Project
12D miR HR13_24
Navigation
Downloads
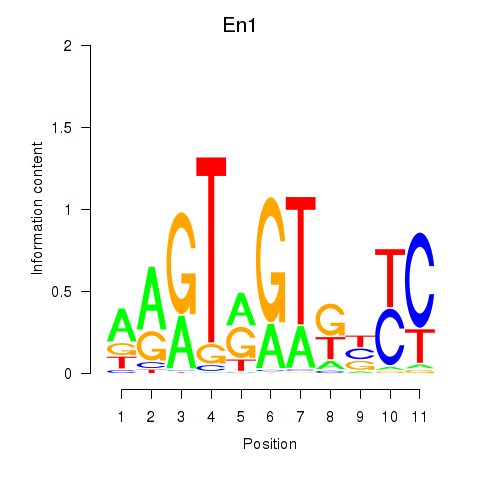
Results for En1
Z-value: 1.57
Transcription factors associated with En1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
En1
|
ENSMUSG00000058665.7 | engrailed 1 |
Activity profile of En1 motif
Sorted Z-values of En1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_71963713 | 3.04 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chr13_-_3945349 | 2.69 |
ENSMUST00000058610.7
|
Ucn3
|
urocortin 3 |
| chr1_-_158814469 | 2.67 |
ENSMUST00000161589.2
|
Pappa2
|
pappalysin 2 |
| chr12_-_119238794 | 2.49 |
ENSMUST00000026360.8
|
Itgb8
|
integrin beta 8 |
| chr4_-_131672133 | 2.12 |
ENSMUST00000144212.1
|
Gm12962
|
predicted gene 12962 |
| chr11_+_115154139 | 2.03 |
ENSMUST00000021076.5
|
Rab37
|
RAB37, member of RAS oncogene family |
| chr7_-_101869012 | 1.91 |
ENSMUST00000123321.1
|
Folr1
|
folate receptor 1 (adult) |
| chr7_-_101868667 | 1.73 |
ENSMUST00000150184.1
|
Folr1
|
folate receptor 1 (adult) |
| chr9_+_46998931 | 1.69 |
ENSMUST00000178065.1
|
Gm4791
|
predicted gene 4791 |
| chr1_+_171155512 | 1.66 |
ENSMUST00000111334.1
|
Mpz
|
myelin protein zero |
| chr10_+_34483400 | 1.60 |
ENSMUST00000019913.7
ENSMUST00000170771.1 |
Frk
|
fyn-related kinase |
| chr7_+_30699783 | 1.57 |
ENSMUST00000013227.7
|
2200002J24Rik
|
RIKEN cDNA 2200002J24 gene |
| chr3_+_141465564 | 1.38 |
ENSMUST00000106236.2
ENSMUST00000075282.3 |
Unc5c
|
unc-5 homolog C (C. elegans) |
| chr19_-_32061438 | 1.38 |
ENSMUST00000096119.4
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr8_+_82863351 | 1.37 |
ENSMUST00000078525.5
|
Rnf150
|
ring finger protein 150 |
| chr2_-_103283760 | 1.34 |
ENSMUST00000111174.1
|
Ehf
|
ets homologous factor |
| chr15_-_75747922 | 1.31 |
ENSMUST00000062002.4
|
Mafa
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein A (avian) |
| chr4_-_106799779 | 1.27 |
ENSMUST00000145061.1
ENSMUST00000102762.3 |
Acot11
|
acyl-CoA thioesterase 11 |
| chr3_-_113532288 | 1.27 |
ENSMUST00000132353.1
|
Amy2a1
|
amylase 2a1 |
| chr9_-_106891870 | 1.27 |
ENSMUST00000160503.1
ENSMUST00000159620.2 ENSMUST00000160978.1 |
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr8_-_24948771 | 1.24 |
ENSMUST00000119720.1
ENSMUST00000121438.2 |
Adam32
|
a disintegrin and metallopeptidase domain 32 |
| chr2_+_90885860 | 1.23 |
ENSMUST00000111466.2
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr15_+_85510812 | 1.22 |
ENSMUST00000079690.2
|
Gm4825
|
predicted pseudogene 4825 |
| chr13_+_67833235 | 1.22 |
ENSMUST00000060609.7
|
Gm10037
|
predicted gene 10037 |
| chr9_-_95815389 | 1.21 |
ENSMUST00000119760.1
|
Pls1
|
plastin 1 (I-isoform) |
| chr3_-_117868821 | 1.18 |
ENSMUST00000167877.1
ENSMUST00000169812.1 |
Snx7
|
sorting nexin 7 |
| chr3_+_141465592 | 1.16 |
ENSMUST00000130636.1
|
Unc5c
|
unc-5 homolog C (C. elegans) |
| chr4_-_42168603 | 1.11 |
ENSMUST00000098121.3
|
Gm13305
|
predicted gene 13305 |
| chr3_+_114904062 | 1.10 |
ENSMUST00000081752.6
|
Olfm3
|
olfactomedin 3 |
| chr17_+_69969217 | 1.08 |
ENSMUST00000060072.5
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr11_+_114727384 | 1.06 |
ENSMUST00000069325.7
|
Dnaic2
|
dynein, axonemal, intermediate chain 2 |
| chr6_+_29859686 | 1.05 |
ENSMUST00000134438.1
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr3_-_145032765 | 1.05 |
ENSMUST00000029919.5
|
Clca3
|
chloride channel calcium activated 3 |
| chr9_-_56928350 | 1.02 |
ENSMUST00000050916.5
|
Snx33
|
sorting nexin 33 |
| chr10_-_118295038 | 1.01 |
ENSMUST00000163808.1
|
Iltifb
|
interleukin 10-related T cell-derived inducible factor beta |
| chr11_+_69991633 | 1.00 |
ENSMUST00000108592.1
|
Gabarap
|
gamma-aminobutyric acid receptor associated protein |
| chr3_-_75270073 | 0.99 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr9_-_106891401 | 0.99 |
ENSMUST00000069036.7
|
Manf
|
mesencephalic astrocyte-derived neurotrophic factor |
| chr4_+_102421518 | 0.99 |
ENSMUST00000106904.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_93452679 | 0.98 |
ENSMUST00000111257.1
ENSMUST00000145553.1 |
Cd82
|
CD82 antigen |
| chr6_+_70726430 | 0.94 |
ENSMUST00000103410.1
|
Igkc
|
immunoglobulin kappa constant |
| chr11_+_93098404 | 0.93 |
ENSMUST00000107859.1
ENSMUST00000042943.6 ENSMUST00000107861.1 ENSMUST00000107858.2 |
Car10
|
carbonic anhydrase 10 |
| chr2_+_120567687 | 0.92 |
ENSMUST00000028743.3
ENSMUST00000116437.1 ENSMUST00000153580.1 ENSMUST00000142278.1 |
Snap23
|
synaptosomal-associated protein 23 |
| chr4_-_129614254 | 0.91 |
ENSMUST00000106037.2
ENSMUST00000179209.1 |
Dcdc2b
|
doublecortin domain containing 2b |
| chr4_-_47010781 | 0.91 |
ENSMUST00000135777.1
|
Gm568
|
predicted gene 568 |
| chr5_-_88527841 | 0.90 |
ENSMUST00000087033.3
|
Igj
|
immunoglobulin joining chain |
| chr3_+_94693556 | 0.88 |
ENSMUST00000090848.3
ENSMUST00000173981.1 ENSMUST00000173849.1 ENSMUST00000174223.1 |
Selenbp2
|
selenium binding protein 2 |
| chr15_+_10177623 | 0.88 |
ENSMUST00000124470.1
|
Prlr
|
prolactin receptor |
| chr11_-_59809774 | 0.87 |
ENSMUST00000047706.2
ENSMUST00000102697.3 |
Flcn
|
folliculin |
| chr12_-_50649190 | 0.87 |
ENSMUST00000002765.7
|
Prkd1
|
protein kinase D1 |
| chr14_+_103513328 | 0.84 |
ENSMUST00000095576.3
|
Scel
|
sciellin |
| chr17_+_34197715 | 0.83 |
ENSMUST00000173441.1
ENSMUST00000025196.8 |
Psmb8
|
proteasome (prosome, macropain) subunit, beta type 8 (large multifunctional peptidase 7) |
| chr19_-_40994133 | 0.82 |
ENSMUST00000117695.1
|
Blnk
|
B cell linker |
| chr3_+_89246397 | 0.82 |
ENSMUST00000168900.1
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr7_-_112159034 | 0.82 |
ENSMUST00000033036.5
|
Dkk3
|
dickkopf homolog 3 (Xenopus laevis) |
| chr11_+_108682602 | 0.82 |
ENSMUST00000106718.3
ENSMUST00000106715.1 ENSMUST00000106724.3 |
Cep112
|
centrosomal protein 112 |
| chr5_+_23787691 | 0.77 |
ENSMUST00000030852.6
ENSMUST00000120869.1 ENSMUST00000117783.1 ENSMUST00000115113.2 |
Rint1
|
RAD50 interactor 1 |
| chr10_-_61273242 | 0.77 |
ENSMUST00000120336.1
|
Adamts14
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 14 |
| chr16_+_17405981 | 0.76 |
ENSMUST00000023449.8
|
Snap29
|
synaptosomal-associated protein 29 |
| chr7_+_125707945 | 0.76 |
ENSMUST00000148701.1
|
D430042O09Rik
|
RIKEN cDNA D430042O09 gene |
| chr16_+_4886100 | 0.75 |
ENSMUST00000070658.8
ENSMUST00000023159.8 |
Mgrn1
|
mahogunin, ring finger 1 |
| chr12_-_83921809 | 0.74 |
ENSMUST00000135962.1
ENSMUST00000155112.1 ENSMUST00000136848.1 ENSMUST00000126943.1 |
Numb
|
numb gene homolog (Drosophila) |
| chr9_+_72958785 | 0.73 |
ENSMUST00000098567.2
ENSMUST00000034734.8 |
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 homolog (human) |
| chr4_-_129558387 | 0.73 |
ENSMUST00000067240.4
|
Lck
|
lymphocyte protein tyrosine kinase |
| chr5_-_120887582 | 0.72 |
ENSMUST00000086368.5
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr2_+_105130883 | 0.72 |
ENSMUST00000111098.1
ENSMUST00000111099.1 |
Wt1
|
Wilms tumor 1 homolog |
| chrX_-_162643575 | 0.71 |
ENSMUST00000101102.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr7_+_27486910 | 0.71 |
ENSMUST00000008528.7
|
Sertad1
|
SERTA domain containing 1 |
| chr3_+_89245952 | 0.70 |
ENSMUST00000040888.5
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr13_-_56895737 | 0.70 |
ENSMUST00000022023.6
ENSMUST00000109871.1 |
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr14_-_51146757 | 0.69 |
ENSMUST00000080126.2
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr1_-_40085823 | 0.69 |
ENSMUST00000181756.1
|
Gm16894
|
predicted gene, 16894 |
| chr5_+_110100414 | 0.69 |
ENSMUST00000141066.1
|
Plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr6_+_142298419 | 0.69 |
ENSMUST00000041993.2
|
Iapp
|
islet amyloid polypeptide |
| chr7_+_30169861 | 0.68 |
ENSMUST00000085668.4
|
Gm5113
|
predicted gene 5113 |
| chr2_-_32847231 | 0.68 |
ENSMUST00000050000.9
|
Stxbp1
|
syntaxin binding protein 1 |
| chr4_-_111902754 | 0.67 |
ENSMUST00000102719.1
ENSMUST00000102721.1 |
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr6_+_71282280 | 0.66 |
ENSMUST00000080949.7
|
Krcc1
|
lysine-rich coiled-coil 1 |
| chr17_-_84682932 | 0.66 |
ENSMUST00000066175.3
|
Abcg5
|
ATP-binding cassette, sub-family G (WHITE), member 5 |
| chr12_+_8012359 | 0.65 |
ENSMUST00000171239.1
|
Apob
|
apolipoprotein B |
| chr2_+_174450678 | 0.65 |
ENSMUST00000016399.5
|
Tubb1
|
tubulin, beta 1 class VI |
| chr1_+_105780693 | 0.65 |
ENSMUST00000027559.6
|
Tnfrsf11a
|
tumor necrosis factor receptor superfamily, member 11a |
| chr6_+_29859662 | 0.64 |
ENSMUST00000128927.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr3_+_107036156 | 0.64 |
ENSMUST00000052718.3
|
Kcna3
|
potassium voltage-gated channel, shaker-related subfamily, member 3 |
| chr1_-_156036473 | 0.63 |
ENSMUST00000097527.3
ENSMUST00000027738.7 |
Tor1aip1
|
torsin A interacting protein 1 |
| chr16_-_52454074 | 0.63 |
ENSMUST00000023312.7
|
Alcam
|
activated leukocyte cell adhesion molecule |
| chr8_+_23153271 | 0.63 |
ENSMUST00000071588.6
|
Nkx6-3
|
NK6 homeobox 3 |
| chr11_-_80779989 | 0.62 |
ENSMUST00000041065.7
ENSMUST00000070997.5 |
Myo1d
|
myosin ID |
| chr12_+_59131286 | 0.62 |
ENSMUST00000176464.1
ENSMUST00000170992.2 ENSMUST00000176322.1 |
Ctage5
|
CTAGE family, member 5 |
| chr15_-_60824942 | 0.62 |
ENSMUST00000100635.3
|
Fam84b
|
family with sequence similarity 84, member B |
| chr9_-_103202113 | 0.61 |
ENSMUST00000035157.8
|
Srprb
|
signal recognition particle receptor, B subunit |
| chr11_-_99986593 | 0.61 |
ENSMUST00000105050.2
|
Krtap16-1
|
keratin associated protein 16-1 |
| chr2_+_120567652 | 0.61 |
ENSMUST00000110711.2
|
Snap23
|
synaptosomal-associated protein 23 |
| chr6_-_142278836 | 0.60 |
ENSMUST00000111825.3
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr5_-_87569023 | 0.60 |
ENSMUST00000113314.2
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr12_+_84069325 | 0.60 |
ENSMUST00000046422.4
ENSMUST00000072505.4 |
Acot5
|
acyl-CoA thioesterase 5 |
| chr1_+_60778743 | 0.60 |
ENSMUST00000130082.1
|
2310016D23Rik
|
RIKEN cDNA 2310016D23 gene |
| chr6_+_124493101 | 0.59 |
ENSMUST00000049124.9
|
C1rl
|
complement component 1, r subcomponent-like |
| chr6_+_87730869 | 0.58 |
ENSMUST00000159570.2
ENSMUST00000032132.8 |
Efcc1
|
EF hand and coiled-coil domain containing 1 |
| chr18_-_15403680 | 0.56 |
ENSMUST00000079081.6
|
Aqp4
|
aquaporin 4 |
| chr16_-_45953493 | 0.56 |
ENSMUST00000136405.1
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr10_+_112165676 | 0.56 |
ENSMUST00000170013.1
|
Caps2
|
calcyphosphine 2 |
| chr11_+_95010277 | 0.55 |
ENSMUST00000124735.1
|
Samd14
|
sterile alpha motif domain containing 14 |
| chr17_+_34914459 | 0.54 |
ENSMUST00000007249.8
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr3_+_40950631 | 0.54 |
ENSMUST00000048490.6
|
Larp1b
|
La ribonucleoprotein domain family, member 1B |
| chr4_-_129227883 | 0.54 |
ENSMUST00000106051.1
|
C77080
|
expressed sequence C77080 |
| chr1_+_57774600 | 0.54 |
ENSMUST00000167971.1
ENSMUST00000170139.1 ENSMUST00000171699.1 ENSMUST00000164302.1 |
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr17_-_33760306 | 0.54 |
ENSMUST00000173860.1
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr9_-_64341288 | 0.54 |
ENSMUST00000068367.7
|
Dis3l
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr10_-_75032528 | 0.54 |
ENSMUST00000159994.1
ENSMUST00000179546.1 ENSMUST00000160450.1 ENSMUST00000160072.1 ENSMUST00000009214.3 ENSMUST00000166088.1 |
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr14_+_44988192 | 0.53 |
ENSMUST00000046891.5
|
Ptger2
|
prostaglandin E receptor 2 (subtype EP2) |
| chr5_-_72752763 | 0.53 |
ENSMUST00000113604.3
|
Txk
|
TXK tyrosine kinase |
| chr1_+_109993982 | 0.53 |
ENSMUST00000027542.6
|
Cdh7
|
cadherin 7, type 2 |
| chr13_-_110357136 | 0.53 |
ENSMUST00000058806.5
|
Gapt
|
Grb2-binding adaptor, transmembrane |
| chr4_-_154025616 | 0.52 |
ENSMUST00000182191.1
ENSMUST00000146543.2 |
Smim1
|
small integral membrane protein 1 |
| chr10_+_127290774 | 0.51 |
ENSMUST00000026475.8
ENSMUST00000139091.1 |
Ddit3
|
DNA-damage inducible transcript 3 |
| chr17_+_47649621 | 0.50 |
ENSMUST00000145314.1
|
Usp49
|
ubiquitin specific peptidase 49 |
| chr3_+_154711125 | 0.50 |
ENSMUST00000051862.6
|
4922501L14Rik
|
RIKEN cDNA 4922501L14 gene |
| chr16_-_38800193 | 0.50 |
ENSMUST00000057767.4
|
Upk1b
|
uroplakin 1B |
| chr10_-_127311740 | 0.50 |
ENSMUST00000037290.5
ENSMUST00000171564.1 |
Mars
|
methionine-tRNA synthetase |
| chr16_-_45953565 | 0.49 |
ENSMUST00000134802.1
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr12_+_11438191 | 0.49 |
ENSMUST00000181268.1
|
4930511A02Rik
|
RIKEN cDNA 4930511A02 gene |
| chr11_+_57645417 | 0.49 |
ENSMUST00000066987.7
ENSMUST00000108846.1 |
Galnt10
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 |
| chr1_-_13660476 | 0.49 |
ENSMUST00000027071.5
|
Lactb2
|
lactamase, beta 2 |
| chr6_+_29859374 | 0.49 |
ENSMUST00000115238.3
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr19_+_60144682 | 0.48 |
ENSMUST00000065383.4
|
E330013P04Rik
|
RIKEN cDNA E330013P04 gene |
| chr9_+_56994932 | 0.48 |
ENSMUST00000034832.6
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr7_+_140218258 | 0.48 |
ENSMUST00000084460.6
|
Cd163l1
|
CD163 molecule-like 1 |
| chr3_-_81975742 | 0.48 |
ENSMUST00000029645.8
|
Tdo2
|
tryptophan 2,3-dioxygenase |
| chr11_-_94507337 | 0.47 |
ENSMUST00000040692.8
|
Mycbpap
|
MYCBP associated protein |
| chr16_-_45844303 | 0.46 |
ENSMUST00000036355.6
|
Phldb2
|
pleckstrin homology-like domain, family B, member 2 |
| chr13_+_3924686 | 0.46 |
ENSMUST00000021639.6
|
Tubal3
|
tubulin, alpha-like 3 |
| chr1_+_88211956 | 0.46 |
ENSMUST00000073049.6
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr2_+_25456830 | 0.45 |
ENSMUST00000114265.2
ENSMUST00000102918.2 |
Clic3
|
chloride intracellular channel 3 |
| chr6_-_137571007 | 0.45 |
ENSMUST00000100841.2
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr16_-_88056176 | 0.45 |
ENSMUST00000072256.5
ENSMUST00000023652.8 ENSMUST00000114137.1 |
Grik1
|
glutamate receptor, ionotropic, kainate 1 |
| chrX_-_6320717 | 0.45 |
ENSMUST00000024049.7
|
Bmp15
|
bone morphogenetic protein 15 |
| chr10_+_118141787 | 0.45 |
ENSMUST00000163238.1
ENSMUST00000020437.5 ENSMUST00000164077.1 ENSMUST00000169817.1 |
Mdm1
|
transformed mouse 3T3 cell double minute 1 |
| chr4_+_43632185 | 0.45 |
ENSMUST00000107874.2
|
Npr2
|
natriuretic peptide receptor 2 |
| chr1_-_52953179 | 0.44 |
ENSMUST00000114492.1
|
1700019D03Rik
|
RIKEN cDNA 1700019D03 gene |
| chr15_+_102028216 | 0.44 |
ENSMUST00000023803.6
|
Krt18
|
keratin 18 |
| chr11_-_79504078 | 0.44 |
ENSMUST00000164465.2
|
Omg
|
oligodendrocyte myelin glycoprotein |
| chr7_+_43824519 | 0.44 |
ENSMUST00000107966.3
ENSMUST00000177514.1 |
Klk6
|
kallikrein related-peptidase 6 |
| chr5_-_120907510 | 0.44 |
ENSMUST00000080322.7
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr9_+_122888471 | 0.43 |
ENSMUST00000063980.6
|
Zkscan7
|
zinc finger with KRAB and SCAN domains 7 |
| chr7_+_43824493 | 0.43 |
ENSMUST00000107968.3
|
Klk6
|
kallikrein related-peptidase 6 |
| chrX_-_21089229 | 0.43 |
ENSMUST00000040667.6
|
Zfp300
|
zinc finger protein 300 |
| chr17_-_33760451 | 0.43 |
ENSMUST00000057373.7
|
Rab11b
|
RAB11B, member RAS oncogene family |
| chr16_+_20548577 | 0.43 |
ENSMUST00000003319.5
|
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr1_+_57774842 | 0.43 |
ENSMUST00000167085.1
|
Spats2l
|
spermatogenesis associated, serine-rich 2-like |
| chr1_+_151571481 | 0.43 |
ENSMUST00000111875.1
|
Fam129a
|
family with sequence similarity 129, member A |
| chr15_-_85581809 | 0.42 |
ENSMUST00000023015.7
|
Wnt7b
|
wingless-related MMTV integration site 7B |
| chrX_+_145119501 | 0.42 |
ENSMUST00000096301.4
|
Zcchc16
|
zinc finger, CCHC domain containing 16 |
| chr4_-_125127817 | 0.42 |
ENSMUST00000036188.7
|
Zc3h12a
|
zinc finger CCCH type containing 12A |
| chr14_-_20618339 | 0.41 |
ENSMUST00000035340.7
|
Usp54
|
ubiquitin specific peptidase 54 |
| chr11_+_72314438 | 0.41 |
ENSMUST00000108504.1
|
Fbxo39
|
F-box protein 39 |
| chr9_+_44326804 | 0.41 |
ENSMUST00000054708.3
|
Dpagt1
|
dolichyl-phosphate (UDP-N-acetylglucosamine) acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr6_-_87809757 | 0.41 |
ENSMUST00000032134.7
|
Rab43
|
RAB43, member RAS oncogene family |
| chr12_+_73997749 | 0.41 |
ENSMUST00000110451.2
|
Syt16
|
synaptotagmin XVI |
| chr16_-_64771146 | 0.40 |
ENSMUST00000076991.6
|
4930453N24Rik
|
RIKEN cDNA 4930453N24 gene |
| chr18_-_35662180 | 0.40 |
ENSMUST00000025209.4
ENSMUST00000096573.2 |
Spata24
|
spermatogenesis associated 24 |
| chr11_-_69369377 | 0.40 |
ENSMUST00000092971.6
ENSMUST00000108661.1 |
Chd3
|
chromodomain helicase DNA binding protein 3 |
| chr8_+_123212857 | 0.40 |
ENSMUST00000060133.6
|
Spata33
|
spermatogenesis associated 33 |
| chr17_-_56290499 | 0.40 |
ENSMUST00000019726.6
|
Plin3
|
perilipin 3 |
| chr11_+_103133303 | 0.39 |
ENSMUST00000107037.1
|
Hexim2
|
hexamethylene bis-acetamide inducible 2 |
| chr16_-_43979050 | 0.39 |
ENSMUST00000165648.1
ENSMUST00000036321.7 |
Zdhhc23
|
zinc finger, DHHC domain containing 23 |
| chr10_-_82285278 | 0.39 |
ENSMUST00000171401.1
|
4932415D10Rik
|
RIKEN cDNA 4932415D10 gene |
| chr4_-_49383576 | 0.39 |
ENSMUST00000107698.1
|
Acnat2
|
acyl-coenzyme A amino acid N-acyltransferase 2 |
| chr15_-_75888754 | 0.39 |
ENSMUST00000184858.1
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr15_+_102407144 | 0.38 |
ENSMUST00000169619.1
|
Sp1
|
trans-acting transcription factor 1 |
| chr11_+_101552188 | 0.38 |
ENSMUST00000147239.1
|
Nbr1
|
neighbor of Brca1 gene 1 |
| chr2_+_4017727 | 0.38 |
ENSMUST00000177457.1
|
Frmd4a
|
FERM domain containing 4A |
| chr14_-_79223876 | 0.38 |
ENSMUST00000040802.4
|
Zfp957
|
zinc finger protein 957 |
| chr2_+_172393794 | 0.38 |
ENSMUST00000099061.2
ENSMUST00000103073.2 |
Cass4
|
Cas scaffolding protein family member 4 |
| chr7_+_75455534 | 0.37 |
ENSMUST00000147005.1
ENSMUST00000166315.1 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr1_+_88138364 | 0.37 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr11_+_101552135 | 0.37 |
ENSMUST00000103099.1
|
Nbr1
|
neighbor of Brca1 gene 1 |
| chr11_-_72796028 | 0.37 |
ENSMUST00000156294.1
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr9_-_64341145 | 0.37 |
ENSMUST00000120760.1
ENSMUST00000168844.2 |
Dis3l
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr2_-_26140468 | 0.36 |
ENSMUST00000133808.1
|
C330006A16Rik
|
RIKEN cDNA C330006A16 gene |
| chr16_-_13903015 | 0.36 |
ENSMUST00000115804.2
|
Pdxdc1
|
pyridoxal-dependent decarboxylase domain containing 1 |
| chr2_-_167062981 | 0.36 |
ENSMUST00000048988.7
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chrX_+_139563316 | 0.36 |
ENSMUST00000113027.1
|
Rnf128
|
ring finger protein 128 |
| chr17_-_42611313 | 0.36 |
ENSMUST00000068355.6
|
Opn5
|
opsin 5 |
| chr16_-_20730544 | 0.36 |
ENSMUST00000076422.5
|
Thpo
|
thrombopoietin |
| chr13_+_22035821 | 0.36 |
ENSMUST00000110455.2
|
Hist1h2bk
|
histone cluster 1, H2bk |
| chr6_-_3968357 | 0.36 |
ENSMUST00000031674.8
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr9_+_72438534 | 0.36 |
ENSMUST00000034746.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr17_-_34287770 | 0.35 |
ENSMUST00000174751.1
ENSMUST00000040655.6 |
H2-Aa
|
histocompatibility 2, class II antigen A, alpha |
| chr16_-_26371828 | 0.35 |
ENSMUST00000023154.2
|
Cldn1
|
claudin 1 |
| chr1_+_24177610 | 0.35 |
ENSMUST00000054588.8
|
Col9a1
|
collagen, type IX, alpha 1 |
| chr3_+_93442330 | 0.35 |
ENSMUST00000064257.5
|
Tchh
|
trichohyalin |
| chr13_+_40859768 | 0.35 |
ENSMUST00000110191.2
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr9_-_58202281 | 0.35 |
ENSMUST00000163897.1
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr1_+_88134786 | 0.35 |
ENSMUST00000113134.1
ENSMUST00000140092.1 |
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr14_+_55559993 | 0.35 |
ENSMUST00000117236.1
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chr11_-_116828000 | 0.35 |
ENSMUST00000047715.5
ENSMUST00000021170.2 |
Mxra7
|
matrix-remodelling associated 7 |
| chr3_+_55140033 | 0.35 |
ENSMUST00000118963.2
ENSMUST00000061099.7 ENSMUST00000153009.1 |
Ccdc169
|
coiled-coil domain containing 169 |
| chr11_+_67586520 | 0.34 |
ENSMUST00000108682.2
|
Gas7
|
growth arrest specific 7 |
| chr15_+_66670749 | 0.34 |
ENSMUST00000065916.7
|
Tg
|
thyroglobulin |
Network of associatons between targets according to the STRING database.
First level regulatory network of En1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.0 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.8 | 2.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.4 | 3.6 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.3 | 2.5 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.3 | 1.2 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.3 | 0.9 | GO:0045796 | negative regulation of intestinal phytosterol absorption(GO:0010949) negative regulation of intestinal cholesterol absorption(GO:0045796) intestinal phytosterol absorption(GO:0060752) negative regulation of intestinal lipid absorption(GO:1904730) |
| 0.3 | 0.9 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.3 | 0.9 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.3 | 2.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.3 | 0.8 | GO:0072425 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.2 | 0.7 | GO:0030862 | neuroblast division in subventricular zone(GO:0021849) positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.2 | 1.0 | GO:0045054 | constitutive secretory pathway(GO:0045054) |
| 0.2 | 0.7 | GO:2001076 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) regulation of metanephric ureteric bud development(GO:2001074) positive regulation of metanephric ureteric bud development(GO:2001076) |
| 0.2 | 0.6 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 1.5 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.2 | 0.8 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.2 | 0.5 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.2 | 1.2 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.2 | 1.2 | GO:1903690 | negative regulation of wound healing, spreading of epidermal cells(GO:1903690) |
| 0.2 | 0.5 | GO:2000016 | cellular response to nitrosative stress(GO:0071500) negative regulation of determination of dorsal identity(GO:2000016) |
| 0.2 | 0.7 | GO:1903294 | regulation of glutamate secretion, neurotransmission(GO:1903294) positive regulation of glutamate secretion, neurotransmission(GO:1903296) |
| 0.2 | 0.7 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.2 | 0.8 | GO:0032348 | negative regulation of aldosterone metabolic process(GO:0032345) negative regulation of aldosterone biosynthetic process(GO:0032348) negative regulation of cortisol biosynthetic process(GO:2000065) |
| 0.2 | 0.5 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.2 | 0.6 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.2 | 0.5 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.2 | 1.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.6 | GO:0032789 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.1 | 1.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.1 | 0.1 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.1 | 0.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.4 | GO:1990868 | response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.1 | 0.4 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.4 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 1.7 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.6 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 0.4 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.6 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.5 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 1.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.4 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.1 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.1 | 0.3 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.3 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.1 | 0.3 | GO:2000229 | negative regulation of NAD(P)H oxidase activity(GO:0033861) pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.1 | 1.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 2.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 0.3 | GO:1904980 | regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.1 | 0.6 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.1 | 0.6 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 1.0 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.5 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.4 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.1 | 0.4 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.1 | 0.7 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 1.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.1 | 0.2 | GO:0034147 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.9 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.1 | 0.4 | GO:0019086 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) late viral transcription(GO:0019086) |
| 0.1 | 0.8 | GO:0070474 | positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.1 | 0.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 1.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 1.5 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.1 | 0.6 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.3 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 0.5 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.1 | 0.2 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.3 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.2 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.1 | 0.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.5 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.1 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.7 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.1 | 0.3 | GO:0038001 | paracrine signaling(GO:0038001) |
| 0.1 | 0.2 | GO:1990859 | cellular response to endothelin(GO:1990859) |
| 0.1 | 0.3 | GO:0097195 | pilomotor reflex(GO:0097195) |
| 0.1 | 0.4 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 0.3 | GO:0060011 | Sertoli cell proliferation(GO:0060011) |
| 0.1 | 0.7 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 3.0 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 0.3 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.3 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.1 | 0.2 | GO:0071879 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.3 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 1.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.1 | 0.3 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.5 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.3 | GO:0006590 | thyroid hormone generation(GO:0006590) |
| 0.0 | 0.3 | GO:0097105 | presynaptic membrane assembly(GO:0097105) |
| 0.0 | 0.3 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.3 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.0 | 0.4 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:1990034 | calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0033382 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.0 | 1.7 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.3 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.0 | 0.5 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.0 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.1 | GO:1903896 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.0 | 0.2 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.0 | 1.3 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.4 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.3 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.3 | GO:1904263 | positive regulation of TORC1 signaling(GO:1904263) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.3 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.3 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.2 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.7 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.3 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0035999 | tetrahydrofolate interconversion(GO:0035999) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.3 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.4 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.4 | GO:0097284 | hepatocyte apoptotic process(GO:0097284) |
| 0.0 | 0.1 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.0 | 0.2 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.5 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.5 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.4 | GO:0003417 | growth plate cartilage development(GO:0003417) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 1.5 | GO:0035383 | acyl-CoA metabolic process(GO:0006637) thioester metabolic process(GO:0035383) |
| 0.0 | 1.8 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 0.2 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.3 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.4 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.6 | GO:0030857 | negative regulation of epithelial cell differentiation(GO:0030857) |
| 0.0 | 0.7 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.0 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0045953 | negative regulation of natural killer cell mediated cytotoxicity(GO:0045953) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0052490 | suppression by virus of host apoptotic process(GO:0019050) negative regulation by symbiont of host apoptotic process(GO:0033668) modulation by virus of host apoptotic process(GO:0039526) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) positive regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902220) |
| 0.0 | 0.3 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.4 | GO:0042036 | negative regulation of cytokine biosynthetic process(GO:0042036) |
| 0.0 | 0.4 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.7 | GO:0060395 | SMAD protein signal transduction(GO:0060395) |
| 0.0 | 0.6 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.3 | 0.8 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.3 | 0.8 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 0.5 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.2 | 0.8 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.2 | 3.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.2 | 1.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 2.7 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 1.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 1.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 0.9 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.3 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.1 | 1.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 0.4 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 1.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 1.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.1 | 0.6 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.1 | 0.7 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.1 | 2.6 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.4 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.3 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 1.5 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.3 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.6 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 1.0 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.8 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.0 | 0.3 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 2.0 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.5 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.7 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 1.0 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.8 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.4 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.7 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.4 | GO:0090545 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 1.1 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.8 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.3 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.5 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.4 | 2.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.3 | 2.7 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) |
| 0.3 | 3.6 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.3 | 2.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.2 | 1.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.2 | 0.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 0.5 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.2 | 0.8 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.7 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.1 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.4 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 1.5 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 1.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.4 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.1 | 0.7 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.1 | 0.4 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.1 | 0.7 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.3 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.1 | 0.7 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.3 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.1 | 0.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.1 | 0.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.7 | GO:0035473 | lipase binding(GO:0035473) |
| 0.1 | 0.3 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.1 | 0.6 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.5 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.5 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.1 | 0.6 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 2.3 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 0.9 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 1.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 0.7 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.8 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 1.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 1.1 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 0.3 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.9 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.1 | 0.2 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 2.3 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.6 | GO:0010857 | calcium-dependent protein kinase activity(GO:0010857) |
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.3 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.5 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 1.5 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 1.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.6 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.3 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.2 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 4.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.0 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 1.3 | GO:0004532 | exoribonuclease activity(GO:0004532) |
| 0.0 | 0.9 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.9 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.6 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) |
| 0.0 | 0.5 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.6 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.5 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.7 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.5 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.3 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0050649 | testosterone 6-beta-hydroxylase activity(GO:0050649) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.9 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.6 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.2 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 0.5 | GO:0004812 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.3 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0035591 | signaling adaptor activity(GO:0035591) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 2.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.8 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.8 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 6.3 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.7 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.0 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.9 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.5 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.0 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.8 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.8 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.7 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.1 | 2.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.5 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.5 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 2.5 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.4 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.5 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 1.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 2.5 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.6 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 1.6 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.2 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.8 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.7 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.8 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.4 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.5 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.0 | REACTOME L1CAM INTERACTIONS | Genes involved in L1CAM interactions |
| 0.0 | 0.4 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.2 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |