Project
12D miR HR13_24
Navigation
Downloads
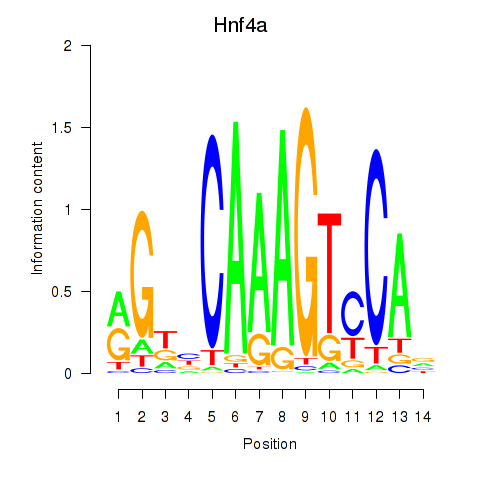
Results for Hnf4a
Z-value: 2.29
Transcription factors associated with Hnf4a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf4a
|
ENSMUSG00000017950.10 | hepatic nuclear factor 4, alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf4a | mm10_v2_chr2_+_163547148_163547188 | -0.29 | 3.8e-01 | Click! |
Activity profile of Hnf4a motif
Sorted Z-values of Hnf4a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_122895072 | 10.43 |
ENSMUST00000023820.5
|
Fabp2
|
fatty acid binding protein 2, intestinal |
| chr9_+_46240696 | 5.69 |
ENSMUST00000034585.6
|
Apoa4
|
apolipoprotein A-IV |
| chr1_-_87101590 | 4.61 |
ENSMUST00000113270.2
|
Alpi
|
alkaline phosphatase, intestinal |
| chr18_+_21072329 | 4.46 |
ENSMUST00000082235.4
|
Mep1b
|
meprin 1 beta |
| chr11_+_115877497 | 4.29 |
ENSMUST00000144032.1
|
Myo15b
|
myosin XVB |
| chr11_+_70505244 | 3.70 |
ENSMUST00000019063.2
|
Tm4sf5
|
transmembrane 4 superfamily member 5 |
| chr16_+_33794345 | 3.68 |
ENSMUST00000023520.6
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr16_+_33794008 | 3.67 |
ENSMUST00000115044.1
|
Muc13
|
mucin 13, epithelial transmembrane |
| chr12_+_7977640 | 3.53 |
ENSMUST00000171271.1
ENSMUST00000037811.6 ENSMUST00000037520.7 |
Apob
|
apolipoprotein B |
| chr6_+_71199827 | 3.42 |
ENSMUST00000067492.7
|
Fabp1
|
fatty acid binding protein 1, liver |
| chr19_+_39060998 | 2.70 |
ENSMUST00000087236.4
|
Cyp2c65
|
cytochrome P450, family 2, subfamily c, polypeptide 65 |
| chr4_+_118527229 | 2.65 |
ENSMUST00000030261.5
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chr7_-_19676749 | 2.57 |
ENSMUST00000003074.9
|
Apoc2
|
apolipoprotein C-II |
| chr15_-_82794236 | 2.56 |
ENSMUST00000006094.4
|
Cyp2d26
|
cytochrome P450, family 2, subfamily d, polypeptide 26 |
| chr9_+_44673227 | 2.56 |
ENSMUST00000034609.4
ENSMUST00000071219.5 |
Treh
|
trehalase (brush-border membrane glycoprotein) |
| chr2_+_173153048 | 2.50 |
ENSMUST00000029017.5
|
Pck1
|
phosphoenolpyruvate carboxykinase 1, cytosolic |
| chr7_-_135528645 | 2.29 |
ENSMUST00000053716.7
|
Clrn3
|
clarin 3 |
| chr2_+_174760619 | 2.21 |
ENSMUST00000029030.2
|
Edn3
|
endothelin 3 |
| chr19_+_39287074 | 2.13 |
ENSMUST00000003137.8
|
Cyp2c29
|
cytochrome P450, family 2, subfamily c, polypeptide 29 |
| chr17_+_36898110 | 2.09 |
ENSMUST00000078438.4
|
Trim31
|
tripartite motif-containing 31 |
| chr3_+_60031754 | 2.08 |
ENSMUST00000029325.3
|
Aadac
|
arylacetamide deacetylase (esterase) |
| chr1_-_82768449 | 1.97 |
ENSMUST00000027331.2
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr14_-_55758458 | 1.93 |
ENSMUST00000001497.7
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B |
| chr9_-_46235631 | 1.76 |
ENSMUST00000118649.1
|
Apoc3
|
apolipoprotein C-III |
| chr16_-_30310773 | 1.63 |
ENSMUST00000061190.6
|
Gp5
|
glycoprotein 5 (platelet) |
| chr19_+_39113898 | 1.61 |
ENSMUST00000087234.2
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr17_+_34377124 | 1.59 |
ENSMUST00000080254.5
|
Btnl1
|
butyrophilin-like 1 |
| chr3_-_152166230 | 1.55 |
ENSMUST00000046614.9
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr15_+_99099412 | 1.54 |
ENSMUST00000061295.6
|
Dnajc22
|
DnaJ (Hsp40) homolog, subfamily C, member 22 |
| chr2_-_27248335 | 1.53 |
ENSMUST00000139312.1
|
Sardh
|
sarcosine dehydrogenase |
| chr19_+_39007019 | 1.46 |
ENSMUST00000025966.4
|
Cyp2c55
|
cytochrome P450, family 2, subfamily c, polypeptide 55 |
| chr6_-_125380793 | 1.45 |
ENSMUST00000042647.6
|
Plekhg6
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
| chr10_-_128960965 | 1.39 |
ENSMUST00000026398.3
|
Mettl7b
|
methyltransferase like 7B |
| chr7_-_45759527 | 1.31 |
ENSMUST00000075571.7
|
Sult2b1
|
sulfotransferase family, cytosolic, 2B, member 1 |
| chrX_+_10252305 | 1.31 |
ENSMUST00000049910.6
|
Otc
|
ornithine transcarbamylase |
| chr15_+_10358525 | 1.27 |
ENSMUST00000110540.1
ENSMUST00000110541.1 ENSMUST00000110542.1 |
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr3_+_107877227 | 1.25 |
ENSMUST00000037375.8
|
Eps8l3
|
EPS8-like 3 |
| chr15_-_78855517 | 1.24 |
ENSMUST00000044584.4
|
Lgals2
|
lectin, galactose-binding, soluble 2 |
| chr15_+_102150502 | 1.23 |
ENSMUST00000023806.6
|
Soat2
|
sterol O-acyltransferase 2 |
| chr6_-_41636389 | 1.21 |
ENSMUST00000031902.5
|
Trpv6
|
transient receptor potential cation channel, subfamily V, member 6 |
| chr7_+_143473736 | 1.19 |
ENSMUST00000052348.5
|
Slc22a18
|
solute carrier family 22 (organic cation transporter), member 18 |
| chr4_+_120854786 | 1.19 |
ENSMUST00000071093.2
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr19_-_39740999 | 1.19 |
ENSMUST00000099472.3
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr8_-_71701796 | 1.17 |
ENSMUST00000034260.7
|
B3gnt3
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 |
| chr17_-_73950172 | 1.11 |
ENSMUST00000024866.4
|
Xdh
|
xanthine dehydrogenase |
| chr18_+_69925466 | 1.08 |
ENSMUST00000043929.4
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr15_+_10358556 | 1.04 |
ENSMUST00000022858.7
|
Agxt2
|
alanine-glyoxylate aminotransferase 2 |
| chr11_-_3504766 | 1.04 |
ENSMUST00000044507.5
|
Inpp5j
|
inositol polyphosphate 5-phosphatase J |
| chr8_-_113848615 | 1.04 |
ENSMUST00000093113.4
|
Adamts18
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 18 |
| chr7_+_29170345 | 1.01 |
ENSMUST00000033886.7
|
Ggn
|
gametogenetin |
| chr17_+_6705830 | 0.97 |
ENSMUST00000159880.1
|
Sytl3
|
synaptotagmin-like 3 |
| chr10_+_79822617 | 0.96 |
ENSMUST00000046833.4
|
Misp
|
mitotic spindle positioning |
| chr2_-_119229849 | 0.96 |
ENSMUST00000110820.2
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr4_+_110397661 | 0.96 |
ENSMUST00000106589.2
ENSMUST00000106587.2 ENSMUST00000106591.1 ENSMUST00000106592.1 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr2_+_58754910 | 0.93 |
ENSMUST00000059102.6
|
Upp2
|
uridine phosphorylase 2 |
| chr2_+_174760781 | 0.92 |
ENSMUST00000140908.1
|
Edn3
|
endothelin 3 |
| chr1_-_136234113 | 0.92 |
ENSMUST00000120339.1
ENSMUST00000048668.8 |
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene |
| chr9_-_32344237 | 0.91 |
ENSMUST00000034533.5
|
Kcnj5
|
potassium inwardly-rectifying channel, subfamily J, member 5 |
| chr4_-_94650092 | 0.90 |
ENSMUST00000107101.1
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr7_-_19950729 | 0.90 |
ENSMUST00000043440.6
|
Igsf23
|
immunoglobulin superfamily, member 23 |
| chr9_+_107975529 | 0.88 |
ENSMUST00000035216.4
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr19_-_11081088 | 0.88 |
ENSMUST00000025636.6
|
Ms4a8a
|
membrane-spanning 4-domains, subfamily A, member 8A |
| chr4_+_118526986 | 0.85 |
ENSMUST00000106367.1
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chr10_-_25536114 | 0.85 |
ENSMUST00000179685.1
|
Smlr1
|
small leucine-rich protein 1 |
| chr18_-_32036941 | 0.84 |
ENSMUST00000134663.1
|
Myo7b
|
myosin VIIB |
| chr4_-_111898695 | 0.84 |
ENSMUST00000102720.1
|
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr5_-_5694269 | 0.81 |
ENSMUST00000148333.1
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr7_+_81114816 | 0.80 |
ENSMUST00000119083.1
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr12_+_58211772 | 0.80 |
ENSMUST00000110671.2
ENSMUST00000044299.2 |
Sstr1
|
somatostatin receptor 1 |
| chr17_-_28486082 | 0.79 |
ENSMUST00000079413.3
|
Fkbp5
|
FK506 binding protein 5 |
| chr3_+_118562129 | 0.79 |
ENSMUST00000039177.7
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chrX_-_162565514 | 0.78 |
ENSMUST00000154424.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr7_+_81114799 | 0.78 |
ENSMUST00000026820.4
|
Slc28a1
|
solute carrier family 28 (sodium-coupled nucleoside transporter), member 1 |
| chr5_+_117120120 | 0.76 |
ENSMUST00000111978.1
|
Taok3
|
TAO kinase 3 |
| chr1_+_24005505 | 0.74 |
ENSMUST00000181961.1
|
Gm26524
|
predicted gene, 26524 |
| chr17_-_91088726 | 0.74 |
ENSMUST00000072671.7
ENSMUST00000174331.1 ENSMUST00000161402.3 ENSMUST00000054059.8 |
Nrxn1
|
neurexin I |
| chr4_+_110397764 | 0.73 |
ENSMUST00000097920.2
ENSMUST00000080744.6 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr18_+_69925542 | 0.72 |
ENSMUST00000080050.5
|
Ccdc68
|
coiled-coil domain containing 68 |
| chr7_-_67222412 | 0.71 |
ENSMUST00000181631.1
|
1700112J16Rik
|
RIKEN cDNA 1700112J16 gene |
| chr3_-_121815212 | 0.71 |
ENSMUST00000029770.5
|
Abcd3
|
ATP-binding cassette, sub-family D (ALD), member 3 |
| chr16_+_37580137 | 0.68 |
ENSMUST00000160847.1
|
Hgd
|
homogentisate 1, 2-dioxygenase |
| chr15_-_100424092 | 0.67 |
ENSMUST00000154676.1
|
Slc11a2
|
solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 |
| chr7_-_25615874 | 0.67 |
ENSMUST00000098663.1
|
Gm7092
|
predicted gene 7092 |
| chr6_-_65144908 | 0.66 |
ENSMUST00000031982.4
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr4_-_94650134 | 0.66 |
ENSMUST00000053419.2
|
Lrrc19
|
leucine rich repeat containing 19 |
| chr19_+_4594312 | 0.64 |
ENSMUST00000113825.2
|
Pcx
|
pyruvate carboxylase |
| chr11_-_3931960 | 0.62 |
ENSMUST00000109990.1
ENSMUST00000020710.4 ENSMUST00000109989.3 ENSMUST00000109991.1 ENSMUST00000109993.2 |
Tcn2
|
transcobalamin 2 |
| chr11_+_118988173 | 0.62 |
ENSMUST00000092373.6
ENSMUST00000106273.1 |
Enpp7
|
ectonucleotide pyrophosphatase/phosphodiesterase 7 |
| chr5_-_5694024 | 0.62 |
ENSMUST00000115425.2
ENSMUST00000115427.1 ENSMUST00000115424.2 ENSMUST00000015797.4 |
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr7_+_112953955 | 0.62 |
ENSMUST00000182858.1
|
Rassf10
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 10 |
| chr1_+_88055467 | 0.61 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr2_-_25500613 | 0.61 |
ENSMUST00000040042.4
|
C8g
|
complement component 8, gamma polypeptide |
| chr9_+_118506226 | 0.61 |
ENSMUST00000084820.4
|
Golga4
|
golgi autoantigen, golgin subfamily a, 4 |
| chr11_+_55213783 | 0.60 |
ENSMUST00000108867.1
|
Slc36a1
|
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr12_+_59131286 | 0.59 |
ENSMUST00000176464.1
ENSMUST00000170992.2 ENSMUST00000176322.1 |
Ctage5
|
CTAGE family, member 5 |
| chr19_-_10604258 | 0.59 |
ENSMUST00000037678.6
|
Dak
|
dihydroxyacetone kinase 2 homolog (yeast) |
| chr11_-_3931789 | 0.58 |
ENSMUST00000109992.1
ENSMUST00000109988.1 |
Tcn2
|
transcobalamin 2 |
| chr12_+_59131473 | 0.58 |
ENSMUST00000177162.1
|
Ctage5
|
CTAGE family, member 5 |
| chr5_+_19907774 | 0.57 |
ENSMUST00000115267.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_-_13491900 | 0.57 |
ENSMUST00000091436.5
|
Cubn
|
cubilin (intrinsic factor-cobalamin receptor) |
| chr2_-_69342600 | 0.56 |
ENSMUST00000102709.1
ENSMUST00000102710.3 ENSMUST00000180142.1 |
Abcb11
|
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr11_+_119314787 | 0.56 |
ENSMUST00000053245.6
|
Card14
|
caspase recruitment domain family, member 14 |
| chr9_-_22002599 | 0.56 |
ENSMUST00000115336.2
ENSMUST00000044926.5 |
Ccdc151
|
coiled-coil domain containing 151 |
| chr5_-_5694559 | 0.56 |
ENSMUST00000115426.2
|
Steap2
|
six transmembrane epithelial antigen of prostate 2 |
| chr4_+_41762309 | 0.53 |
ENSMUST00000108042.2
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr14_-_63245219 | 0.53 |
ENSMUST00000118022.1
ENSMUST00000067417.3 |
Gata4
|
GATA binding protein 4 |
| chr19_+_45998131 | 0.53 |
ENSMUST00000181820.1
|
4930505N22Rik
|
RIKEN cDNA 4930505N22 gene |
| chr2_-_119229885 | 0.52 |
ENSMUST00000076084.5
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr3_-_104818539 | 0.51 |
ENSMUST00000106774.1
ENSMUST00000106775.1 ENSMUST00000166979.1 ENSMUST00000136148.1 |
Mov10
|
Moloney leukemia virus 10 |
| chr2_+_91096744 | 0.51 |
ENSMUST00000132741.2
|
Spi1
|
spleen focus forming virus (SFFV) proviral integration oncogene |
| chr16_-_34095983 | 0.50 |
ENSMUST00000114973.1
ENSMUST00000114964.1 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr19_+_46152505 | 0.49 |
ENSMUST00000026254.7
|
Gbf1
|
golgi-specific brefeldin A-resistance factor 1 |
| chr3_-_104818224 | 0.49 |
ENSMUST00000002297.5
|
Mov10
|
Moloney leukemia virus 10 |
| chrX_-_16817339 | 0.49 |
ENSMUST00000040820.6
|
Maob
|
monoamine oxidase B |
| chr4_-_97778042 | 0.48 |
ENSMUST00000146447.1
|
E130114P18Rik
|
RIKEN cDNA E130114P18 gene |
| chr11_-_61267177 | 0.48 |
ENSMUST00000066277.3
ENSMUST00000074127.7 ENSMUST00000108715.2 |
Aldh3a2
|
aldehyde dehydrogenase family 3, subfamily A2 |
| chr16_-_30267524 | 0.48 |
ENSMUST00000064856.7
|
Cpn2
|
carboxypeptidase N, polypeptide 2 |
| chr6_+_86195214 | 0.47 |
ENSMUST00000032066.9
|
Tgfa
|
transforming growth factor alpha |
| chr19_+_8850785 | 0.47 |
ENSMUST00000096257.2
|
Lrrn4cl
|
LRRN4 C-terminal like |
| chr11_+_7197780 | 0.47 |
ENSMUST00000020704.7
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr6_-_122602404 | 0.47 |
ENSMUST00000112586.1
|
Apobec1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr1_+_88055377 | 0.46 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr3_-_104818266 | 0.45 |
ENSMUST00000168015.1
|
Mov10
|
Moloney leukemia virus 10 |
| chr5_-_24447587 | 0.45 |
ENSMUST00000127194.1
ENSMUST00000115033.1 ENSMUST00000123167.1 ENSMUST00000030799.8 |
Tmub1
|
transmembrane and ubiquitin-like domain containing 1 |
| chr6_-_122602345 | 0.43 |
ENSMUST00000147760.1
ENSMUST00000112585.1 |
Apobec1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr5_+_19907502 | 0.43 |
ENSMUST00000101558.3
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr19_-_8405060 | 0.42 |
ENSMUST00000064507.5
ENSMUST00000120540.1 ENSMUST00000096269.4 |
Slc22a30
|
solute carrier family 22, member 30 |
| chr12_+_59130767 | 0.42 |
ENSMUST00000175877.1
|
Ctage5
|
CTAGE family, member 5 |
| chr7_-_99695628 | 0.42 |
ENSMUST00000145381.1
|
Slco2b1
|
solute carrier organic anion transporter family, member 2b1 |
| chr13_+_67833235 | 0.42 |
ENSMUST00000060609.7
|
Gm10037
|
predicted gene 10037 |
| chr12_+_59130994 | 0.41 |
ENSMUST00000177460.1
|
Ctage5
|
CTAGE family, member 5 |
| chr11_+_101582236 | 0.40 |
ENSMUST00000039581.7
ENSMUST00000100403.2 ENSMUST00000107194.1 ENSMUST00000128614.1 |
Tmem106a
|
transmembrane protein 106A |
| chr1_+_171214013 | 0.40 |
ENSMUST00000111328.1
|
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr7_+_123214808 | 0.40 |
ENSMUST00000033035.6
|
Slc5a11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11 |
| chr1_+_193301953 | 0.40 |
ENSMUST00000016315.9
|
Lamb3
|
laminin, beta 3 |
| chr2_+_58755177 | 0.40 |
ENSMUST00000102755.3
|
Upp2
|
uridine phosphorylase 2 |
| chr4_-_126163470 | 0.39 |
ENSMUST00000097891.3
|
Sh3d21
|
SH3 domain containing 21 |
| chr16_-_34514084 | 0.39 |
ENSMUST00000089655.5
|
Kalrn
|
kalirin, RhoGEF kinase |
| chr16_-_34513944 | 0.39 |
ENSMUST00000151491.1
ENSMUST00000114960.2 |
Kalrn
|
kalirin, RhoGEF kinase |
| chr6_+_83156401 | 0.38 |
ENSMUST00000032106.4
|
1700003E16Rik
|
RIKEN cDNA 1700003E16 gene |
| chr4_+_131873608 | 0.38 |
ENSMUST00000053819.3
|
Srsf4
|
serine/arginine-rich splicing factor 4 |
| chr7_+_29170204 | 0.38 |
ENSMUST00000098609.2
|
Ggn
|
gametogenetin |
| chr11_+_3895223 | 0.38 |
ENSMUST00000055931.4
ENSMUST00000109996.1 |
Dusp18
|
dual specificity phosphatase 18 |
| chr10_+_70204675 | 0.38 |
ENSMUST00000020090.1
|
2310015B20Rik
|
RIKEN cDNA 2310015B20 gene |
| chr19_+_5088534 | 0.37 |
ENSMUST00000025811.4
|
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr1_+_157526127 | 0.37 |
ENSMUST00000111700.1
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr2_-_110305730 | 0.35 |
ENSMUST00000046233.2
|
Bbox1
|
butyrobetaine (gamma), 2-oxoglutarate dioxygenase 1 (gamma-butyrobetaine hydroxylase) |
| chr15_-_82620907 | 0.35 |
ENSMUST00000109515.1
|
Cyp2d34
|
cytochrome P450, family 2, subfamily d, polypeptide 34 |
| chr11_-_75439551 | 0.35 |
ENSMUST00000128330.1
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr4_-_83324239 | 0.34 |
ENSMUST00000048274.4
ENSMUST00000102823.3 |
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr7_+_31059342 | 0.34 |
ENSMUST00000039775.7
|
Lgi4
|
leucine-rich repeat LGI family, member 4 |
| chrX_+_36328353 | 0.34 |
ENSMUST00000016383.3
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chrX_+_6415736 | 0.34 |
ENSMUST00000143641.3
|
Shroom4
|
shroom family member 4 |
| chr4_-_138326234 | 0.34 |
ENSMUST00000105817.3
ENSMUST00000030536.6 |
Pink1
|
PTEN induced putative kinase 1 |
| chr9_+_22003035 | 0.33 |
ENSMUST00000115331.2
ENSMUST00000003493.7 |
Prkcsh
|
protein kinase C substrate 80K-H |
| chr7_+_25268387 | 0.33 |
ENSMUST00000169392.1
|
Cic
|
capicua homolog (Drosophila) |
| chr2_-_127729872 | 0.33 |
ENSMUST00000028856.2
|
Mall
|
mal, T cell differentiation protein-like |
| chr14_+_79515618 | 0.33 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr19_+_46356880 | 0.33 |
ENSMUST00000086969.6
ENSMUST00000128455.1 |
Tmem180
|
transmembrane protein 180 |
| chr16_-_22439719 | 0.33 |
ENSMUST00000079601.6
|
Etv5
|
ets variant gene 5 |
| chr17_-_15826521 | 0.32 |
ENSMUST00000170578.1
|
Rgmb
|
RGM domain family, member B |
| chr6_+_125145235 | 0.32 |
ENSMUST00000119527.1
ENSMUST00000088276.6 ENSMUST00000051171.7 ENSMUST00000117675.1 |
Iffo1
|
intermediate filament family orphan 1 |
| chr10_-_128919259 | 0.32 |
ENSMUST00000149961.1
ENSMUST00000026406.7 |
Rdh5
|
retinol dehydrogenase 5 |
| chr7_-_105600103 | 0.31 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chrX_+_48623737 | 0.31 |
ENSMUST00000114936.1
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr7_-_66689474 | 0.31 |
ENSMUST00000068980.3
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr4_-_139833524 | 0.31 |
ENSMUST00000030508.7
|
Pax7
|
paired box gene 7 |
| chr2_+_158028733 | 0.30 |
ENSMUST00000152452.1
|
Rprd1b
|
regulation of nuclear pre-mRNA domain containing 1B |
| chr7_+_28833975 | 0.29 |
ENSMUST00000066723.8
|
Lgals4
|
lectin, galactose binding, soluble 4 |
| chr19_+_42052228 | 0.28 |
ENSMUST00000164518.2
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
| chr11_+_104282371 | 0.27 |
ENSMUST00000106988.1
ENSMUST00000106989.1 |
Mapt
|
microtubule-associated protein tau |
| chr7_+_123214776 | 0.27 |
ENSMUST00000131933.1
|
Slc5a11
|
solute carrier family 5 (sodium/glucose cotransporter), member 11 |
| chr4_-_118409219 | 0.27 |
ENSMUST00000075406.5
|
Szt2
|
seizure threshold 2 |
| chr4_-_152131865 | 0.27 |
ENSMUST00000105653.1
|
Espn
|
espin |
| chr1_+_178319130 | 0.27 |
ENSMUST00000027781.6
|
Cox20
|
COX20 Cox2 chaperone |
| chr11_+_103774150 | 0.26 |
ENSMUST00000000127.5
|
Wnt3
|
wingless-related MMTV integration site 3 |
| chr17_-_35910032 | 0.26 |
ENSMUST00000141662.1
ENSMUST00000056034.6 ENSMUST00000077494.6 ENSMUST00000149277.1 ENSMUST00000061052.5 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr10_-_20724696 | 0.26 |
ENSMUST00000170265.1
|
Pde7b
|
phosphodiesterase 7B |
| chr18_+_63708689 | 0.26 |
ENSMUST00000072726.5
|
Wdr7
|
WD repeat domain 7 |
| chr6_-_83775767 | 0.25 |
ENSMUST00000014892.6
|
Tex261
|
testis expressed gene 261 |
| chr9_-_77347816 | 0.25 |
ENSMUST00000184138.1
ENSMUST00000184006.1 ENSMUST00000185144.1 ENSMUST00000034910.9 |
Mlip
|
muscular LMNA-interacting protein |
| chr4_-_83324135 | 0.25 |
ENSMUST00000030205.7
|
Ttc39b
|
tetratricopeptide repeat domain 39B |
| chr17_+_34605855 | 0.25 |
ENSMUST00000037489.8
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) |
| chr12_+_17690793 | 0.24 |
ENSMUST00000071858.3
|
Hpcal1
|
hippocalcin-like 1 |
| chr13_-_64248495 | 0.24 |
ENSMUST00000109769.2
|
Cdc14b
|
CDC14 cell division cycle 14B |
| chr1_+_171225054 | 0.24 |
ENSMUST00000111321.1
ENSMUST00000005824.5 ENSMUST00000111320.1 ENSMUST00000111319.1 |
Apoa2
|
apolipoprotein A-II |
| chr7_-_28598140 | 0.24 |
ENSMUST00000040531.8
ENSMUST00000108283.1 |
Samd4b
Pak4
|
sterile alpha motif domain containing 4B p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr2_-_77170534 | 0.23 |
ENSMUST00000111833.2
|
Ccdc141
|
coiled-coil domain containing 141 |
| chr11_-_8664499 | 0.23 |
ENSMUST00000020695.6
|
Tns3
|
tensin 3 |
| chr4_+_126609818 | 0.23 |
ENSMUST00000097886.3
ENSMUST00000164362.1 |
5730409E04Rik
|
RIKEN cDNA 5730409E04Rik gene |
| chr5_-_103977326 | 0.23 |
ENSMUST00000120320.1
|
Hsd17b13
|
hydroxysteroid (17-beta) dehydrogenase 13 |
| chr13_+_64248649 | 0.21 |
ENSMUST00000181403.1
|
1810034E14Rik
|
RIKEN cDNA 1810034E14 gene |
| chr3_-_84304762 | 0.20 |
ENSMUST00000107692.1
|
Trim2
|
tripartite motif-containing 2 |
| chr1_+_175631996 | 0.20 |
ENSMUST00000040250.8
|
Kmo
|
kynurenine 3-monooxygenase (kynurenine 3-hydroxylase) |
| chr2_+_71117923 | 0.20 |
ENSMUST00000028403.2
|
Cybrd1
|
cytochrome b reductase 1 |
| chr16_-_17928136 | 0.20 |
ENSMUST00000003622.8
|
Slc25a1
|
solute carrier family 25 (mitochondrial carrier, citrate transporter), member 1 |
| chr4_-_42853888 | 0.20 |
ENSMUST00000107979.1
|
Gm12429
|
predicted gene 12429 |
| chr4_+_134397380 | 0.20 |
ENSMUST00000105870.1
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr5_+_90561102 | 0.19 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr6_+_124663027 | 0.19 |
ENSMUST00000004381.7
|
Lpcat3
|
lysophosphatidylcholine acyltransferase 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf4a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 5.7 | GO:0034443 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) multicellular organism lipid catabolic process(GO:0044240) |
| 0.8 | 2.5 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.8 | 2.3 | GO:0042853 | glycine biosynthetic process, by transamination of glyoxylate(GO:0019265) L-alanine catabolic process(GO:0042853) |
| 0.7 | 4.3 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.6 | 2.6 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.6 | 9.1 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.5 | 1.6 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.5 | 3.1 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.5 | 1.5 | GO:1901053 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.5 | 7.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.4 | 1.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.4 | 3.5 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.4 | 1.7 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.4 | 2.1 | GO:0010898 | positive regulation of triglyceride catabolic process(GO:0010898) |
| 0.4 | 1.6 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.4 | 1.4 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.3 | 13.1 | GO:0050892 | intestinal absorption(GO:0050892) |
| 0.3 | 1.8 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.3 | 1.2 | GO:0030311 | poly-N-acetyllactosamine metabolic process(GO:0030309) poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.3 | 1.9 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.3 | 1.3 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 0.7 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.2 | 0.9 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.2 | 0.6 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.2 | 2.0 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.2 | 1.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.2 | 0.6 | GO:0046618 | drug export(GO:0046618) |
| 0.2 | 0.5 | GO:1905204 | septum secundum development(GO:0003285) cardiac muscle tissue regeneration(GO:0061026) negative regulation of connective tissue replacement(GO:1905204) |
| 0.2 | 1.2 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.2 | 0.9 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 1.3 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.4 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.1 | 0.7 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.1 | 0.7 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 1.5 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.4 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.1 | 0.9 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 1.2 | GO:0035898 | parathyroid hormone secretion(GO:0035898) calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 0.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.5 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 2.5 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.1 | 0.5 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.1 | 0.6 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.1 | 0.5 | GO:0045915 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.1 | 2.2 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 2.9 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.1 | 1.1 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:1901668 | regulation of superoxide dismutase activity(GO:1901668) |
| 0.1 | 1.0 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 0.3 | GO:1903384 | neuron intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:0036482) positive regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902958) regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903383) negative regulation of hydrogen peroxide-induced neuron intrinsic apoptotic signaling pathway(GO:1903384) |
| 0.1 | 4.5 | GO:1901998 | toxin transport(GO:1901998) |
| 0.1 | 0.5 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
| 0.1 | 0.3 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 0.5 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.2 | GO:0006842 | tricarboxylic acid transport(GO:0006842) citrate transport(GO:0015746) |
| 0.1 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.1 | 0.7 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.1 | 0.4 | GO:0015747 | urate transport(GO:0015747) |
| 0.1 | 1.9 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.1 | 0.3 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 2.2 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.0 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.3 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 1.2 | GO:0015893 | drug transport(GO:0015893) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0035963 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.0 | 0.2 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.3 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.3 | GO:0014816 | skeletal muscle satellite cell differentiation(GO:0014816) |
| 0.0 | 0.8 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.5 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.4 | GO:0048207 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.0 | 0.8 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.6 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.3 | GO:1900034 | regulation of cellular response to heat(GO:1900034) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.8 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.1 | GO:0001192 | maintenance of transcriptional fidelity during DNA-templated transcription elongation(GO:0001192) maintenance of transcriptional fidelity during DNA-templated transcription elongation from RNA polymerase II promoter(GO:0001193) |
| 0.0 | 0.5 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.2 | GO:1904668 | mitotic spindle midzone assembly(GO:0051256) positive regulation of ubiquitin protein ligase activity(GO:1904668) |
| 0.0 | 0.6 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 1.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:1901146 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.0 | 4.7 | GO:0071772 | response to BMP(GO:0071772) cellular response to BMP stimulus(GO:0071773) |
| 0.0 | 0.2 | GO:0043928 | exonucleolytic nuclear-transcribed mRNA catabolic process involved in deadenylation-dependent decay(GO:0043928) |
| 0.0 | 0.2 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.0 | 1.6 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.6 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0090043 | regulation of tubulin deacetylation(GO:0090043) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.6 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.0 | GO:0061386 | closure of optic fissure(GO:0061386) |
| 0.0 | 0.4 | GO:0048025 | negative regulation of mRNA splicing, via spliceosome(GO:0048025) |
| 0.0 | 0.2 | GO:0010039 | response to iron ion(GO:0010039) |
| 0.0 | 0.2 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 13.9 | GO:0045179 | apical cortex(GO:0045179) |
| 0.8 | 13.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.2 | 1.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.6 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.5 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.6 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.5 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.1 | 2.0 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.6 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.3 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 6.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 1.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.5 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 6.2 | GO:0016324 | apical plasma membrane(GO:0016324) |
| 0.0 | 2.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 1.2 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 5.5 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.1 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 1.1 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 1.2 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0036038 | MKS complex(GO:0036038) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.1 | 6.4 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 2.1 | 14.6 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 1.0 | 5.9 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.9 | 1.8 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.9 | 2.6 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.8 | 3.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.7 | 4.6 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.5 | 1.6 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.5 | 1.5 | GO:0008480 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.5 | 2.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.4 | 3.5 | GO:0035473 | lipase binding(GO:0035473) |
| 0.4 | 1.2 | GO:0047223 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,3-N-acetylglucosaminyltransferase activity(GO:0047223) |
| 0.3 | 1.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.3 | 1.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.3 | 0.9 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.3 | 1.3 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.3 | 4.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.2 | 2.0 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.2 | 1.1 | GO:0004854 | xanthine dehydrogenase activity(GO:0004854) oxidoreductase activity, acting on CH or CH2 groups, NAD or NADP as acceptor(GO:0016726) |
| 0.2 | 0.9 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 1.5 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.2 | 0.6 | GO:0005302 | hydrogen:amino acid symporter activity(GO:0005280) L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.2 | 1.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.7 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.2 | 0.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 1.5 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.2 | 0.9 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 1.0 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.5 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 1.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.1 | 1.2 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.1 | 2.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 2.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.1 | 0.8 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.1 | 0.5 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.7 | GO:0009374 | biotin binding(GO:0009374) |
| 0.1 | 1.0 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.1 | 0.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.1 | 0.3 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.0 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.1 | 0.5 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 1.8 | GO:0015238 | drug transmembrane transporter activity(GO:0015238) |
| 0.1 | 1.7 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.6 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 5.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.6 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.8 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 1.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 1.1 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 1.5 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.4 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.8 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 2.6 | GO:0004553 | hydrolase activity, hydrolyzing O-glycosyl compounds(GO:0004553) |
| 0.0 | 0.4 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 1.2 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.2 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.5 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.6 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0004535 | poly(A)-specific ribonuclease activity(GO:0004535) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 1.6 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.5 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.8 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.2 | GO:0005262 | calcium channel activity(GO:0005262) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.1 | 4.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.1 | 3.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 3.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 3.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 5.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.6 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.5 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.5 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.3 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 1.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.5 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.3 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.4 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.4 | 7.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.3 | 8.1 | REACTOME CHYLOMICRON MEDIATED LIPID TRANSPORT | Genes involved in Chylomicron-mediated lipid transport |
| 0.3 | 4.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.2 | 2.5 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.2 | 1.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.9 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.1 | 1.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.6 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 1.2 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 1.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 1.0 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 3.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.5 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 3.0 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.6 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.9 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.6 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.5 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.6 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 1.4 | REACTOME G ALPHA Q SIGNALLING EVENTS | Genes involved in G alpha (q) signalling events |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |