005548.6
|
Hmox1
|
heme oxygenase (decycling) 1 |
| chr10_-_121626316 | 0.24 |
ENSMUST00000039810.7
|
Xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr7_+_44467980 | 0.22 |
ENSMUST00000035844.4
|
Josd2
|
Josephin domain containing 2 |
| chr9_-_44344159 | 0.22 |
ENSMUST00000077353.7
|
Hmbs
|
hydroxymethylbilane synthase |
| chr8_+_94386438 | 0.22 |
ENSMUST00000161576.1
|
Herpud1
|
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr9_-_71771535 | 0.22 |
ENSMUST00000122065.1
ENSMUST00000121322.1 ENSMUST00000072899.2 |
Cgnl1
|
cingulin-like 1 |
| chr7_-_138846202 | 0.22 |
ENSMUST00000118810.1
ENSMUST00000075667.4 ENSMUST00000119664.1 |
Mapk1ip1
|
mitogen-activated protein kinase 1 interacting protein 1 |
| chr5_+_145140362 | 0.21 |
ENSMUST00000162594.1
ENSMUST00000162308.1 ENSMUST00000159018.1 ENSMUST00000160075.1 |
Bud31
|
BUD31 homolog (yeast) |
| chr14_-_66868572 | 0.20 |
ENSMUST00000022629.8
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr3_-_127408986 | 0.20 |
ENSMUST00000182588.1
ENSMUST00000182959.1 ENSMUST00000182452.1 |
Ank2
|
ankyrin 2, brain |
| chr10_+_102158858 | 0.20 |
ENSMUST00000138522.1
ENSMUST00000163753.1 ENSMUST00000138016.1 |
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr7_+_138846579 | 0.18 |
ENSMUST00000155672.1
|
Ppp2r2d
|
protein phosphatase 2, regulatory subunit B, delta isoform |
| chr4_+_129189760 | 0.17 |
ENSMUST00000106054.2
ENSMUST00000001365.2 |
Yars
|
tyrosyl-tRNA synthetase |
| chr1_+_130800902 | 0.16 |
ENSMUST00000112477.2
ENSMUST00000027670.3 |
Fcamr
|
Fc receptor, IgA, IgM, high affinity |
| chr8_+_123477859 | 0.16 |
ENSMUST00000001520.7
|
Afg3l1
|
AFG3(ATPase family gene 3)-like 1 (yeast) |
| chr7_+_126565919 | 0.16 |
ENSMUST00000180459.1
|
G730046D07Rik
|
RIKEN cDNA G730046D07 gene |
| chr15_+_82147238 | 0.16 |
ENSMUST00000023100.6
|
Srebf2
|
sterol regulatory element binding factor 2 |
| chr7_+_144896523 | 0.15 |
ENSMUST00000033389.5
|
Fgf15
|
fibroblast growth factor 15 |
| chr15_-_83510861 | 0.14 |
ENSMUST00000109479.1
ENSMUST00000109480.1 ENSMUST00000016897.4 |
Ttll1
|
tubulin tyrosine ligase-like 1 |
| chr4_+_117835387 | 0.14 |
ENSMUST00000169885.1
|
Slc6a9
|
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr11_-_75422524 | 0.13 |
ENSMUST00000125982.1
ENSMUST00000137103.1 |
Serpinf1
|
serine (or cysteine) peptidase inhibitor, clade F, member 1 |
| chr1_-_22315792 | 0.11 |
ENSMUST00000164877.1
|
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr3_-_127408937 | 0.11 |
ENSMUST00000183095.1
ENSMUST00000182610.1 |
Ank2
|
ankyrin 2, brain |
| chr15_-_50889691 | 0.11 |
ENSMUST00000165201.2
ENSMUST00000184458.1 |
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr7_+_125603420 | 0.10 |
ENSMUST00000033000.6
|
Il21r
|
interleukin 21 receptor |
| chr16_-_34095983 | 0.09 |
ENSMUST00000114973.1
ENSMUST00000114964.1 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr17_+_34187545 | 0.09 |
ENSMUST00000170086.1
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr8_+_25808474 | 0.09 |
ENSMUST00000033979.4
|
Star
|
steroidogenic acute regulatory protein |
| chr19_-_4793851 | 0.09 |
ENSMUST00000178615.1
ENSMUST00000179189.1 ENSMUST00000164376.2 ENSMUST00000164209.2 ENSMUST00000180248.1 |
Rbm4
|
RNA binding motif protein 4 |
| chr17_+_34187789 | 0.09 |
ENSMUST00000041633.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr5_-_32133045 | 0.08 |
ENSMUST00000031308.6
|
Gm10463
|
predicted gene 10463 |
| chr11_+_96271453 | 0.08 |
ENSMUST00000000010.8
ENSMUST00000174042.1 |
Hoxb9
|
homeobox B9 |
| chr3_-_127409014 | 0.08 |
ENSMUST00000182008.1
ENSMUST00000182711.1 ENSMUST00000182547.1 |
Ank2
|
ankyrin 2, brain |
| chr15_-_83510793 | 0.07 |
ENSMUST00000154401.1
|
Ttll1
|
tubulin tyrosine ligase-like 1 |
| chr8_+_105701142 | 0.07 |
ENSMUST00000098444.2
|
Pard6a
|
par-6 (partitioning defective 6,) homolog alpha (C. elegans) |
| chr5_-_90223923 | 0.05 |
ENSMUST00000118816.1
ENSMUST00000048363.7 |
Cox18
|
cytochrome c oxidase assembly protein 18 |
| chr7_-_126502094 | 0.05 |
ENSMUST00000179818.1
|
Atxn2l
|
ataxin 2-like |
| chr3_-_127409044 | 0.05 |
ENSMUST00000182704.1
|
Ank2
|
ankyrin 2, brain |
| chr5_-_31180110 | 0.05 |
ENSMUST00000043161.6
ENSMUST00000088010.5 |
Gtf3c2
|
general transcription factor IIIC, polypeptide 2, beta |
| chr5_-_31179901 | 0.04 |
ENSMUST00000101411.2
ENSMUST00000140793.1 |
Gtf3c2
|
general transcription factor IIIC, polypeptide 2, beta |
| chr3_+_88616133 | 0.04 |
ENSMUST00000176500.1
ENSMUST00000177498.1 |
Arhgef2
|
rho/rac guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_134455524 | 0.04 |
ENSMUST00000112232.1
ENSMUST00000027725.4 ENSMUST00000116528.1 |
Klhl12
|
kelch-like 12 |
| chr1_-_191183244 | 0.03 |
ENSMUST00000027941.8
|
Atf3
|
activating transcription factor 3 |
| chr2_+_172393794 | 0.02 |
ENSMUST00000099061.2
ENSMUST00000103073.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr7_-_126566364 | 0.01 |
ENSMUST00000032992.5
|
Eif3c
|
eukaryotic translation initiation factor 3, subunit C |
| chr4_-_108833608 | 0.01 |
ENSMUST00000102742.4
|
Btf3l4
|
basic transcription factor 3-like 4 |
| chr11_+_115462464 | 0.01 |
ENSMUST00000106532.3
ENSMUST00000092445.5 ENSMUST00000153466.1 |
Slc16a5
|
solute carrier family 16 (monocarboxylic acid transporters), member 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Atf4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 4.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.9 | 2.8 | GO:0009397 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.6 | 2.3 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.5 | 2.3 | GO:1904721 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.4 | 2.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.3 | 1.0 | GO:0007161 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.3 | 1.0 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.3 | 4.1 | GO:0006751 | glutathione catabolic process(GO:0006751) |
| 0.3 | 2.3 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.3 | 1.0 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.2 | 0.6 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.7 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.5 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.4 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.8 | GO:2000194 | regulation of female gonad development(GO:2000194) |
| 0.1 | 0.5 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.1 | 0.6 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.3 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.8 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.5 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.9 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.4 | GO:1903912 | negative regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:1903912) |
| 0.1 | 0.3 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.2 | GO:0071431 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.1 | 0.6 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.6 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.1 | 0.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.4 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.5 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.5 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 1.5 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) |
| 0.0 | 0.3 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.2 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.0 | 0.1 | GO:0070859 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.3 | GO:0034374 | low-density lipoprotein particle remodeling(GO:0034374) |
| 0.0 | 0.2 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.0 | 0.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 2.1 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.2 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 1.0 | GO:0045776 | negative regulation of blood pressure(GO:0045776) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.0 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.4 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.2 | GO:0034982 | mitochondrial protein processing(GO:0034982) |
| 0.0 | 0.1 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 1.0 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 0.5 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.5 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.9 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.3 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 4.8 | GO:0016605 | PML body(GO:0016605) |
| 0.0 | 0.2 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.0 | 0.8 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.4 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.6 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.0 | 2.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 2.1 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.0 | 0.2 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 4.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.5 | GO:0031227 | intrinsic component of endoplasmic reticulum membrane(GO:0031227) |
| 0.0 | 1.6 | GO:0032993 | protein-DNA complex(GO:0032993) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 4.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.9 | 2.8 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.8 | 4.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.4 | 2.3 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 2.3 | GO:0022858 | L-alanine transmembrane transporter activity(GO:0015180) alanine transmembrane transporter activity(GO:0022858) |
| 0.2 | 0.9 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 0.6 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 0.5 | GO:0004816 | asparagine-tRNA ligase activity(GO:0004816) |
| 0.2 | 1.0 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.2 | 0.7 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.2 | 0.5 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.1 | 0.4 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.6 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.4 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 1.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.1 | 0.3 | GO:0003692 | left-handed Z-DNA binding(GO:0003692) |
| 0.1 | 1.0 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 2.4 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.3 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.5 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.1 | 0.2 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.2 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 2.1 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.2 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.3 | GO:0031489 | myosin V binding(GO:0031489) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.6 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 5.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 1.0 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 1.0 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.6 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.6 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.9 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 0.1 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 3.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |