Project
12D miR HR13_24
Navigation
Downloads
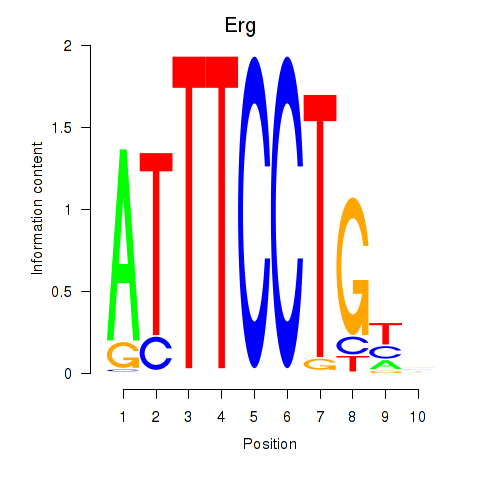
Results for Erg
Z-value: 1.84
Transcription factors associated with Erg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Erg
|
ENSMUSG00000040732.12 | ETS transcription factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Erg | mm10_v2_chr16_-_95586585_95586595 | 0.88 | 3.0e-04 | Click! |
Activity profile of Erg motif
Sorted Z-values of Erg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_44333135 | 4.12 |
ENSMUST00000047446.6
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr16_-_44332925 | 4.09 |
ENSMUST00000136381.1
|
Sidt1
|
SID1 transmembrane family, member 1 |
| chr17_+_34048280 | 3.74 |
ENSMUST00000143354.1
|
Col11a2
|
collagen, type XI, alpha 2 |
| chr15_-_98807910 | 3.46 |
ENSMUST00000075444.6
|
Ddn
|
dendrin |
| chr1_+_40515362 | 3.36 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr9_-_54661870 | 2.88 |
ENSMUST00000034822.5
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr8_+_69808672 | 2.69 |
ENSMUST00000036074.8
ENSMUST00000123453.1 |
Gmip
|
Gem-interacting protein |
| chr13_-_19619820 | 2.69 |
ENSMUST00000002885.6
|
Epdr1
|
ependymin related protein 1 (zebrafish) |
| chr17_-_32403551 | 2.55 |
ENSMUST00000135618.1
ENSMUST00000063824.7 |
Rasal3
|
RAS protein activator like 3 |
| chr2_+_118598209 | 2.53 |
ENSMUST00000038341.7
|
Bub1b
|
budding uninhibited by benzimidazoles 1 homolog, beta (S. cerevisiae) |
| chr8_+_75214502 | 2.48 |
ENSMUST00000132133.1
|
Rasd2
|
RASD family, member 2 |
| chr17_-_32403526 | 2.44 |
ENSMUST00000137458.1
|
Rasal3
|
RAS protein activator like 3 |
| chr15_-_64382908 | 2.39 |
ENSMUST00000177374.1
ENSMUST00000023008.9 ENSMUST00000110115.2 ENSMUST00000110114.3 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr6_+_21986887 | 2.35 |
ENSMUST00000151315.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr15_-_64382736 | 2.25 |
ENSMUST00000176384.1
ENSMUST00000175799.1 |
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain1 |
| chr6_+_21986438 | 2.01 |
ENSMUST00000115382.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chrX_+_110814390 | 1.87 |
ENSMUST00000078229.3
|
Pou3f4
|
POU domain, class 3, transcription factor 4 |
| chr18_+_5593566 | 1.82 |
ENSMUST00000160910.1
|
Zeb1
|
zinc finger E-box binding homeobox 1 |
| chr16_-_63864114 | 1.75 |
ENSMUST00000064405.6
|
Epha3
|
Eph receptor A3 |
| chr4_-_43523388 | 1.74 |
ENSMUST00000107913.3
ENSMUST00000030184.5 |
Tpm2
|
tropomyosin 2, beta |
| chr9_+_70207342 | 1.71 |
ENSMUST00000034745.7
|
Myo1e
|
myosin IE |
| chr2_+_55437100 | 1.70 |
ENSMUST00000112633.2
ENSMUST00000112632.1 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr17_+_32403006 | 1.69 |
ENSMUST00000065921.5
|
A530088E08Rik
|
RIKEN cDNA A530088E08 gene |
| chr6_+_129397297 | 1.65 |
ENSMUST00000032262.7
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr9_-_54661666 | 1.63 |
ENSMUST00000128624.1
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr3_+_103832562 | 1.56 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr14_+_3049285 | 1.56 |
ENSMUST00000166494.1
|
Gm2897
|
predicted gene 2897 |
| chr4_+_126556935 | 1.55 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chr12_-_28623282 | 1.55 |
ENSMUST00000036136.7
|
Colec11
|
collectin sub-family member 11 |
| chr3_+_103832741 | 1.55 |
ENSMUST00000106822.1
|
Bcl2l15
|
BCLl2-like 15 |
| chr7_-_102759465 | 1.54 |
ENSMUST00000168007.1
ENSMUST00000060187.7 |
Olfr78
|
olfactory receptor 78 |
| chr7_-_27181149 | 1.53 |
ENSMUST00000071986.6
ENSMUST00000121848.1 |
Mia
|
melanoma inhibitory activity |
| chr3_+_124321031 | 1.49 |
ENSMUST00000058994.4
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr3_-_33083016 | 1.48 |
ENSMUST00000078226.3
ENSMUST00000108224.1 |
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr7_-_116334132 | 1.47 |
ENSMUST00000170953.1
|
Rps13
|
ribosomal protein S13 |
| chr11_+_61485431 | 1.46 |
ENSMUST00000064783.3
ENSMUST00000040522.6 |
Mfap4
|
microfibrillar-associated protein 4 |
| chr10_+_7667503 | 1.45 |
ENSMUST00000040135.8
|
Nup43
|
nucleoporin 43 |
| chr12_+_75308308 | 1.41 |
ENSMUST00000118602.1
ENSMUST00000118966.1 ENSMUST00000055390.5 |
Rhoj
|
ras homolog gene family, member J |
| chrX_+_164373363 | 1.38 |
ENSMUST00000033751.7
|
Figf
|
c-fos induced growth factor |
| chr16_-_38713235 | 1.38 |
ENSMUST00000023487.4
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr2_-_73486456 | 1.36 |
ENSMUST00000141264.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr9_+_73113426 | 1.33 |
ENSMUST00000169399.1
ENSMUST00000034738.7 |
Rsl24d1
|
ribosomal L24 domain containing 1 |
| chr1_-_87394721 | 1.32 |
ENSMUST00000113212.3
|
Kcnj13
|
potassium inwardly-rectifying channel, subfamily J, member 13 |
| chr4_-_43523746 | 1.32 |
ENSMUST00000150592.1
|
Tpm2
|
tropomyosin 2, beta |
| chr2_-_32083783 | 1.30 |
ENSMUST00000056406.6
|
Fam78a
|
family with sequence similarity 78, member A |
| chr6_+_88465409 | 1.28 |
ENSMUST00000032165.9
|
Ruvbl1
|
RuvB-like protein 1 |
| chr4_-_43523595 | 1.27 |
ENSMUST00000107914.3
|
Tpm2
|
tropomyosin 2, beta |
| chr3_-_10208569 | 1.27 |
ENSMUST00000029041.4
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr6_+_129397478 | 1.27 |
ENSMUST00000112081.2
ENSMUST00000112079.2 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr16_-_38433145 | 1.26 |
ENSMUST00000002926.6
|
Pla1a
|
phospholipase A1 member A |
| chr4_+_136462250 | 1.26 |
ENSMUST00000084593.2
|
6030445D17Rik
|
RIKEN cDNA 6030445D17 gene |
| chr17_-_14694223 | 1.25 |
ENSMUST00000170872.1
|
Thbs2
|
thrombospondin 2 |
| chr4_-_45108038 | 1.24 |
ENSMUST00000107809.2
ENSMUST00000107808.2 ENSMUST00000107807.1 ENSMUST00000107810.2 |
Tomm5
|
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr6_+_134035691 | 1.23 |
ENSMUST00000081028.6
ENSMUST00000111963.1 |
Etv6
|
ets variant gene 6 (TEL oncogene) |
| chr7_-_62420139 | 1.23 |
ENSMUST00000094340.3
|
Mkrn3
|
makorin, ring finger protein, 3 |
| chr6_+_65778988 | 1.23 |
ENSMUST00000031976.7
ENSMUST00000081219.7 ENSMUST00000031973.6 ENSMUST00000172638.1 |
Prdm5
|
PR domain containing 5 |
| chr11_-_90687572 | 1.23 |
ENSMUST00000107869.2
ENSMUST00000154599.1 ENSMUST00000107868.1 ENSMUST00000020849.2 |
Tom1l1
|
target of myb1-like 1 (chicken) |
| chr7_+_105640448 | 1.22 |
ENSMUST00000058333.3
|
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr16_-_95459245 | 1.19 |
ENSMUST00000176345.1
ENSMUST00000121809.2 ENSMUST00000118113.1 ENSMUST00000122199.1 |
Erg
|
avian erythroblastosis virus E-26 (v-ets) oncogene related |
| chr9_+_32116040 | 1.18 |
ENSMUST00000174641.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr12_-_34528844 | 1.17 |
ENSMUST00000110819.2
|
Hdac9
|
histone deacetylase 9 |
| chrX_-_36902877 | 1.16 |
ENSMUST00000057093.6
|
Nkrf
|
NF-kappaB repressing factor |
| chr9_+_7445822 | 1.14 |
ENSMUST00000034497.6
|
Mmp3
|
matrix metallopeptidase 3 |
| chr3_-_51396502 | 1.12 |
ENSMUST00000108046.1
|
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr3_-_130730310 | 1.11 |
ENSMUST00000062601.7
|
Rpl34
|
ribosomal protein L34 |
| chr3_-_51396528 | 1.11 |
ENSMUST00000038154.5
|
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr7_+_105640522 | 1.10 |
ENSMUST00000106785.1
ENSMUST00000106786.1 ENSMUST00000106780.1 ENSMUST00000106784.1 |
Timm10b
|
translocase of inner mitochondrial membrane 10B |
| chr11_-_97150025 | 1.10 |
ENSMUST00000118375.1
|
Tbkbp1
|
TBK1 binding protein 1 |
| chr3_-_130730375 | 1.09 |
ENSMUST00000079085.6
|
Rpl34
|
ribosomal protein L34 |
| chr3_-_51396716 | 1.08 |
ENSMUST00000141156.1
|
Mgarp
|
mitochondria localized glutamic acid rich protein |
| chr5_-_137858034 | 1.08 |
ENSMUST00000110978.2
|
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr2_-_73453918 | 1.08 |
ENSMUST00000102679.1
|
Wipf1
|
WAS/WASL interacting protein family, member 1 |
| chr1_+_171388954 | 1.06 |
ENSMUST00000056449.8
|
Arhgap30
|
Rho GTPase activating protein 30 |
| chr8_-_119605199 | 1.05 |
ENSMUST00000093099.6
|
Taf1c
|
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, C |
| chr2_-_5714490 | 1.04 |
ENSMUST00000044009.7
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr6_+_4003926 | 1.03 |
ENSMUST00000031670.8
|
Gng11
|
guanine nucleotide binding protein (G protein), gamma 11 |
| chr2_+_30078213 | 1.02 |
ENSMUST00000150770.1
|
Pkn3
|
protein kinase N3 |
| chr14_-_31168587 | 1.02 |
ENSMUST00000036618.7
|
Stab1
|
stabilin 1 |
| chr19_-_41896132 | 1.02 |
ENSMUST00000038677.3
|
Rrp12
|
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr5_-_96164147 | 1.01 |
ENSMUST00000137207.1
|
Cnot6l
|
CCR4-NOT transcription complex, subunit 6-like |
| chr7_+_24907618 | 1.01 |
ENSMUST00000151121.1
|
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr19_+_47854970 | 1.00 |
ENSMUST00000026050.7
|
Gsto1
|
glutathione S-transferase omega 1 |
| chr9_-_20952838 | 1.00 |
ENSMUST00000004202.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr13_-_115101909 | 1.00 |
ENSMUST00000061673.7
|
Itga1
|
integrin alpha 1 |
| chr19_-_46338632 | 0.99 |
ENSMUST00000051234.8
ENSMUST00000167861.1 |
Cuedc2
|
CUE domain containing 2 |
| chr6_+_42349826 | 0.98 |
ENSMUST00000070635.6
|
Zyx
|
zyxin |
| chr16_+_36934976 | 0.97 |
ENSMUST00000023531.8
|
Hcls1
|
hematopoietic cell specific Lyn substrate 1 |
| chr13_+_109632760 | 0.96 |
ENSMUST00000135275.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr9_-_114564315 | 0.96 |
ENSMUST00000111816.2
|
Trim71
|
tripartite motif-containing 71 |
| chr11_-_103363431 | 0.96 |
ENSMUST00000092557.5
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr4_-_116627921 | 0.96 |
ENSMUST00000030456.7
|
Nasp
|
nuclear autoantigenic sperm protein (histone-binding) |
| chrX_-_150440887 | 0.96 |
ENSMUST00000163233.1
|
Tmem29
|
transmembrane protein 29 |
| chr12_+_31265234 | 0.95 |
ENSMUST00000169088.1
|
Lamb1
|
laminin B1 |
| chr12_+_98268626 | 0.94 |
ENSMUST00000075072.4
|
Gpr65
|
G-protein coupled receptor 65 |
| chr18_-_14682756 | 0.94 |
ENSMUST00000040964.6
ENSMUST00000092041.3 ENSMUST00000040924.7 |
Ss18
|
synovial sarcoma translocation, Chromosome 18 |
| chr14_-_5389049 | 0.94 |
ENSMUST00000177986.1
|
Gm3500
|
predicted gene 3500 |
| chr5_+_65863563 | 0.93 |
ENSMUST00000031106.4
|
Rhoh
|
ras homolog gene family, member H |
| chr4_-_127313980 | 0.93 |
ENSMUST00000053753.7
|
Gja4
|
gap junction protein, alpha 4 |
| chr15_-_103215285 | 0.92 |
ENSMUST00000122182.1
ENSMUST00000108813.3 ENSMUST00000127191.1 |
Cbx5
|
chromobox 5 |
| chr18_-_36783146 | 0.92 |
ENSMUST00000001416.6
|
Hars
|
histidyl-tRNA synthetase |
| chr11_-_55419898 | 0.91 |
ENSMUST00000108858.1
ENSMUST00000141530.1 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr7_-_126369543 | 0.91 |
ENSMUST00000032997.6
|
Lat
|
linker for activation of T cells |
| chr5_+_99979061 | 0.91 |
ENSMUST00000046721.1
|
4930524J08Rik
|
RIKEN cDNA 4930524J08 gene |
| chr6_+_60944472 | 0.91 |
ENSMUST00000129603.1
|
Mmrn1
|
multimerin 1 |
| chr3_+_68494208 | 0.90 |
ENSMUST00000182719.1
|
Schip1
|
schwannomin interacting protein 1 |
| chr8_-_41215146 | 0.90 |
ENSMUST00000034003.4
|
Fgl1
|
fibrinogen-like protein 1 |
| chr5_+_92387846 | 0.90 |
ENSMUST00000138687.1
ENSMUST00000124509.1 |
Art3
|
ADP-ribosyltransferase 3 |
| chr5_-_121836852 | 0.90 |
ENSMUST00000086310.1
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr1_-_131200089 | 0.89 |
ENSMUST00000068564.8
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5 |
| chr5_+_104202609 | 0.89 |
ENSMUST00000066708.5
|
Dmp1
|
dentin matrix protein 1 |
| chr14_-_7568566 | 0.89 |
ENSMUST00000163790.1
|
Gm3558
|
predicted gene 3558 |
| chr19_+_55253369 | 0.89 |
ENSMUST00000043150.4
|
Acsl5
|
acyl-CoA synthetase long-chain family member 5 |
| chr5_-_137072254 | 0.88 |
ENSMUST00000077523.3
ENSMUST00000041388.4 |
Serpine1
|
serine (or cysteine) peptidase inhibitor, clade E, member 1 |
| chr4_+_126556994 | 0.86 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chr1_-_106714217 | 0.86 |
ENSMUST00000112751.1
|
Bcl2
|
B cell leukemia/lymphoma 2 |
| chr9_-_43116514 | 0.85 |
ENSMUST00000061833.4
|
Tmem136
|
transmembrane protein 136 |
| chr15_-_76195710 | 0.85 |
ENSMUST00000023226.6
|
Plec
|
plectin |
| chr7_+_92819892 | 0.85 |
ENSMUST00000107180.1
ENSMUST00000107179.1 |
Rab30
|
RAB30, member RAS oncogene family |
| chr9_+_20868628 | 0.84 |
ENSMUST00000043911.7
|
A230050P20Rik
|
RIKEN cDNA A230050P20 gene |
| chr8_-_95142477 | 0.84 |
ENSMUST00000034240.7
ENSMUST00000169748.1 |
Kifc3
|
kinesin family member C3 |
| chr3_+_26331150 | 0.84 |
ENSMUST00000099182.2
|
A830092H15Rik
|
RIKEN cDNA A830092H15 gene |
| chr2_-_148443543 | 0.82 |
ENSMUST00000099269.3
|
Cd93
|
CD93 antigen |
| chr6_+_42350000 | 0.82 |
ENSMUST00000164375.1
|
Zyx
|
zyxin |
| chr10_-_68278713 | 0.81 |
ENSMUST00000020106.7
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr6_-_16898441 | 0.81 |
ENSMUST00000031533.7
|
Tfec
|
transcription factor EC |
| chr1_-_133921393 | 0.80 |
ENSMUST00000048432.5
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr7_+_109519139 | 0.80 |
ENSMUST00000143107.1
|
Rpl27a
|
ribosomal protein L27A |
| chr5_+_92387673 | 0.80 |
ENSMUST00000145072.1
|
Art3
|
ADP-ribosyltransferase 3 |
| chr4_-_136053343 | 0.79 |
ENSMUST00000102536.4
|
Rpl11
|
ribosomal protein L11 |
| chrX_-_75843185 | 0.79 |
ENSMUST00000137192.1
|
Pls3
|
plastin 3 (T-isoform) |
| chr8_-_121083085 | 0.79 |
ENSMUST00000182264.1
|
Fendrr
|
Foxf1 adjacent non-coding developmental regulatory RNA |
| chr3_+_10088173 | 0.78 |
ENSMUST00000061419.7
|
Gm9833
|
predicted gene 9833 |
| chr3_-_32365608 | 0.78 |
ENSMUST00000168566.1
|
Zmat3
|
zinc finger matrin type 3 |
| chr8_-_36952414 | 0.78 |
ENSMUST00000163663.2
|
Dlc1
|
deleted in liver cancer 1 |
| chr12_+_31265279 | 0.78 |
ENSMUST00000002979.8
ENSMUST00000170495.1 |
Lamb1
|
laminin B1 |
| chr8_+_70501116 | 0.77 |
ENSMUST00000127983.1
|
Crlf1
|
cytokine receptor-like factor 1 |
| chr4_+_86053887 | 0.77 |
ENSMUST00000107178.2
ENSMUST00000048885.5 ENSMUST00000141889.1 ENSMUST00000120678.1 |
Adamtsl1
|
ADAMTS-like 1 |
| chr7_-_28302238 | 0.76 |
ENSMUST00000108315.3
|
Dll3
|
delta-like 3 (Drosophila) |
| chr5_-_148392810 | 0.76 |
ENSMUST00000138257.1
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr16_+_57549232 | 0.76 |
ENSMUST00000159414.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr6_+_54039935 | 0.76 |
ENSMUST00000114403.1
|
Chn2
|
chimerin (chimaerin) 2 |
| chr2_-_156887056 | 0.76 |
ENSMUST00000029164.2
|
Sla2
|
Src-like-adaptor 2 |
| chr5_-_136198908 | 0.75 |
ENSMUST00000149151.1
ENSMUST00000151786.1 |
Prkrip1
|
Prkr interacting protein 1 (IL11 inducible) |
| chr3_+_129532386 | 0.75 |
ENSMUST00000071402.2
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr3_+_68869563 | 0.74 |
ENSMUST00000054551.2
|
1110032F04Rik
|
RIKEN cDNA 1110032F04 gene |
| chr2_-_156887172 | 0.74 |
ENSMUST00000109561.3
|
Sla2
|
Src-like-adaptor 2 |
| chr13_-_98316967 | 0.73 |
ENSMUST00000022163.8
ENSMUST00000152704.1 |
Btf3
|
basic transcription factor 3 |
| chr5_+_90931196 | 0.73 |
ENSMUST00000071652.4
|
Mthfd2l
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like |
| chr9_+_44407629 | 0.73 |
ENSMUST00000080300.7
|
Rps25
|
ribosomal protein S25 |
| chr1_+_132008285 | 0.73 |
ENSMUST00000146432.1
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr4_+_118620799 | 0.72 |
ENSMUST00000030501.8
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr7_+_101378183 | 0.72 |
ENSMUST00000084895.5
|
Arap1
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
| chr1_+_17601893 | 0.72 |
ENSMUST00000088476.2
|
Pi15
|
peptidase inhibitor 15 |
| chr17_+_80224441 | 0.72 |
ENSMUST00000069486.6
|
Gemin6
|
gem (nuclear organelle) associated protein 6 |
| chr15_+_5116589 | 0.72 |
ENSMUST00000045356.7
|
Rpl37
|
ribosomal protein L37 |
| chr15_+_8109313 | 0.72 |
ENSMUST00000163765.1
|
Nup155
|
nucleoporin 155 |
| chr15_-_78495059 | 0.72 |
ENSMUST00000089398.1
|
Il2rb
|
interleukin 2 receptor, beta chain |
| chr1_-_193264006 | 0.71 |
ENSMUST00000161737.1
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr3_-_95891938 | 0.71 |
ENSMUST00000036360.6
ENSMUST00000090476.3 |
BC028528
|
cDNA sequence BC028528 |
| chr3_-_32365643 | 0.70 |
ENSMUST00000029199.5
|
Zmat3
|
zinc finger matrin type 3 |
| chr2_+_84840612 | 0.70 |
ENSMUST00000111625.1
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr1_+_62703667 | 0.70 |
ENSMUST00000114155.1
ENSMUST00000027112.6 ENSMUST00000063594.6 ENSMUST00000114157.2 |
Nrp2
|
neuropilin 2 |
| chr19_+_11770415 | 0.69 |
ENSMUST00000167199.1
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chrX_-_75843063 | 0.68 |
ENSMUST00000114057.1
|
Pls3
|
plastin 3 (T-isoform) |
| chr12_+_102949450 | 0.68 |
ENSMUST00000179002.1
|
Unc79
|
unc-79 homolog (C. elegans) |
| chr3_+_116594959 | 0.68 |
ENSMUST00000029571.8
|
Sass6
|
spindle assembly 6 homolog (C. elegans) |
| chrX_+_93675088 | 0.67 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr4_+_8690399 | 0.67 |
ENSMUST00000127476.1
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr6_+_124712279 | 0.67 |
ENSMUST00000004375.9
|
Phb2
|
prohibitin 2 |
| chr5_+_91517615 | 0.66 |
ENSMUST00000040576.9
|
Parm1
|
prostate androgen-regulated mucin-like protein 1 |
| chr14_-_33185066 | 0.66 |
ENSMUST00000061753.8
ENSMUST00000130509.2 |
Wdfy4
|
WD repeat and FYVE domain containing 4 |
| chr9_+_122923050 | 0.66 |
ENSMUST00000051667.7
ENSMUST00000148851.1 |
Zfp105
|
zinc finger protein 105 |
| chr9_-_89738414 | 0.66 |
ENSMUST00000060700.1
|
Ankrd34c
|
ankyrin repeat domain 34C |
| chr2_+_121357714 | 0.65 |
ENSMUST00000125812.1
ENSMUST00000078222.2 ENSMUST00000125221.1 ENSMUST00000150271.1 |
Ckmt1
|
creatine kinase, mitochondrial 1, ubiquitous |
| chr2_+_30078584 | 0.65 |
ENSMUST00000045246.7
|
Pkn3
|
protein kinase N3 |
| chr5_-_113650390 | 0.64 |
ENSMUST00000047936.6
|
Cmklr1
|
chemokine-like receptor 1 |
| chr2_-_26021532 | 0.64 |
ENSMUST00000136750.1
|
Ubac1
|
ubiquitin associated domain containing 1 |
| chr6_+_134981998 | 0.64 |
ENSMUST00000167323.1
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr15_+_55557399 | 0.64 |
ENSMUST00000022998.7
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chr3_+_37639945 | 0.63 |
ENSMUST00000108109.1
ENSMUST00000038569.1 |
Spry1
|
sprouty homolog 1 (Drosophila) |
| chr6_+_134035953 | 0.63 |
ENSMUST00000164648.1
|
Etv6
|
ets variant gene 6 (TEL oncogene) |
| chr7_-_127218390 | 0.63 |
ENSMUST00000142356.1
ENSMUST00000106314.1 |
Sept1
|
septin 1 |
| chr14_+_44102654 | 0.62 |
ENSMUST00000074839.6
|
Ear2
|
eosinophil-associated, ribonuclease A family, member 2 |
| chr2_-_130284422 | 0.62 |
ENSMUST00000028892.4
|
Idh3b
|
isocitrate dehydrogenase 3 (NAD+) beta |
| chr9_-_66514567 | 0.62 |
ENSMUST00000056890.8
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr1_+_189728264 | 0.61 |
ENSMUST00000097442.2
|
Ptpn14
|
protein tyrosine phosphatase, non-receptor type 14 |
| chr14_+_27238018 | 0.61 |
ENSMUST00000049206.5
|
Arhgef3
|
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr3_-_82074639 | 0.60 |
ENSMUST00000029635.8
|
Gucy1b3
|
guanylate cyclase 1, soluble, beta 3 |
| chr9_+_107551516 | 0.60 |
ENSMUST00000093786.2
ENSMUST00000122225.1 |
Rassf1
|
Ras association (RalGDS/AF-6) domain family member 1 |
| chr15_-_55557748 | 0.60 |
ENSMUST00000172387.1
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr7_-_126200413 | 0.60 |
ENSMUST00000163959.1
|
Xpo6
|
exportin 6 |
| chr2_+_147012996 | 0.59 |
ENSMUST00000028921.5
|
Xrn2
|
5'-3' exoribonuclease 2 |
| chr10_+_128035339 | 0.58 |
ENSMUST00000092048.5
ENSMUST00000073868.7 |
Naca
|
nascent polypeptide-associated complex alpha polypeptide |
| chr5_-_121836810 | 0.58 |
ENSMUST00000118580.1
ENSMUST00000040308.7 |
Sh2b3
|
SH2B adaptor protein 3 |
| chr10_+_128232065 | 0.58 |
ENSMUST00000055539.4
ENSMUST00000105244.1 ENSMUST00000105243.2 ENSMUST00000125289.1 ENSMUST00000105242.1 |
Timeless
|
timeless circadian clock 1 |
| chr11_-_52282564 | 0.58 |
ENSMUST00000086844.3
|
Tcf7
|
transcription factor 7, T cell specific |
| chr8_+_4243264 | 0.58 |
ENSMUST00000110996.1
|
Map2k7
|
mitogen-activated protein kinase kinase 7 |
| chr7_+_87246649 | 0.58 |
ENSMUST00000068829.5
ENSMUST00000032781.7 |
Nox4
|
NADPH oxidase 4 |
| chr7_-_126200474 | 0.58 |
ENSMUST00000168189.1
|
Xpo6
|
exportin 6 |
| chr2_+_164805082 | 0.58 |
ENSMUST00000052107.4
|
Zswim3
|
zinc finger SWIM-type containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Erg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 5.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.9 | 4.6 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.9 | 8.2 | GO:0033227 | dsRNA transport(GO:0033227) |
| 0.8 | 2.5 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.7 | 3.7 | GO:0060023 | soft palate development(GO:0060023) |
| 0.6 | 1.7 | GO:0097155 | fasciculation of sensory neuron axon(GO:0097155) |
| 0.6 | 3.3 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.5 | 1.5 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.4 | 1.3 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.4 | 1.7 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.3 | 0.9 | GO:2000556 | regulation of T-helper 1 cell cytokine production(GO:2000554) positive regulation of T-helper 1 cell cytokine production(GO:2000556) |
| 0.3 | 0.9 | GO:0070173 | regulation of enamel mineralization(GO:0070173) |
| 0.3 | 1.2 | GO:0051311 | spindle assembly involved in female meiosis(GO:0007056) meiotic metaphase plate congression(GO:0051311) |
| 0.3 | 0.9 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.3 | 1.7 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.3 | 1.9 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.3 | 1.0 | GO:1990401 | embryonic lung development(GO:1990401) |
| 0.2 | 1.0 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.2 | 1.0 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.2 | 1.7 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.2 | 2.4 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.2 | 0.7 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.2 | 0.9 | GO:0019740 | regulation of nitrogen utilization(GO:0006808) nitrogen utilization(GO:0019740) regulation of cellular pH reduction(GO:0032847) |
| 0.2 | 0.6 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) ganglion morphogenesis(GO:0061552) dorsal root ganglion morphogenesis(GO:1904835) |
| 0.2 | 1.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 0.2 | 0.6 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.2 | 2.3 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.2 | 0.6 | GO:0099543 | trans-synaptic signaling by soluble gas(GO:0099543) |
| 0.2 | 0.4 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.2 | 1.2 | GO:2000504 | positive regulation of blood vessel remodeling(GO:2000504) |
| 0.2 | 1.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 0.6 | GO:1904975 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.2 | 1.7 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.2 | 1.5 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.2 | 0.5 | GO:0040010 | positive regulation of growth rate(GO:0040010) |
| 0.2 | 0.5 | GO:0032685 | negative regulation of granulocyte macrophage colony-stimulating factor production(GO:0032685) |
| 0.2 | 0.8 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.2 | 0.8 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.1 | 0.9 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.4 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.1 | 1.8 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 1.5 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.1 | 0.8 | GO:1904073 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.8 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.5 | GO:0086053 | AV node cell to bundle of His cell communication by electrical coupling(GO:0086053) |
| 0.1 | 1.9 | GO:2001054 | negative regulation of mesenchymal cell apoptotic process(GO:2001054) |
| 0.1 | 1.1 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.5 | GO:2000439 | positive regulation of monocyte extravasation(GO:2000439) |
| 0.1 | 0.9 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.1 | 0.5 | GO:0010726 | positive regulation of hydrogen peroxide metabolic process(GO:0010726) |
| 0.1 | 5.3 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.1 | 0.7 | GO:0006547 | histidine metabolic process(GO:0006547) 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.1 | 0.4 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.1 | 1.4 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 1.5 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.6 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.6 | GO:2000671 | regulation of motor neuron apoptotic process(GO:2000671) |
| 0.1 | 1.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 1.1 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.1 | 0.3 | GO:0045645 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 1.0 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.1 | 0.4 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.7 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.3 | GO:1990166 | protein localization to site of double-strand break(GO:1990166) |
| 0.1 | 1.5 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.1 | 0.3 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) |
| 0.1 | 0.3 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 2.5 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.5 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.5 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.1 | 0.5 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 0.6 | GO:0002681 | somatic diversification of T cell receptor genes(GO:0002568) somatic recombination of T cell receptor gene segments(GO:0002681) T cell receptor V(D)J recombination(GO:0033153) |
| 0.1 | 0.1 | GO:0035744 | T-helper 1 cell cytokine production(GO:0035744) |
| 0.1 | 0.4 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.1 | 0.6 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.1 | 0.4 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.1 | 1.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.6 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.1 | 1.0 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 0.3 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.1 | 0.2 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 0.6 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.3 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.1 | 0.5 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 0.4 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.4 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.2 | GO:0045918 | negative regulation of cytolysis(GO:0045918) |
| 0.1 | 0.4 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.1 | 0.1 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) |
| 0.1 | 0.2 | GO:0048866 | endodermal cell fate determination(GO:0007493) stem cell fate specification(GO:0048866) regulation of cardiac cell fate specification(GO:2000043) |
| 0.1 | 2.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.2 | GO:0032962 | positive regulation of inositol trisphosphate biosynthetic process(GO:0032962) |
| 0.1 | 0.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) negative regulation of sensory perception of pain(GO:1904057) |
| 0.1 | 0.3 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.1 | 0.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.9 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.1 | 0.8 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.1 | 1.0 | GO:0061158 | 3'-UTR-mediated mRNA destabilization(GO:0061158) |
| 0.1 | 2.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.5 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 1.5 | GO:0043486 | histone exchange(GO:0043486) |
| 0.1 | 0.2 | GO:0009946 | proximal/distal axis specification(GO:0009946) |
| 0.1 | 0.6 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.2 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.1 | 0.7 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.4 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.1 | 0.3 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.1 | 0.5 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 0.2 | GO:0010710 | calcium-independent cell-matrix adhesion(GO:0007161) regulation of collagen catabolic process(GO:0010710) |
| 0.1 | 0.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 1.0 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.1 | 1.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.1 | 0.2 | GO:1904925 | termination of signal transduction(GO:0023021) positive regulation of macromitophagy(GO:1901526) positive regulation of protein lipidation(GO:1903061) positive regulation of neuronal action potential(GO:1904457) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 1.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 0.3 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.2 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.4 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.1 | 0.2 | GO:0043323 | positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.4 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.2 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.3 | GO:0031087 | nuclear-transcribed mRNA catabolic process, deadenylation-independent decay(GO:0031086) deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.1 | GO:0035602 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) coronal suture morphogenesis(GO:0060365) fibroblast growth factor receptor signaling pathway involved in mammary gland specification(GO:0060595) mammary gland bud formation(GO:0060615) branch elongation involved in salivary gland morphogenesis(GO:0060667) mesenchymal cell differentiation involved in lung development(GO:0060915) |
| 0.0 | 0.2 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.4 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.7 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.5 | GO:0032482 | Rab protein signal transduction(GO:0032482) |
| 0.0 | 0.3 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.8 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.3 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.3 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.3 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.4 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.5 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.3 | GO:2000002 | negative regulation of DNA damage checkpoint(GO:2000002) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.6 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 1.0 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.3 | GO:0010897 | negative regulation of triglyceride catabolic process(GO:0010897) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 1.2 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.5 | GO:0001771 | immunological synapse formation(GO:0001771) |
| 0.0 | 0.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.9 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0010561 | negative regulation of glycoprotein biosynthetic process(GO:0010561) |
| 0.0 | 1.2 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.9 | GO:0097150 | neuronal stem cell population maintenance(GO:0097150) |
| 0.0 | 0.4 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 1.2 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.0 | 0.3 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.4 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.6 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.9 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 1.9 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) |
| 0.0 | 1.1 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.6 | GO:0080184 | response to stilbenoid(GO:0035634) response to phenylpropanoid(GO:0080184) |
| 0.0 | 0.3 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 1.0 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.6 | GO:0001946 | lymphangiogenesis(GO:0001946) |
| 0.0 | 0.1 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.6 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.4 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0044857 | plasma membrane raft assembly(GO:0044854) plasma membrane raft organization(GO:0044857) caveola assembly(GO:0070836) |
| 0.0 | 0.2 | GO:1900272 | negative regulation of long-term synaptic potentiation(GO:1900272) |
| 0.0 | 0.3 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.5 | GO:0032695 | negative regulation of interleukin-12 production(GO:0032695) |
| 0.0 | 0.2 | GO:0044154 | histone H4-K12 acetylation(GO:0043983) histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 1.2 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.6 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.5 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.2 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.5 | GO:0006379 | mRNA cleavage(GO:0006379) |
| 0.0 | 0.8 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 1.8 | GO:0007229 | integrin-mediated signaling pathway(GO:0007229) |
| 0.0 | 0.7 | GO:0007099 | centriole replication(GO:0007099) |
| 0.0 | 0.0 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) regulation of neutrophil activation(GO:1902563) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0032824 | peptidoglycan metabolic process(GO:0000270) natural killer cell differentiation involved in immune response(GO:0002325) peptidoglycan catabolic process(GO:0009253) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.7 | GO:0034340 | response to type I interferon(GO:0034340) |
| 0.0 | 0.3 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.7 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.0 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.2 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.3 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.7 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:2001138 | regulation of phospholipid transport(GO:2001138) positive regulation of phospholipid transport(GO:2001140) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.1 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.7 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.5 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.7 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.2 | GO:0006817 | phosphate ion transport(GO:0006817) |
| 0.0 | 0.8 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 1.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 3.3 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.7 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.0 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.0 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.0 | 0.1 | GO:0010873 | positive regulation of cholesterol esterification(GO:0010873) |
| 0.0 | 0.2 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.4 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.6 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.2 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.8 | GO:0016525 | negative regulation of angiogenesis(GO:0016525) |
| 0.0 | 0.3 | GO:2000401 | regulation of lymphocyte migration(GO:2000401) |
| 0.0 | 0.2 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.4 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.3 | GO:0002026 | regulation of the force of heart contraction(GO:0002026) |
| 0.0 | 0.2 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.4 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.3 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.5 | 2.3 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.4 | 1.5 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.4 | 4.6 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.3 | 5.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.3 | 1.1 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.3 | 1.8 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.2 | 1.7 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 1.7 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.9 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.2 | 1.0 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.2 | 2.5 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.2 | 0.8 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.2 | 0.9 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.1 | 0.6 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.1 | 0.7 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.1 | 1.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 1.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.5 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 3.3 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.1 | 0.7 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.1 | 0.7 | GO:0098536 | deuterosome(GO:0098536) |
| 0.1 | 0.2 | GO:0098799 | outer mitochondrial membrane protein complex(GO:0098799) |
| 0.1 | 1.9 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.1 | 1.2 | GO:0072687 | meiotic spindle(GO:0072687) |
| 0.1 | 0.5 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.1 | 1.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 5.9 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.1 | 3.3 | GO:0002102 | podosome(GO:0002102) |
| 0.1 | 0.2 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.8 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 1.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 3.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 1.2 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.1 | 5.6 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.1 | 0.5 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.1 | 1.9 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.9 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.5 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 1.0 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.2 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.3 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.6 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0005940 | septin ring(GO:0005940) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 1.3 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 2.0 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 3.9 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.1 | GO:0008623 | CHRAC(GO:0008623) |
| 0.0 | 1.0 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0005687 | U4 snRNP(GO:0005687) U6 snRNP(GO:0005688) |
| 0.0 | 0.8 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 0.3 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.5 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.3 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 2.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 3.8 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0030123 | AP-3 adaptor complex(GO:0030123) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 1.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.4 | GO:1902562 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 2.0 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 1.0 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 3.2 | GO:0005777 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.6 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.8 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 2.1 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.6 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 0.3 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 1.6 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.2 | GO:0051033 | nucleic acid transmembrane transporter activity(GO:0051032) RNA transmembrane transporter activity(GO:0051033) |
| 0.4 | 2.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.4 | 3.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.3 | 4.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.3 | 1.0 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.3 | 1.7 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 1.1 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.2 | 1.5 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.2 | 0.7 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.2 | 0.7 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.2 | 0.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 1.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.2 | 1.7 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.2 | 1.3 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.2 | 1.3 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.2 | 0.6 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) 5'-3' exoribonuclease activity(GO:0004534) |
| 0.2 | 1.7 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.2 | 0.6 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.4 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.1 | 0.5 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 2.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.5 | GO:0086077 | gap junction channel activity involved in AV node cell-bundle of His cell electrical coupling(GO:0086077) |
| 0.1 | 1.5 | GO:0070181 | small ribosomal subunit rRNA binding(GO:0070181) |
| 0.1 | 0.7 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.1 | 0.5 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.5 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.1 | 1.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 1.7 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.1 | 0.6 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.3 | GO:0052593 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 5.4 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.1 | 0.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.5 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.1 | 1.5 | GO:0001163 | RNA polymerase I regulatory region sequence-specific DNA binding(GO:0001163) RNA polymerase I CORE element sequence-specific DNA binding(GO:0001164) |
| 0.1 | 4.5 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.8 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 0.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 5.2 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.1 | 0.8 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.1 | 2.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.3 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.1 | 0.8 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 1.0 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.4 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.1 | 0.4 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.1 | 0.2 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.1 | 1.5 | GO:0005537 | mannose binding(GO:0005537) |
| 0.1 | 0.4 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 1.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.1 | 0.5 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.9 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.1 | 1.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.9 | GO:0102391 | long-chain fatty acid-CoA ligase activity(GO:0004467) decanoate--CoA ligase activity(GO:0102391) |
| 0.1 | 1.3 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.1 | 0.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 1.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.1 | 0.5 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 0.4 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 1.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.7 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.8 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.1 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.2 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.1 | 0.9 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.6 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 1.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.1 | 0.2 | GO:0034211 | GTP-dependent protein kinase activity(GO:0034211) |
| 0.1 | 1.2 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.8 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.5 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 1.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 7.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 1.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 14.4 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.0 | 0.8 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 1.8 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 1.5 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.3 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.4 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 1.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 1.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.7 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.7 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.5 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.7 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.4 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.3 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 1.1 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.8 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0099530 | PLC activating G-protein coupled glutamate receptor activity(GO:0001639) G-protein coupled receptor activity involved in regulation of postsynaptic membrane potential(GO:0099530) |
| 0.0 | 0.4 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.5 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) peptidoglycan receptor activity(GO:0016019) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.8 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.0 | 1.5 | GO:0004540 | ribonuclease activity(GO:0004540) |
| 0.0 | 0.4 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0003951 | NAD+ kinase activity(GO:0003951) |
| 0.0 | 0.4 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.8 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.4 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.4 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 2.6 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.9 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.2 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.4 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 2.2 | GO:0008168 | methyltransferase activity(GO:0008168) |
| 0.0 | 0.2 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.3 | GO:0019210 | kinase inhibitor activity(GO:0019210) |
| 0.0 | 0.3 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.3 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.1 | 4.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 1.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 1.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 3.8 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 5.6 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.1 | 4.9 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.1 | 4.9 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.1 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 1.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.1 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.1 | 2.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.6 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.9 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.6 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.4 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 1.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 2.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 4.5 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 1.2 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.6 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 1.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 1.2 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.0 | 1.4 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 0.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.1 | 1.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.1 | 5.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.6 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.1 | 0.5 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 0.1 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.1 | 1.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.8 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.3 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.1 | 1.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 0.5 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.1 | 7.5 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.1 | 2.2 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.1 | 1.6 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.1 | 0.5 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.1 | 3.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 2.5 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 1.3 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.0 | 2.2 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 1.0 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 1.0 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.9 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.2 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.9 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.0 | 0.6 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 1.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 6.5 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 1.4 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.7 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 1.1 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.6 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 1.4 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.0 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.7 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.3 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.6 | REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | Genes involved in Nitric oxide stimulates guanylate cyclase |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 1.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |