Project
12D miR HR13_24
Navigation
Downloads
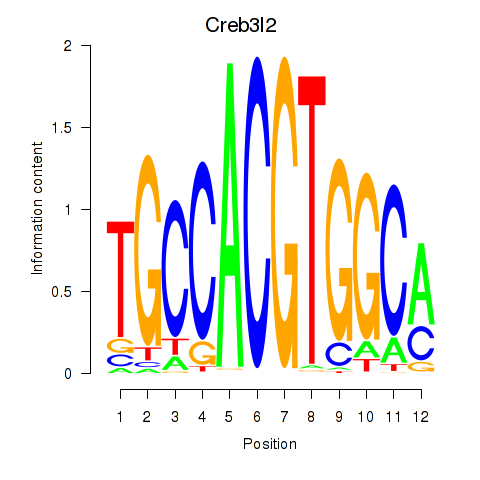
Results for Creb3l2
Z-value: 1.68
Transcription factors associated with Creb3l2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Creb3l2
|
ENSMUSG00000038648.5 | cAMP responsive element binding protein 3-like 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l2 | mm10_v2_chr6_-_37442095_37442154 | 0.47 | 1.4e-01 | Click! |
Activity profile of Creb3l2 motif
Sorted Z-values of Creb3l2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_92131494 | 4.98 |
ENSMUST00000099326.3
ENSMUST00000146492.1 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr2_+_180725263 | 4.97 |
ENSMUST00000094218.3
|
Slc17a9
|
solute carrier family 17, member 9 |
| chr4_+_104766308 | 3.86 |
ENSMUST00000031663.3
|
C8b
|
complement component 8, beta polypeptide |
| chr4_+_104766334 | 3.77 |
ENSMUST00000065072.6
|
C8b
|
complement component 8, beta polypeptide |
| chr18_+_65800543 | 3.06 |
ENSMUST00000025394.6
ENSMUST00000153193.1 |
Sec11c
|
SEC11 homolog C (S. cerevisiae) |
| chr5_-_139814231 | 2.58 |
ENSMUST00000044002.4
|
Tmem184a
|
transmembrane protein 184a |
| chr2_+_160880642 | 2.40 |
ENSMUST00000109456.2
|
Lpin3
|
lipin 3 |
| chr4_-_106617232 | 2.23 |
ENSMUST00000106788.1
|
BC055111
|
cDNA sequence BC055111 |
| chr4_+_133553370 | 2.06 |
ENSMUST00000042706.2
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr14_-_70443442 | 2.05 |
ENSMUST00000000793.5
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr2_+_129198757 | 1.90 |
ENSMUST00000028880.3
|
Slc20a1
|
solute carrier family 20, member 1 |
| chr14_-_70443219 | 1.86 |
ENSMUST00000180358.1
|
Polr3d
|
polymerase (RNA) III (DNA directed) polypeptide D |
| chr17_-_26199008 | 1.86 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr9_+_21955747 | 1.85 |
ENSMUST00000053583.5
|
Swsap1
|
SWIM type zinc finger 7 associated protein 1 |
| chr8_-_106136890 | 1.84 |
ENSMUST00000115979.2
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chr19_-_6840590 | 1.84 |
ENSMUST00000170516.2
ENSMUST00000025903.5 |
Rps6ka4
|
ribosomal protein S6 kinase, polypeptide 4 |
| chr5_-_139814025 | 1.83 |
ENSMUST00000146780.1
|
Tmem184a
|
transmembrane protein 184a |
| chr8_+_14911663 | 1.81 |
ENSMUST00000084207.5
ENSMUST00000161162.1 ENSMUST00000110800.2 |
Arhgef10
|
Rho guanine nucleotide exchange factor (GEF) 10 |
| chr3_+_106482427 | 1.81 |
ENSMUST00000029508.4
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr7_+_30169861 | 1.78 |
ENSMUST00000085668.4
|
Gm5113
|
predicted gene 5113 |
| chr15_-_83350151 | 1.76 |
ENSMUST00000067215.7
|
Arfgap3
|
ADP-ribosylation factor GTPase activating protein 3 |
| chr10_-_20725023 | 1.74 |
ENSMUST00000020165.7
|
Pde7b
|
phosphodiesterase 7B |
| chrX_+_50841434 | 1.70 |
ENSMUST00000114887.2
|
2610018G03Rik
|
RIKEN cDNA 2610018G03 gene |
| chr7_+_78913765 | 1.67 |
ENSMUST00000038142.8
|
Isg20
|
interferon-stimulated protein |
| chr13_-_119408985 | 1.64 |
ENSMUST00000099149.3
ENSMUST00000069902.6 ENSMUST00000109204.1 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr3_+_123267445 | 1.46 |
ENSMUST00000047923.7
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr6_-_13871459 | 1.39 |
ENSMUST00000155856.1
|
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr4_-_106800249 | 1.38 |
ENSMUST00000148688.1
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr17_-_56276750 | 1.36 |
ENSMUST00000058136.8
|
Ticam1
|
toll-like receptor adaptor molecule 1 |
| chr6_-_13871477 | 1.36 |
ENSMUST00000139231.1
|
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr9_-_43239816 | 1.31 |
ENSMUST00000034512.5
|
Oaf
|
OAF homolog (Drosophila) |
| chr11_+_54438188 | 1.31 |
ENSMUST00000046835.7
|
Fnip1
|
folliculin interacting protein 1 |
| chr5_+_129941949 | 1.29 |
ENSMUST00000051758.7
ENSMUST00000073945.4 |
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr18_-_40219324 | 1.26 |
ENSMUST00000025364.4
|
Yipf5
|
Yip1 domain family, member 5 |
| chrX_+_139563316 | 1.23 |
ENSMUST00000113027.1
|
Rnf128
|
ring finger protein 128 |
| chr7_+_19094594 | 1.18 |
ENSMUST00000049454.5
|
Six5
|
sine oculis-related homeobox 5 |
| chr15_-_34495180 | 1.18 |
ENSMUST00000022946.5
|
Hrsp12
|
heat-responsive protein 12 |
| chr11_-_120572822 | 1.15 |
ENSMUST00000168360.1
|
P4hb
|
prolyl 4-hydroxylase, beta polypeptide |
| chr7_+_78914216 | 1.14 |
ENSMUST00000120331.2
|
Isg20
|
interferon-stimulated protein |
| chr8_-_105255100 | 1.08 |
ENSMUST00000093217.2
ENSMUST00000161745.2 ENSMUST00000136822.2 |
B3gnt9
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| chr15_-_102136225 | 1.07 |
ENSMUST00000154032.1
|
Spryd3
|
SPRY domain containing 3 |
| chr6_+_87042838 | 1.07 |
ENSMUST00000113658.1
ENSMUST00000113657.1 ENSMUST00000113655.1 ENSMUST00000032057.7 |
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1 |
| chr4_-_119422355 | 1.07 |
ENSMUST00000106316.1
ENSMUST00000030385.6 |
Ppcs
|
phosphopantothenoylcysteine synthetase |
| chr12_+_17544873 | 1.06 |
ENSMUST00000171737.1
|
Odc1
|
ornithine decarboxylase, structural 1 |
| chr2_-_180334665 | 1.04 |
ENSMUST00000015771.2
|
Gata5
|
GATA binding protein 5 |
| chr7_+_27195781 | 1.04 |
ENSMUST00000108379.1
ENSMUST00000179391.1 |
BC024978
|
cDNA sequence BC024978 |
| chr9_-_87255536 | 1.01 |
ENSMUST00000093802.4
|
4922501C03Rik
|
RIKEN cDNA 4922501C03 gene |
| chr3_-_108086590 | 0.99 |
ENSMUST00000102638.1
ENSMUST00000102637.1 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr15_+_99591028 | 0.98 |
ENSMUST00000169082.1
|
Aqp5
|
aquaporin 5 |
| chr3_-_27710413 | 0.96 |
ENSMUST00000046157.4
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr4_+_155993305 | 0.95 |
ENSMUST00000105578.1
|
Sdf4
|
stromal cell derived factor 4 |
| chr7_-_30169701 | 0.95 |
ENSMUST00000062181.7
|
Zfp146
|
zinc finger protein 146 |
| chr9_+_89909775 | 0.91 |
ENSMUST00000034912.4
ENSMUST00000034909.4 |
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr6_-_83317589 | 0.89 |
ENSMUST00000005810.6
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NAD+ dependent), methenyltetrahydrofolate cyclohydrolase |
| chr11_-_103954015 | 0.89 |
ENSMUST00000103075.4
|
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr18_+_75018709 | 0.88 |
ENSMUST00000039608.7
|
Dym
|
dymeclin |
| chr3_+_40540751 | 0.88 |
ENSMUST00000091186.3
|
Intu
|
inturned planar cell polarity effector homolog (Drosophila) |
| chr8_-_124663368 | 0.87 |
ENSMUST00000034464.6
|
2310022B05Rik
|
RIKEN cDNA 2310022B05 gene |
| chr9_-_25151772 | 0.86 |
ENSMUST00000008573.7
|
Herpud2
|
HERPUD family member 2 |
| chr10_+_61680302 | 0.85 |
ENSMUST00000020285.8
|
Sar1a
|
SAR1 gene homolog A (S. cerevisiae) |
| chr5_+_143403819 | 0.84 |
ENSMUST00000110731.2
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr2_+_144368961 | 0.84 |
ENSMUST00000028911.8
ENSMUST00000147747.1 ENSMUST00000183618.1 |
Csrp2bp
Pet117
|
cysteine and glycine-rich protein 2 binding protein PET117 homolog (S. cerevisiae) |
| chr11_+_20201406 | 0.83 |
ENSMUST00000020358.5
ENSMUST00000109602.1 ENSMUST00000109601.1 |
Rab1
|
RAB1, member RAS oncogene family |
| chr3_-_101604580 | 0.82 |
ENSMUST00000036493.6
|
Atp1a1
|
ATPase, Na+/K+ transporting, alpha 1 polypeptide |
| chr11_-_120643643 | 0.78 |
ENSMUST00000141254.1
ENSMUST00000170556.1 ENSMUST00000151876.1 ENSMUST00000026133.8 ENSMUST00000139706.1 |
Pycr1
|
pyrroline-5-carboxylate reductase 1 |
| chr11_-_100759740 | 0.77 |
ENSMUST00000107361.2
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr19_+_46573362 | 0.77 |
ENSMUST00000026011.6
|
Sfxn2
|
sideroflexin 2 |
| chr15_-_97767798 | 0.77 |
ENSMUST00000129223.2
ENSMUST00000126854.2 ENSMUST00000135080.1 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr11_-_100759942 | 0.76 |
ENSMUST00000107363.2
|
Kcnh4
|
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr8_+_111033890 | 0.75 |
ENSMUST00000034441.7
|
Aars
|
alanyl-tRNA synthetase |
| chr5_-_45857473 | 0.75 |
ENSMUST00000016026.7
ENSMUST00000067997.6 ENSMUST00000045586.6 |
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr15_-_97767644 | 0.72 |
ENSMUST00000128775.2
ENSMUST00000134885.2 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr17_-_26939464 | 0.72 |
ENSMUST00000025027.8
ENSMUST00000114935.1 |
Cuta
|
cutA divalent cation tolerance homolog (E. coli) |
| chr6_+_13871517 | 0.72 |
ENSMUST00000181090.1
ENSMUST00000181225.1 |
1110019D14Rik
|
RIKEN cDNA 1110019D14 gene |
| chr15_-_3995708 | 0.70 |
ENSMUST00000046633.8
|
AW549877
|
expressed sequence AW549877 |
| chr15_+_34238026 | 0.70 |
ENSMUST00000022867.3
|
Laptm4b
|
lysosomal-associated protein transmembrane 4B |
| chr10_-_52382074 | 0.70 |
ENSMUST00000020008.8
ENSMUST00000105475.2 |
Gopc
|
golgi associated PDZ and coiled-coil motif containing |
| chr14_-_55643800 | 0.70 |
ENSMUST00000122358.1
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr3_-_63964659 | 0.69 |
ENSMUST00000161659.1
|
Slc33a1
|
solute carrier family 33 (acetyl-CoA transporter), member 1 |
| chr12_+_84451485 | 0.67 |
ENSMUST00000137170.1
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr2_-_130424242 | 0.66 |
ENSMUST00000089581.4
|
Pced1a
|
PC-esterase domain containing 1A |
| chr14_-_55643720 | 0.66 |
ENSMUST00000138085.1
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr11_-_97500340 | 0.64 |
ENSMUST00000056955.1
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr2_-_92370999 | 0.63 |
ENSMUST00000176810.1
ENSMUST00000090582.4 |
Gyltl1b
|
glycosyltransferase-like 1B |
| chr5_+_137641334 | 0.63 |
ENSMUST00000177466.1
ENSMUST00000166099.2 |
Sap25
|
sin3 associated polypeptide |
| chr2_+_150786735 | 0.63 |
ENSMUST00000045441.7
|
Pygb
|
brain glycogen phosphorylase |
| chr18_+_75367529 | 0.59 |
ENSMUST00000026999.3
|
Smad7
|
SMAD family member 7 |
| chr13_+_90089705 | 0.59 |
ENSMUST00000012566.8
|
Tmem167
|
transmembrane protein 167 |
| chr11_+_76672456 | 0.54 |
ENSMUST00000056184.1
|
Bhlha9
|
basic helix-loop-helix family, member a9 |
| chr11_+_44518959 | 0.54 |
ENSMUST00000019333.3
|
Rnf145
|
ring finger protein 145 |
| chr2_+_132846638 | 0.54 |
ENSMUST00000028835.6
ENSMUST00000110122.3 |
Crls1
|
cardiolipin synthase 1 |
| chr6_+_56832059 | 0.54 |
ENSMUST00000031795.7
|
Fkbp9
|
FK506 binding protein 9 |
| chr6_+_87887814 | 0.54 |
ENSMUST00000113607.3
ENSMUST00000049966.5 |
Copg1
|
coatomer protein complex, subunit gamma 1 |
| chr2_-_92370968 | 0.53 |
ENSMUST00000176774.1
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr13_-_90089060 | 0.51 |
ENSMUST00000161396.1
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr2_-_92371039 | 0.51 |
ENSMUST00000068586.6
|
Gyltl1b
|
glycosyltransferase-like 1B |
| chr13_-_55321928 | 0.50 |
ENSMUST00000035242.7
|
Rab24
|
RAB24, member RAS oncogene family |
| chr14_-_55643523 | 0.49 |
ENSMUST00000132338.1
|
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr10_+_128909866 | 0.49 |
ENSMUST00000026407.7
|
Cd63
|
CD63 antigen |
| chr15_+_79516396 | 0.48 |
ENSMUST00000010974.7
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr10_+_80172934 | 0.48 |
ENSMUST00000041882.6
|
1600002K03Rik
|
RIKEN cDNA 1600002K03 gene |
| chr12_-_84450944 | 0.47 |
ENSMUST00000085192.5
|
Aldh6a1
|
aldehyde dehydrogenase family 6, subfamily A1 |
| chr4_+_148039097 | 0.47 |
ENSMUST00000141283.1
|
Mthfr
|
5,10-methylenetetrahydrofolate reductase |
| chr2_+_70661556 | 0.47 |
ENSMUST00000112201.1
ENSMUST00000028509.4 ENSMUST00000133432.1 ENSMUST00000112205.1 |
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr11_+_95712673 | 0.46 |
ENSMUST00000107717.1
|
Zfp652
|
zinc finger protein 652 |
| chr14_-_55643251 | 0.46 |
ENSMUST00000120041.1
ENSMUST00000121937.1 ENSMUST00000133707.1 ENSMUST00000002391.8 ENSMUST00000121791.1 |
Tm9sf1
|
transmembrane 9 superfamily member 1 |
| chr3_-_106483435 | 0.45 |
ENSMUST00000164330.1
|
2010016I18Rik
|
RIKEN cDNA 2010016I18 gene |
| chr1_+_119526125 | 0.45 |
ENSMUST00000183952.1
|
TMEM185B
|
Transmembrane protein 185B |
| chr11_-_59571813 | 0.44 |
ENSMUST00000071943.2
|
Olfr222
|
olfactory receptor 222 |
| chr13_-_90089556 | 0.43 |
ENSMUST00000022115.7
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr9_-_67832325 | 0.43 |
ENSMUST00000054500.5
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr14_-_78536762 | 0.42 |
ENSMUST00000123853.1
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr3_+_96697100 | 0.42 |
ENSMUST00000107077.3
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chr8_+_94601928 | 0.41 |
ENSMUST00000060389.8
ENSMUST00000121101.1 |
Rspry1
|
ring finger and SPRY domain containing 1 |
| chr14_-_78536854 | 0.41 |
ENSMUST00000022593.5
|
Akap11
|
A kinase (PRKA) anchor protein 11 |
| chr10_+_78574492 | 0.40 |
ENSMUST00000105384.3
|
Ilvbl
|
ilvB (bacterial acetolactate synthase)-like |
| chr11_-_96075581 | 0.39 |
ENSMUST00000107686.1
ENSMUST00000107684.1 |
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) |
| chr4_+_148039035 | 0.39 |
ENSMUST00000097788.4
|
Mthfr
|
5,10-methylenetetrahydrofolate reductase |
| chr11_-_96075655 | 0.38 |
ENSMUST00000090541.5
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit c1 (subunit 9) |
| chr19_+_8819401 | 0.38 |
ENSMUST00000096753.3
|
Hnrnpul2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr3_+_144570409 | 0.37 |
ENSMUST00000082437.3
|
Sep15
|
selenoprotein |
| chr5_+_30869623 | 0.36 |
ENSMUST00000114716.1
|
Tmem214
|
transmembrane protein 214 |
| chr7_-_16874845 | 0.35 |
ENSMUST00000181501.1
|
9330104G04Rik
|
RIKEN cDNA 9330104G04 gene |
| chr6_-_22356176 | 0.34 |
ENSMUST00000081288.7
|
Fam3c
|
family with sequence similarity 3, member C |
| chr12_+_69241832 | 0.34 |
ENSMUST00000063445.6
|
Klhdc1
|
kelch domain containing 1 |
| chr6_-_22356068 | 0.33 |
ENSMUST00000163963.1
ENSMUST00000165576.1 |
Fam3c
|
family with sequence similarity 3, member C |
| chr3_+_96697076 | 0.32 |
ENSMUST00000162778.2
ENSMUST00000064900.9 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr10_+_111164794 | 0.32 |
ENSMUST00000105275.1
ENSMUST00000095310.1 |
Osbpl8
|
oxysterol binding protein-like 8 |
| chr17_+_35424842 | 0.31 |
ENSMUST00000174699.1
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr5_+_30869579 | 0.31 |
ENSMUST00000046349.7
|
Tmem214
|
transmembrane protein 214 |
| chr10_-_34418465 | 0.29 |
ENSMUST00000099973.3
ENSMUST00000105512.1 ENSMUST00000047885.7 |
Nt5dc1
|
5'-nucleotidase domain containing 1 |
| chr11_+_5099406 | 0.29 |
ENSMUST00000134267.1
ENSMUST00000036320.5 ENSMUST00000150632.1 |
Rhbdd3
|
rhomboid domain containing 3 |
| chr10_-_19014549 | 0.29 |
ENSMUST00000146388.1
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chr2_+_153875045 | 0.28 |
ENSMUST00000028983.2
|
Bpifb2
|
BPI fold containing family B, member 2 |
| chr11_+_100415722 | 0.27 |
ENSMUST00000107400.2
|
Fkbp10
|
FK506 binding protein 10 |
| chr7_-_31115227 | 0.25 |
ENSMUST00000168884.1
ENSMUST00000108102.2 |
Hpn
|
hepsin |
| chr14_+_26638074 | 0.24 |
ENSMUST00000022429.2
|
Arf4
|
ADP-ribosylation factor 4 |
| chr13_-_90089513 | 0.24 |
ENSMUST00000160232.1
|
Xrcc4
|
X-ray repair complementing defective repair in Chinese hamster cells 4 |
| chr10_-_81060134 | 0.24 |
ENSMUST00000005067.5
|
Sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr4_-_129742275 | 0.23 |
ENSMUST00000066257.5
|
Khdrbs1
|
KH domain containing, RNA binding, signal transduction associated 1 |
| chr6_-_38299236 | 0.23 |
ENSMUST00000058524.2
|
Zc3hav1l
|
zinc finger CCCH-type, antiviral 1-like |
| chr10_-_19015347 | 0.23 |
ENSMUST00000019997.4
|
Tnfaip3
|
tumor necrosis factor, alpha-induced protein 3 |
| chrX_+_157818435 | 0.23 |
ENSMUST00000087157.4
|
Klhl34
|
kelch-like 34 |
| chr1_+_75142775 | 0.23 |
ENSMUST00000097694.4
|
Fam134a
|
family with sequence similarity 134, member A |
| chr6_-_101377342 | 0.23 |
ENSMUST00000151175.1
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr3_-_88177671 | 0.22 |
ENSMUST00000181837.1
|
1700113A16Rik
|
RIKEN cDNA 1700113A16 gene |
| chr16_+_5050012 | 0.22 |
ENSMUST00000052449.5
|
Ubn1
|
ubinuclein 1 |
| chr13_-_24206281 | 0.21 |
ENSMUST00000123076.1
|
Lrrc16a
|
leucine rich repeat containing 16A |
| chr5_+_63812447 | 0.21 |
ENSMUST00000081747.3
|
0610040J01Rik
|
RIKEN cDNA 0610040J01 gene |
| chr14_+_26638237 | 0.20 |
ENSMUST00000112318.3
|
Arf4
|
ADP-ribosylation factor 4 |
| chr11_-_51756378 | 0.20 |
ENSMUST00000109092.1
ENSMUST00000064297.4 ENSMUST00000109097.2 |
Sec24a
|
Sec24 related gene family, member A (S. cerevisiae) |
| chr14_-_26638183 | 0.20 |
ENSMUST00000166902.1
|
4930570N19Rik
|
RIKEN cDNA 4930570N19 gene |
| chr9_-_115310421 | 0.20 |
ENSMUST00000035010.8
|
Stt3b
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) |
| chr4_+_44756553 | 0.19 |
ENSMUST00000107824.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr11_-_5099223 | 0.19 |
ENSMUST00000079949.6
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr17_+_35424870 | 0.19 |
ENSMUST00000113879.3
|
H2-Q6
|
histocompatibility 2, Q region locus 6 |
| chr5_+_108268897 | 0.18 |
ENSMUST00000031190.4
|
Dr1
|
down-regulator of transcription 1 |
| chr4_+_44756609 | 0.17 |
ENSMUST00000143385.1
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr10_-_119240006 | 0.16 |
ENSMUST00000020315.6
|
Cand1
|
cullin associated and neddylation disassociated 1 |
| chr3_-_89773221 | 0.16 |
ENSMUST00000038450.1
|
4632404H12Rik
|
RIKEN cDNA 4632404H12 gene |
| chr7_+_30421724 | 0.15 |
ENSMUST00000108176.1
|
Nfkbid
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, delta |
| chr19_+_6975048 | 0.15 |
ENSMUST00000070850.6
|
Ppp1r14b
|
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
| chr2_-_155473788 | 0.14 |
ENSMUST00000109670.1
ENSMUST00000123293.1 |
Ncoa6
|
nuclear receptor coactivator 6 |
| chr4_-_132075250 | 0.12 |
ENSMUST00000105970.1
ENSMUST00000105975.1 |
Epb4.1
|
erythrocyte protein band 4.1 |
| chr1_-_36683115 | 0.11 |
ENSMUST00000170295.1
ENSMUST00000114981.1 |
Fam178b
|
family with sequence similarity 178, member B |
| chr5_+_31054821 | 0.11 |
ENSMUST00000174367.1
ENSMUST00000170329.1 ENSMUST00000031049.6 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr3_-_20155069 | 0.11 |
ENSMUST00000184552.1
ENSMUST00000178328.1 |
Gyg
|
glycogenin |
| chr15_+_10714836 | 0.10 |
ENSMUST00000180604.1
|
4930556M19Rik
|
RIKEN cDNA 4930556M19 gene |
| chr5_-_31154152 | 0.09 |
ENSMUST00000114632.1
ENSMUST00000114631.2 ENSMUST00000067186.6 ENSMUST00000137165.1 ENSMUST00000131391.1 ENSMUST00000141823.1 ENSMUST00000154241.1 |
Mpv17
|
MpV17 mitochondrial inner membrane protein |
| chrX_+_7822289 | 0.09 |
ENSMUST00000009875.4
|
Kcnd1
|
potassium voltage-gated channel, Shal-related family, member 1 |
| chr17_-_56074932 | 0.08 |
ENSMUST00000019722.5
|
Ubxn6
|
UBX domain protein 6 |
| chr5_-_100416115 | 0.08 |
ENSMUST00000182886.1
ENSMUST00000094578.4 |
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr11_+_70000578 | 0.08 |
ENSMUST00000019362.8
|
Dvl2
|
dishevelled 2, dsh homolog (Drosophila) |
| chr11_-_5099053 | 0.08 |
ENSMUST00000093365.5
ENSMUST00000073308.4 |
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr10_+_20148457 | 0.07 |
ENSMUST00000020173.8
|
Map7
|
microtubule-associated protein 7 |
| chr9_-_106887000 | 0.07 |
ENSMUST00000055843.7
|
Rbm15b
|
RNA binding motif protein 15B |
| chr11_-_5099036 | 0.06 |
ENSMUST00000102930.3
|
Ewsr1
|
Ewing sarcoma breakpoint region 1 |
| chr4_+_128654686 | 0.05 |
ENSMUST00000030588.6
ENSMUST00000136377.1 |
Phc2
|
polyhomeotic-like 2 (Drosophila) |
| chr17_-_24163668 | 0.05 |
ENSMUST00000040735.5
|
Amdhd2
|
amidohydrolase domain containing 2 |
| chr10_+_61648552 | 0.05 |
ENSMUST00000020286.6
|
Ppa1
|
pyrophosphatase (inorganic) 1 |
| chr11_-_55033398 | 0.04 |
ENSMUST00000108883.3
ENSMUST00000102727.2 |
Anxa6
|
annexin A6 |
| chr5_+_31054766 | 0.04 |
ENSMUST00000013773.5
ENSMUST00000114646.1 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr5_-_35679416 | 0.03 |
ENSMUST00000114233.2
|
Htra3
|
HtrA serine peptidase 3 |
| chr2_+_164833781 | 0.03 |
ENSMUST00000143780.1
|
Ctsa
|
cathepsin A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Creb3l2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 4.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.6 | 1.8 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.6 | 2.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.5 | 7.6 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.4 | 1.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.4 | 1.5 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.4 | 1.1 | GO:0009446 | putrescine biosynthetic process(GO:0009446) |
| 0.3 | 1.4 | GO:0035666 | TRIF-dependent toll-like receptor signaling pathway(GO:0035666) |
| 0.3 | 1.0 | GO:0070625 | zymogen granule exocytosis(GO:0070625) |
| 0.3 | 0.8 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.2 | 0.9 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.2 | 1.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 0.8 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.2 | 0.8 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.2 | 1.3 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.2 | 3.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.2 | 5.9 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.2 | 0.7 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) |
| 0.2 | 0.5 | GO:0034148 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.2 | 1.3 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) regulation of pro-B cell differentiation(GO:2000973) |
| 0.2 | 0.5 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 1.2 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.1 | 1.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 1.1 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 1.0 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.1 | 0.6 | GO:2000317 | negative regulation of T-helper 17 type immune response(GO:2000317) negative regulation of T-helper 17 cell differentiation(GO:2000320) |
| 0.1 | 1.0 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 5.0 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 0.3 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 1.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 0.5 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.1 | 3.9 | GO:0006383 | transcription from RNA polymerase III promoter(GO:0006383) |
| 0.1 | 1.2 | GO:0014842 | regulation of skeletal muscle satellite cell proliferation(GO:0014842) |
| 0.1 | 0.3 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.2 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.2 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 1.9 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.1 | 0.9 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.1 | 0.5 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.1 | 0.2 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.1 | 1.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.1 | 1.9 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 1.0 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.1 | 1.0 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.8 | GO:0033617 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.1 | 1.8 | GO:0032292 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.1 | 1.8 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.7 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.5 | GO:0051683 | establishment of Golgi localization(GO:0051683) |
| 0.0 | 0.8 | GO:1903206 | negative regulation of hydrogen peroxide-induced cell death(GO:1903206) |
| 0.0 | 0.9 | GO:0046653 | tetrahydrofolate metabolic process(GO:0046653) |
| 0.0 | 1.2 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.4 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 1.4 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.9 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.8 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.6 | GO:0009251 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.1 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.0 | 1.1 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 2.1 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 0.8 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.7 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0044340 | canonical Wnt signaling pathway involved in regulation of cell proliferation(GO:0044340) |
| 0.0 | 0.2 | GO:0070245 | positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 1.9 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.5 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 1.3 | GO:0007030 | Golgi organization(GO:0007030) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 7.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.4 | 3.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.4 | 1.5 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.3 | 1.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 1.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.2 | 3.9 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.2 | 1.4 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.1 | 2.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.1 | 1.0 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 0.5 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.8 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.1 | 4.3 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 1.8 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.7 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.1 | 2.8 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.8 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 1.9 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.6 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.5 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.6 | 1.9 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.3 | 1.3 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.3 | 0.9 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.3 | 2.9 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.3 | 0.9 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.3 | 0.8 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.2 | 1.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.2 | 1.2 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.2 | 0.6 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.2 | 3.9 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.2 | 0.8 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.2 | 1.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 0.6 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.8 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.1 | 1.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 0.7 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 2.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 0.8 | GO:0008556 | sodium:potassium-exchanging ATPase activity(GO:0005391) potassium-transporting ATPase activity(GO:0008556) steroid hormone binding(GO:1990239) |
| 0.1 | 1.3 | GO:0048038 | quinone binding(GO:0048038) |
| 0.1 | 1.0 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.1 | 7.7 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.9 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 1.7 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.2 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 1.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.8 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.8 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.5 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 1.8 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.7 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 1.6 | GO:0050661 | NADP binding(GO:0050661) |
| 0.0 | 0.8 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 1.1 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 1.1 | GO:0008378 | galactosyltransferase activity(GO:0008378) |
| 0.0 | 0.7 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.5 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.5 | GO:0016790 | thiolester hydrolase activity(GO:0016790) |
| 0.0 | 1.9 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 1.5 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 2.2 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.0 | 0.7 | GO:0005507 | copper ion binding(GO:0005507) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 0.7 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 1.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 1.4 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 1.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 2.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.9 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.0 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.2 | 5.9 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.2 | 3.9 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 3.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.4 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.1 | 1.0 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.1 | 1.2 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.1 | 1.1 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.0 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.9 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 1.1 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.1 | 2.8 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 1.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.8 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.6 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 1.8 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.6 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.7 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 1.1 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.6 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 1.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.8 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.5 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 1.7 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |