Project
12D miR HR13_24
Navigation
Downloads
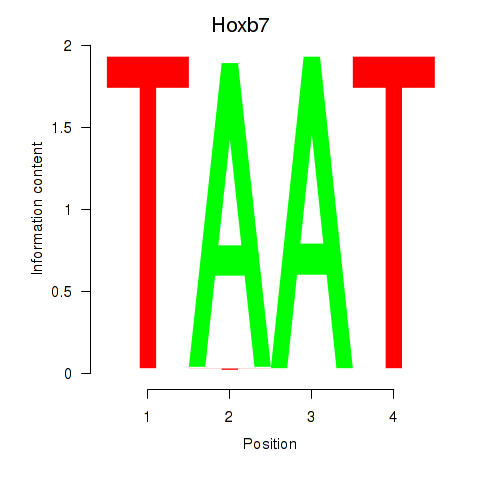
Results for Hoxb7
Z-value: 2.20
Transcription factors associated with Hoxb7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hoxb7
|
ENSMUSG00000038721.8 | homeobox B7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hoxb7 | mm10_v2_chr11_+_96286623_96286653 | -0.65 | 2.9e-02 | Click! |
Activity profile of Hoxb7 motif
Sorted Z-values of Hoxb7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_102589687 | 4.82 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr2_-_62483637 | 4.39 |
ENSMUST00000136686.1
ENSMUST00000102733.3 |
Gcg
|
glucagon |
| chr13_-_18382041 | 3.96 |
ENSMUST00000139064.2
ENSMUST00000175703.2 |
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr8_+_54954728 | 3.93 |
ENSMUST00000033915.7
|
Gpm6a
|
glycoprotein m6a |
| chr5_-_123141067 | 3.60 |
ENSMUST00000162697.1
ENSMUST00000160321.1 ENSMUST00000159637.1 |
AI480526
|
expressed sequence AI480526 |
| chr6_+_96113146 | 3.46 |
ENSMUST00000122120.1
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chr13_-_3945349 | 3.04 |
ENSMUST00000058610.7
|
Ucn3
|
urocortin 3 |
| chr14_-_110755100 | 2.84 |
ENSMUST00000078386.2
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr17_-_90088343 | 2.78 |
ENSMUST00000173917.1
|
Nrxn1
|
neurexin I |
| chr8_-_38661508 | 2.66 |
ENSMUST00000118896.1
|
Sgcz
|
sarcoglycan zeta |
| chr10_+_73821857 | 2.66 |
ENSMUST00000177128.1
ENSMUST00000064562.7 ENSMUST00000129404.2 ENSMUST00000105426.3 ENSMUST00000131321.2 ENSMUST00000126920.2 ENSMUST00000147189.2 ENSMUST00000105424.3 ENSMUST00000092420.6 ENSMUST00000105429.3 ENSMUST00000131724.2 ENSMUST00000152655.2 ENSMUST00000151116.2 ENSMUST00000155701.2 ENSMUST00000152819.2 ENSMUST00000125517.2 ENSMUST00000124046.1 ENSMUST00000149977.2 ENSMUST00000146682.1 ENSMUST00000177107.1 |
Pcdh15
|
protocadherin 15 |
| chr5_+_90518932 | 2.64 |
ENSMUST00000113179.2
ENSMUST00000128740.1 |
Afm
|
afamin |
| chr11_+_29718563 | 2.63 |
ENSMUST00000060992.5
|
Rtn4
|
reticulon 4 |
| chr9_+_46998931 | 2.61 |
ENSMUST00000178065.1
|
Gm4791
|
predicted gene 4791 |
| chr3_+_115080965 | 2.60 |
ENSMUST00000051309.8
|
Olfm3
|
olfactomedin 3 |
| chr6_+_29859662 | 2.52 |
ENSMUST00000128927.2
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr1_+_109993982 | 2.51 |
ENSMUST00000027542.6
|
Cdh7
|
cadherin 7, type 2 |
| chr6_+_29859686 | 2.40 |
ENSMUST00000134438.1
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr6_-_52226165 | 2.25 |
ENSMUST00000114425.2
|
Hoxa9
|
homeobox A9 |
| chr14_+_58072686 | 2.16 |
ENSMUST00000022545.7
|
Fgf9
|
fibroblast growth factor 9 |
| chr10_+_5639210 | 2.13 |
ENSMUST00000019906.4
|
Vip
|
vasoactive intestinal polypeptide |
| chr13_+_42680565 | 2.11 |
ENSMUST00000128646.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr4_+_82065855 | 2.09 |
ENSMUST00000151038.1
|
Gm5860
|
predicted gene 5860 |
| chr4_-_148500449 | 2.06 |
ENSMUST00000030840.3
|
Angptl7
|
angiopoietin-like 7 |
| chr8_-_8639363 | 2.00 |
ENSMUST00000152698.1
|
Efnb2
|
ephrin B2 |
| chr4_+_19280850 | 1.99 |
ENSMUST00000102999.1
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr17_+_70561739 | 1.97 |
ENSMUST00000097288.2
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr11_+_69966896 | 1.95 |
ENSMUST00000151515.1
|
Cldn7
|
claudin 7 |
| chr10_-_115185015 | 1.94 |
ENSMUST00000006949.8
|
Tph2
|
tryptophan hydroxylase 2 |
| chr17_+_70522083 | 1.91 |
ENSMUST00000148486.1
ENSMUST00000133717.1 |
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_+_21367532 | 1.90 |
ENSMUST00000055946.7
|
Gpr158
|
G protein-coupled receptor 158 |
| chr3_-_59220150 | 1.90 |
ENSMUST00000170388.1
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr10_+_34483400 | 1.89 |
ENSMUST00000019913.7
ENSMUST00000170771.1 |
Frk
|
fyn-related kinase |
| chr14_-_108914237 | 1.87 |
ENSMUST00000100322.2
|
Slitrk1
|
SLIT and NTRK-like family, member 1 |
| chr3_+_60081861 | 1.83 |
ENSMUST00000029326.5
|
Sucnr1
|
succinate receptor 1 |
| chr1_-_79440039 | 1.82 |
ENSMUST00000049972.4
|
Scg2
|
secretogranin II |
| chr13_+_89540636 | 1.82 |
ENSMUST00000022108.7
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr10_-_64090265 | 1.82 |
ENSMUST00000105439.1
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr1_-_158958367 | 1.81 |
ENSMUST00000159861.2
|
Pappa2
|
pappalysin 2 |
| chr14_-_48665098 | 1.81 |
ENSMUST00000118578.1
|
Otx2
|
orthodenticle homolog 2 (Drosophila) |
| chr16_-_44016387 | 1.80 |
ENSMUST00000036174.3
|
Gramd1c
|
GRAM domain containing 1C |
| chr13_+_42681513 | 1.79 |
ENSMUST00000149235.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr19_+_22692613 | 1.77 |
ENSMUST00000099564.2
ENSMUST00000099566.3 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr19_+_26749726 | 1.76 |
ENSMUST00000175842.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr15_+_18818895 | 1.70 |
ENSMUST00000166873.2
|
Cdh10
|
cadherin 10 |
| chr9_+_53301571 | 1.67 |
ENSMUST00000051014.1
|
Exph5
|
exophilin 5 |
| chr4_+_82065924 | 1.66 |
ENSMUST00000161588.1
|
Gm5860
|
predicted gene 5860 |
| chr5_+_149678224 | 1.60 |
ENSMUST00000100404.3
|
B3galtl
|
beta 1,3-galactosyltransferase-like |
| chr17_+_69439326 | 1.60 |
ENSMUST00000169935.1
|
A330050F15Rik
|
RIKEN cDNA A330050F15 gene |
| chr12_+_21417872 | 1.59 |
ENSMUST00000180671.1
|
Gm4419
|
predicted gene 4419 |
| chr11_+_34314757 | 1.59 |
ENSMUST00000165963.1
ENSMUST00000093192.3 |
Fam196b
|
family with sequence similarity 196, member B |
| chr5_+_66968416 | 1.59 |
ENSMUST00000038188.7
|
Limch1
|
LIM and calponin homology domains 1 |
| chr6_-_13839916 | 1.59 |
ENSMUST00000060442.7
|
Gpr85
|
G protein-coupled receptor 85 |
| chr2_-_94264713 | 1.58 |
ENSMUST00000129661.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chr11_-_37235882 | 1.58 |
ENSMUST00000102801.1
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr19_+_26750939 | 1.57 |
ENSMUST00000175953.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chrX_+_164436987 | 1.57 |
ENSMUST00000036858.4
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr10_+_24076500 | 1.55 |
ENSMUST00000051133.5
|
Taar8a
|
trace amine-associated receptor 8A |
| chr7_-_118443549 | 1.55 |
ENSMUST00000081574.4
|
Syt17
|
synaptotagmin XVII |
| chr9_+_120929216 | 1.52 |
ENSMUST00000130466.1
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chrX_+_164438039 | 1.52 |
ENSMUST00000033755.5
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr1_-_158814469 | 1.51 |
ENSMUST00000161589.2
|
Pappa2
|
pappalysin 2 |
| chr6_+_142298419 | 1.51 |
ENSMUST00000041993.2
|
Iapp
|
islet amyloid polypeptide |
| chr15_+_25773985 | 1.51 |
ENSMUST00000125667.1
|
Myo10
|
myosin X |
| chr4_+_102421518 | 1.49 |
ENSMUST00000106904.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr16_-_88056176 | 1.47 |
ENSMUST00000072256.5
ENSMUST00000023652.8 ENSMUST00000114137.1 |
Grik1
|
glutamate receptor, ionotropic, kainate 1 |
| chr2_+_70562147 | 1.45 |
ENSMUST00000148210.1
|
Gad1
|
glutamate decarboxylase 1 |
| chrX_+_159303266 | 1.44 |
ENSMUST00000112491.1
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr19_+_55894508 | 1.44 |
ENSMUST00000142291.1
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr6_+_80018877 | 1.43 |
ENSMUST00000147663.1
ENSMUST00000128718.1 ENSMUST00000126005.1 ENSMUST00000133918.1 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr15_+_10314102 | 1.42 |
ENSMUST00000127467.1
|
Prlr
|
prolactin receptor |
| chr9_+_43310763 | 1.41 |
ENSMUST00000034511.5
|
Trim29
|
tripartite motif-containing 29 |
| chr10_-_24101951 | 1.41 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr6_+_29859374 | 1.41 |
ENSMUST00000115238.3
|
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr6_+_96115249 | 1.40 |
ENSMUST00000075080.5
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chr1_-_126830632 | 1.40 |
ENSMUST00000112583.1
ENSMUST00000094609.3 |
Nckap5
|
NCK-associated protein 5 |
| chr16_-_23890805 | 1.40 |
ENSMUST00000004480.3
|
Sst
|
somatostatin |
| chr7_-_5125937 | 1.39 |
ENSMUST00000147835.2
|
Rasl2-9
|
RAS-like, family 2, locus 9 |
| chr10_-_53647080 | 1.39 |
ENSMUST00000169866.1
|
Fam184a
|
family with sequence similarity 184, member A |
| chrX_+_164140447 | 1.39 |
ENSMUST00000073973.4
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr12_-_84698769 | 1.38 |
ENSMUST00000095550.2
|
Syndig1l
|
synapse differentiation inducing 1 like |
| chr4_+_104766334 | 1.38 |
ENSMUST00000065072.6
|
C8b
|
complement component 8, beta polypeptide |
| chr6_+_97991776 | 1.38 |
ENSMUST00000043628.6
|
Mitf
|
microphthalmia-associated transcription factor |
| chr12_+_59095653 | 1.36 |
ENSMUST00000021384.4
|
Mia2
|
melanoma inhibitory activity 2 |
| chr13_-_91388079 | 1.35 |
ENSMUST00000181054.1
|
A830009L08Rik
|
RIKEN cDNA A830009L08 gene |
| chr10_+_42583787 | 1.35 |
ENSMUST00000105497.1
ENSMUST00000144806.1 |
Ostm1
|
osteopetrosis associated transmembrane protein 1 |
| chr4_+_104766308 | 1.33 |
ENSMUST00000031663.3
|
C8b
|
complement component 8, beta polypeptide |
| chr15_-_58364148 | 1.33 |
ENSMUST00000068515.7
|
Anxa13
|
annexin A13 |
| chr14_+_27000362 | 1.32 |
ENSMUST00000035433.8
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr13_+_42866247 | 1.31 |
ENSMUST00000131942.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr2_+_38341068 | 1.31 |
ENSMUST00000133661.1
|
Lhx2
|
LIM homeobox protein 2 |
| chr5_+_66968559 | 1.31 |
ENSMUST00000127184.1
|
Limch1
|
LIM and calponin homology domains 1 |
| chr10_+_115817247 | 1.30 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr8_-_67974567 | 1.30 |
ENSMUST00000098696.3
ENSMUST00000038959.9 ENSMUST00000093469.4 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr9_-_62070606 | 1.30 |
ENSMUST00000034785.7
|
Glce
|
glucuronyl C5-epimerase |
| chr2_-_35100677 | 1.28 |
ENSMUST00000045776.4
ENSMUST00000113050.3 |
AI182371
|
expressed sequence AI182371 |
| chr3_+_41563356 | 1.28 |
ENSMUST00000163764.1
|
Phf17
|
PHD finger protein 17 |
| chr3_+_138065052 | 1.27 |
ENSMUST00000163080.2
|
1110002E22Rik
|
RIKEN cDNA 1110002E22 gene |
| chr3_-_85741389 | 1.26 |
ENSMUST00000094148.4
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr4_-_137118135 | 1.25 |
ENSMUST00000154285.1
|
Gm13001
|
predicted gene 13001 |
| chr16_-_22439719 | 1.25 |
ENSMUST00000079601.6
|
Etv5
|
ets variant gene 5 |
| chr10_+_73821937 | 1.25 |
ENSMUST00000134009.2
ENSMUST00000125006.2 ENSMUST00000177420.1 |
Pcdh15
|
protocadherin 15 |
| chr5_+_19907774 | 1.24 |
ENSMUST00000115267.2
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr17_+_17402672 | 1.22 |
ENSMUST00000115576.2
|
Lix1
|
limb expression 1 homolog (chicken) |
| chr1_-_166002613 | 1.22 |
ENSMUST00000177358.1
ENSMUST00000160908.1 ENSMUST00000027850.8 ENSMUST00000160260.2 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr5_+_90561102 | 1.21 |
ENSMUST00000094615.4
|
5830473C10Rik
|
RIKEN cDNA 5830473C10 gene |
| chr1_-_56969864 | 1.19 |
ENSMUST00000177424.1
|
Satb2
|
special AT-rich sequence binding protein 2 |
| chr1_+_110099295 | 1.19 |
ENSMUST00000134301.1
|
Cdh7
|
cadherin 7, type 2 |
| chr19_+_55741810 | 1.16 |
ENSMUST00000111657.3
ENSMUST00000061496.9 ENSMUST00000041717.7 ENSMUST00000111662.4 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr9_-_77045788 | 1.16 |
ENSMUST00000034911.6
|
Tinag
|
tubulointerstitial nephritis antigen |
| chr6_+_80019008 | 1.16 |
ENSMUST00000126399.1
ENSMUST00000136421.1 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chrX_+_103356464 | 1.16 |
ENSMUST00000116547.2
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr19_-_59170978 | 1.15 |
ENSMUST00000172821.2
|
Vax1
|
ventral anterior homeobox containing gene 1 |
| chr17_+_70522149 | 1.15 |
ENSMUST00000140728.1
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr7_-_90129339 | 1.14 |
ENSMUST00000181189.1
|
2310010J17Rik
|
RIKEN cDNA 2310010J17 gene |
| chr3_+_138277489 | 1.14 |
ENSMUST00000004232.9
|
Adh1
|
alcohol dehydrogenase 1 (class I) |
| chr4_+_108879063 | 1.13 |
ENSMUST00000106650.2
|
Rab3b
|
RAB3B, member RAS oncogene family |
| chr14_+_75455957 | 1.12 |
ENSMUST00000164848.1
|
Siah3
|
seven in absentia homolog 3 (Drosophila) |
| chr1_-_89933290 | 1.11 |
ENSMUST00000036954.7
|
Gbx2
|
gastrulation brain homeobox 2 |
| chr2_-_103303179 | 1.09 |
ENSMUST00000090475.3
|
Ehf
|
ets homologous factor |
| chr8_+_47822143 | 1.09 |
ENSMUST00000079639.2
|
Cldn24
|
claudin 24 |
| chr14_-_12345847 | 1.08 |
ENSMUST00000022262.4
|
Fezf2
|
Fez family zinc finger 2 |
| chr14_+_41105359 | 1.07 |
ENSMUST00000047286.6
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr13_-_52981027 | 1.07 |
ENSMUST00000071065.7
|
Nfil3
|
nuclear factor, interleukin 3, regulated |
| chr4_-_110292719 | 1.07 |
ENSMUST00000106601.1
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) |
| chr2_+_70562007 | 1.06 |
ENSMUST00000094934.4
|
Gad1
|
glutamate decarboxylase 1 |
| chr1_-_9298499 | 1.06 |
ENSMUST00000132064.1
|
Sntg1
|
syntrophin, gamma 1 |
| chr19_+_37436707 | 1.05 |
ENSMUST00000128184.1
|
Hhex
|
hematopoietically expressed homeobox |
| chr4_+_108879130 | 1.05 |
ENSMUST00000106651.2
|
Rab3b
|
RAB3B, member RAS oncogene family |
| chr2_-_103303158 | 1.05 |
ENSMUST00000111176.2
|
Ehf
|
ets homologous factor |
| chr13_-_97747373 | 1.05 |
ENSMUST00000123535.1
|
5330416C01Rik
|
RIKEN cDNA 5330416C01 gene |
| chr5_-_123140135 | 1.05 |
ENSMUST00000160099.1
|
AI480526
|
expressed sequence AI480526 |
| chr2_-_63184253 | 1.04 |
ENSMUST00000075052.3
ENSMUST00000112454.1 |
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7 |
| chr2_+_125136692 | 1.04 |
ENSMUST00000099452.2
|
Ctxn2
|
cortexin 2 |
| chr9_-_21312255 | 1.04 |
ENSMUST00000115433.3
ENSMUST00000003397.7 |
Ap1m2
|
adaptor protein complex AP-1, mu 2 subunit |
| chr2_+_69219971 | 1.03 |
ENSMUST00000005364.5
ENSMUST00000112317.2 |
G6pc2
|
glucose-6-phosphatase, catalytic, 2 |
| chr5_-_67099235 | 1.03 |
ENSMUST00000012664.8
|
Phox2b
|
paired-like homeobox 2b |
| chr10_-_80421847 | 1.02 |
ENSMUST00000156244.1
|
Tcf3
|
transcription factor 3 |
| chrX_-_104857228 | 1.01 |
ENSMUST00000033575.5
|
Magee2
|
melanoma antigen, family E, 2 |
| chr1_+_177445660 | 1.01 |
ENSMUST00000077225.6
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr6_-_116716888 | 1.01 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chrX_-_17319316 | 1.01 |
ENSMUST00000026014.7
|
Efhc2
|
EF-hand domain (C-terminal) containing 2 |
| chr16_-_22161450 | 1.01 |
ENSMUST00000115379.1
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr10_-_109009055 | 1.00 |
ENSMUST00000156979.1
|
Syt1
|
synaptotagmin I |
| chr13_-_102905740 | 0.99 |
ENSMUST00000167462.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chrX_-_139085211 | 0.99 |
ENSMUST00000033626.8
ENSMUST00000060824.3 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr12_-_86892540 | 0.99 |
ENSMUST00000181290.1
|
Gm26698
|
predicted gene, 26698 |
| chr18_+_36952621 | 0.98 |
ENSMUST00000115661.2
|
Pcdha2
|
protocadherin alpha 2 |
| chr9_-_50746501 | 0.98 |
ENSMUST00000034564.1
|
2310030G06Rik
|
RIKEN cDNA 2310030G06 gene |
| chr16_-_22439570 | 0.98 |
ENSMUST00000170393.1
|
Etv5
|
ets variant gene 5 |
| chr5_+_19907502 | 0.98 |
ENSMUST00000101558.3
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr2_-_94264745 | 0.97 |
ENSMUST00000134563.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chr4_-_42168603 | 0.97 |
ENSMUST00000098121.3
|
Gm13305
|
predicted gene 13305 |
| chr11_-_42000532 | 0.97 |
ENSMUST00000070735.3
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2 |
| chr5_+_88487982 | 0.96 |
ENSMUST00000031222.8
|
Enam
|
enamelin |
| chr10_-_53638269 | 0.96 |
ENSMUST00000164393.1
|
Fam184a
|
family with sequence similarity 184, member A |
| chr2_-_7396192 | 0.95 |
ENSMUST00000137733.2
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr17_-_51810866 | 0.95 |
ENSMUST00000176669.1
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr18_+_50051702 | 0.94 |
ENSMUST00000134348.1
ENSMUST00000153873.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr6_-_13871477 | 0.94 |
ENSMUST00000139231.1
|
2610001J05Rik
|
RIKEN cDNA 2610001J05 gene |
| chr1_+_81077204 | 0.94 |
ENSMUST00000123720.1
|
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr6_+_29853746 | 0.93 |
ENSMUST00000064872.6
ENSMUST00000152581.1 ENSMUST00000176265.1 ENSMUST00000154079.1 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr8_+_105305572 | 0.93 |
ENSMUST00000109375.2
|
Elmo3
|
engulfment and cell motility 3 |
| chrX_+_134295225 | 0.93 |
ENSMUST00000037687.7
|
Tmem35
|
transmembrane protein 35 |
| chr8_+_12395287 | 0.93 |
ENSMUST00000180353.1
|
Sox1
|
SRY-box containing gene 1 |
| chr11_-_109298121 | 0.92 |
ENSMUST00000020920.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr6_-_142322978 | 0.92 |
ENSMUST00000081380.3
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr6_+_97807014 | 0.92 |
ENSMUST00000043637.7
|
Mitf
|
microphthalmia-associated transcription factor |
| chr6_+_56017489 | 0.92 |
ENSMUST00000052827.4
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr11_-_96747419 | 0.92 |
ENSMUST00000181758.1
|
2010300F17Rik
|
RIKEN cDNA 2010300F17 gene |
| chrX_+_107816477 | 0.91 |
ENSMUST00000143975.1
ENSMUST00000144695.1 ENSMUST00000167154.1 |
Fam46d
|
family with sequence similarity 46, member D |
| chr13_+_94875600 | 0.91 |
ENSMUST00000022195.10
|
Otp
|
orthopedia homolog (Drosophila) |
| chr15_-_50890396 | 0.91 |
ENSMUST00000185183.1
|
Trps1
|
trichorhinophalangeal syndrome I (human) |
| chr7_+_91090728 | 0.91 |
ENSMUST00000074273.3
|
Dlg2
|
discs, large homolog 2 (Drosophila) |
| chr14_-_48662740 | 0.91 |
ENSMUST00000122009.1
|
Otx2
|
orthodenticle homolog 2 (Drosophila) |
| chr4_-_94928789 | 0.90 |
ENSMUST00000030309.5
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr1_+_81077274 | 0.90 |
ENSMUST00000068275.5
ENSMUST00000113494.2 |
Nyap2
|
neuronal tyrosine-phophorylated phosphoinositide 3-kinase adaptor 2 |
| chr3_+_159839729 | 0.90 |
ENSMUST00000068952.5
|
Wls
|
wntless homolog (Drosophila) |
| chr18_+_76059458 | 0.89 |
ENSMUST00000167921.1
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr3_-_113532288 | 0.89 |
ENSMUST00000132353.1
|
Amy2a1
|
amylase 2a1 |
| chr10_+_112165676 | 0.89 |
ENSMUST00000170013.1
|
Caps2
|
calcyphosphine 2 |
| chr15_-_34356421 | 0.89 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr1_-_162866502 | 0.89 |
ENSMUST00000046049.7
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr5_-_123865491 | 0.89 |
ENSMUST00000057145.5
|
Niacr1
|
niacin receptor 1 |
| chrX_-_162565514 | 0.88 |
ENSMUST00000154424.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr4_-_45532470 | 0.88 |
ENSMUST00000147448.1
|
Shb
|
src homology 2 domain-containing transforming protein B |
| chr3_+_106113229 | 0.88 |
ENSMUST00000079132.5
ENSMUST00000139086.1 |
Chia
|
chitinase, acidic |
| chr7_+_91090697 | 0.87 |
ENSMUST00000107196.2
|
Dlg2
|
discs, large homolog 2 (Drosophila) |
| chr4_+_114821722 | 0.87 |
ENSMUST00000137570.1
|
Gm12830
|
predicted gene 12830 |
| chr4_-_35845204 | 0.87 |
ENSMUST00000164772.1
ENSMUST00000065173.2 |
Lingo2
|
leucine rich repeat and Ig domain containing 2 |
| chr10_+_101681487 | 0.87 |
ENSMUST00000179929.1
ENSMUST00000127504.1 |
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr14_+_33954020 | 0.87 |
ENSMUST00000035695.8
|
Rbp3
|
retinol binding protein 3, interstitial |
| chr4_+_33924632 | 0.87 |
ENSMUST00000057188.6
|
Cnr1
|
cannabinoid receptor 1 (brain) |
| chr19_-_34166037 | 0.87 |
ENSMUST00000025686.7
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr8_+_76902277 | 0.86 |
ENSMUST00000109912.1
ENSMUST00000128862.1 ENSMUST00000109911.1 |
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr18_+_51117754 | 0.86 |
ENSMUST00000116639.2
|
Prr16
|
proline rich 16 |
| chr17_+_69969073 | 0.86 |
ENSMUST00000133983.1
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1 |
| chr4_-_110290884 | 0.86 |
ENSMUST00000142722.1
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) |
| chrX_+_159708593 | 0.85 |
ENSMUST00000080394.6
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr14_-_93888732 | 0.85 |
ENSMUST00000068992.2
|
Pcdh9
|
protocadherin 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hoxb7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:0050973 | detection of mechanical stimulus involved in equilibrioception(GO:0050973) |
| 0.9 | 7.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.8 | 3.1 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.7 | 6.5 | GO:0010908 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.6 | 1.9 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.6 | 2.6 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.6 | 4.4 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.6 | 3.0 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.6 | 4.2 | GO:0048852 | diencephalon morphogenesis(GO:0048852) |
| 0.6 | 1.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.6 | 5.0 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.5 | 2.2 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) regulation of vesicle size(GO:0097494) |
| 0.5 | 1.6 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.5 | 1.0 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) |
| 0.5 | 2.4 | GO:0010578 | regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010578) positive regulation of adenylate cyclase activity involved in G-protein coupled receptor signaling pathway(GO:0010579) |
| 0.5 | 6.0 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.5 | 5.9 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.5 | 2.7 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.4 | 1.7 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.4 | 0.4 | GO:0072199 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) |
| 0.4 | 2.5 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.4 | 2.4 | GO:0060478 | acrosomal vesicle exocytosis(GO:0060478) |
| 0.4 | 1.5 | GO:0014054 | positive regulation of gamma-aminobutyric acid secretion(GO:0014054) |
| 0.4 | 2.6 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 2.8 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.3 | 1.4 | GO:0097168 | mesenchymal stem cell proliferation(GO:0097168) |
| 0.3 | 1.4 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.3 | 0.3 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.3 | 1.4 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.3 | 0.7 | GO:1900377 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.3 | 3.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.3 | 0.3 | GO:2000974 | negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.3 | 1.0 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.3 | 1.0 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.3 | 0.9 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.3 | 1.9 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 0.9 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.3 | 1.5 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.3 | 0.9 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 0.9 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.3 | 1.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 1.1 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.3 | 1.1 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.3 | 1.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.3 | 0.3 | GO:0061047 | positive regulation of branching involved in lung morphogenesis(GO:0061047) |
| 0.3 | 0.8 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.3 | 1.3 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 | 2.0 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.2 | 0.5 | GO:0090481 | pyrimidine nucleotide-sugar transmembrane transport(GO:0090481) |
| 0.2 | 2.7 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.2 | 0.7 | GO:0003345 | proepicardium cell migration involved in pericardium morphogenesis(GO:0003345) |
| 0.2 | 0.5 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.2 | 0.7 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.2 | 1.8 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.2 | 0.2 | GO:0048880 | sensory system development(GO:0048880) |
| 0.2 | 1.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.2 | 0.7 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.2 | 0.4 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in spinal cord motor neuron fate specification(GO:0021912) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.2 | 0.6 | GO:0060809 | mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.2 | 0.6 | GO:0043056 | forward locomotion(GO:0043056) |
| 0.2 | 1.0 | GO:0021764 | amygdala development(GO:0021764) |
| 0.2 | 1.9 | GO:1901162 | serotonin biosynthetic process(GO:0042427) primary amino compound biosynthetic process(GO:1901162) |
| 0.2 | 1.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.6 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.2 | 1.0 | GO:0015676 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.2 | 0.2 | GO:0007468 | regulation of rhodopsin gene expression(GO:0007468) |
| 0.2 | 0.4 | GO:2000722 | regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.2 | 1.7 | GO:0098907 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) regulation of SA node cell action potential(GO:0098907) |
| 0.2 | 2.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.2 | 0.8 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 1.3 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 0.7 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 | 1.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.2 | 1.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.2 | 0.7 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.2 | 0.7 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.2 | 1.6 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.2 | 0.2 | GO:0060060 | post-embryonic retina morphogenesis in camera-type eye(GO:0060060) |
| 0.2 | 2.6 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.2 | 0.3 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.2 | 2.6 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.2 | 1.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.2 | 1.2 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.2 | 1.0 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.2 | 1.2 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.2 | 0.7 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.2 | 1.6 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.2 | 0.3 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.2 | 1.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.2 | 0.5 | GO:1904980 | regulation of endosome organization(GO:1904978) positive regulation of endosome organization(GO:1904980) |
| 0.2 | 1.2 | GO:0097646 | dimeric G-protein coupled receptor signaling pathway(GO:0038042) calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.2 | 0.6 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.2 | 0.2 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.2 | 0.5 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.2 | 0.8 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 1.2 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.4 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.1 | 0.3 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.6 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.1 | 0.3 | GO:0035947 | regulation of gluconeogenesis by regulation of transcription from RNA polymerase II promoter(GO:0035947) |
| 0.1 | 0.4 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.1 | 0.4 | GO:0090274 | reduction of food intake in response to dietary excess(GO:0002023) positive regulation of somatostatin secretion(GO:0090274) |
| 0.1 | 0.3 | GO:0061026 | cardiac muscle tissue regeneration(GO:0061026) |
| 0.1 | 0.8 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.1 | 1.7 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.7 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 1.5 | GO:0060573 | ventral spinal cord interneuron specification(GO:0021521) cell fate specification involved in pattern specification(GO:0060573) |
| 0.1 | 0.8 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.5 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.8 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.8 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.1 | 0.1 | GO:0035434 | copper ion transmembrane transport(GO:0035434) |
| 0.1 | 0.7 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.1 | 1.5 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.3 | GO:0021502 | neural fold elevation formation(GO:0021502) |
| 0.1 | 0.4 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.5 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.1 | 0.4 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.1 | 0.8 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.4 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.1 | 0.4 | GO:0090361 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.1 | 0.5 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.1 | 0.5 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 1.9 | GO:0045161 | neuronal ion channel clustering(GO:0045161) |
| 0.1 | 0.4 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.1 | 0.8 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 1.5 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 3.5 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.1 | 3.5 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.1 | 0.4 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.1 | 0.5 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 0.9 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.1 | 0.3 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.1 | 0.8 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.1 | 0.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.5 | GO:0035482 | gastric motility(GO:0035482) |
| 0.1 | 0.2 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.1 | 0.4 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.3 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.1 | 0.1 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.1 | 0.3 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.1 | 0.3 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.1 | 0.3 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.1 | 2.7 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.1 | 0.3 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.8 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.1 | 0.3 | GO:0072343 | pancreatic stellate cell proliferation(GO:0072343) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.1 | 0.6 | GO:0072318 | clathrin coat disassembly(GO:0072318) |
| 0.1 | 0.5 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 1.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.4 | GO:2000359 | regulation of binding of sperm to zona pellucida(GO:2000359) |
| 0.1 | 0.5 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.1 | 4.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.1 | 0.8 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.5 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.1 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 0.4 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.7 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.1 | 0.1 | GO:1904349 | positive regulation of small intestine smooth muscle contraction(GO:1904349) |
| 0.1 | 1.4 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.6 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.5 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.1 | 0.5 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.1 | 0.3 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.1 | 0.3 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 1.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.1 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.3 | GO:0006780 | uroporphyrinogen III biosynthetic process(GO:0006780) |
| 0.1 | 1.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 1.0 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.1 | 0.5 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.9 | GO:0090394 | negative regulation of excitatory postsynaptic potential(GO:0090394) |
| 0.1 | 0.7 | GO:0046874 | quinolinate metabolic process(GO:0046874) |
| 0.1 | 0.8 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.1 | 0.2 | GO:0002304 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 0.5 | GO:0098961 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.1 | 0.4 | GO:2000224 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) regulation of testosterone biosynthetic process(GO:2000224) |
| 0.1 | 3.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.1 | 0.2 | GO:0072282 | metanephric nephron tubule morphogenesis(GO:0072282) |
| 0.1 | 0.3 | GO:0072362 | regulation of glycolytic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072362) |
| 0.1 | 0.2 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.9 | GO:0015677 | copper ion import(GO:0015677) |
| 0.1 | 0.6 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.1 | 0.3 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.1 | 0.5 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.1 | 0.3 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.1 | 0.8 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.1 | 0.2 | GO:0070429 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.1 | 0.3 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.1 | 0.3 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.4 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.2 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.1 | 0.4 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.4 | GO:0021860 | pyramidal neuron development(GO:0021860) |
| 0.1 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.1 | 0.2 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.1 | 0.8 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.1 | GO:0002386 | immune response in mucosal-associated lymphoid tissue(GO:0002386) |
| 0.1 | 0.5 | GO:0051177 | meiotic sister chromatid cohesion(GO:0051177) |
| 0.1 | 1.2 | GO:1902692 | regulation of neuroblast proliferation(GO:1902692) |
| 0.1 | 0.2 | GO:0038001 | autocrine signaling(GO:0035425) paracrine signaling(GO:0038001) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.3 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.1 | 0.5 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.3 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.4 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.1 | 0.3 | GO:0043091 | regulation of amino acid import(GO:0010958) L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.1 | 0.7 | GO:0006901 | vesicle coating(GO:0006901) vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.1 | GO:0045199 | maintenance of epithelial cell apical/basal polarity(GO:0045199) |
| 0.1 | 1.8 | GO:1903861 | positive regulation of dendrite extension(GO:1903861) |
| 0.1 | 0.5 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.1 | 0.1 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.1 | 0.1 | GO:0090325 | regulation of locomotion involved in locomotory behavior(GO:0090325) negative regulation of locomotion involved in locomotory behavior(GO:0090327) |
| 0.1 | 0.2 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.2 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.3 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.1 | 0.3 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.1 | 0.4 | GO:0072092 | ureteric bud invasion(GO:0072092) metanephric renal vesicle formation(GO:0072093) |
| 0.1 | 1.0 | GO:0000054 | ribosomal subunit export from nucleus(GO:0000054) ribosome localization(GO:0033750) establishment of ribosome localization(GO:0033753) |
| 0.1 | 1.1 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.1 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.5 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.1 | 0.8 | GO:0070431 | nucleotide-binding oligomerization domain containing signaling pathway(GO:0070423) nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070431) |
| 0.1 | 0.2 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.1 | 0.5 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.1 | 0.8 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.2 | GO:1904017 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.1 | 0.3 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.1 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.1 | 0.5 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 2.7 | GO:0003407 | neural retina development(GO:0003407) |
| 0.1 | 0.4 | GO:0042436 | tryptophan catabolic process(GO:0006569) tryptophan catabolic process to kynurenine(GO:0019441) indole-containing compound catabolic process(GO:0042436) indolalkylamine catabolic process(GO:0046218) kynurenine metabolic process(GO:0070189) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 1.7 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.9 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.1 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.2 | GO:0055071 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.3 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.2 | GO:0001868 | regulation of complement activation, lectin pathway(GO:0001868) negative regulation of complement activation, lectin pathway(GO:0001869) |
| 0.1 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.3 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.1 | 0.6 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.1 | 0.6 | GO:0071257 | cellular response to electrical stimulus(GO:0071257) |
| 0.1 | 0.5 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.1 | 0.1 | GO:0061317 | canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:0061317) regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901295) regulation of canonical Wnt signaling pathway involved in heart development(GO:1905066) |
| 0.1 | 0.4 | GO:0090520 | sphingolipid mediated signaling pathway(GO:0090520) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.2 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.1 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.1 | 0.4 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.6 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.3 | GO:0060178 | regulation of exocyst localization(GO:0060178) |
| 0.1 | 0.3 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.1 | 0.2 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.1 | 0.7 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.1 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.1 | 0.2 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) |
| 0.1 | 1.0 | GO:0031069 | hair follicle morphogenesis(GO:0031069) |
| 0.1 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.1 | 0.3 | GO:0046601 | positive regulation of centriole replication(GO:0046601) |
| 0.1 | 0.5 | GO:0035871 | protein K11-linked deubiquitination(GO:0035871) |
| 0.1 | 2.7 | GO:0007189 | adenylate cyclase-activating G-protein coupled receptor signaling pathway(GO:0007189) |
| 0.1 | 0.5 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.1 | 1.9 | GO:1900047 | negative regulation of blood coagulation(GO:0030195) negative regulation of hemostasis(GO:1900047) |
| 0.1 | 0.3 | GO:0034454 | microtubule anchoring at centrosome(GO:0034454) |
| 0.1 | 0.2 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.3 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.5 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.1 | GO:0048818 | positive regulation of hair follicle maturation(GO:0048818) positive regulation of anagen(GO:0051885) |
| 0.0 | 0.5 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 1.3 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 1.6 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 0.2 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.2 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.2 | GO:0046898 | suppression by virus of host apoptotic process(GO:0019050) modulation by virus of host apoptotic process(GO:0039526) response to cycloheximide(GO:0046898) |
| 0.0 | 0.3 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.3 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.7 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.9 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.1 | GO:0071871 | response to epinephrine(GO:0071871) cellular response to epinephrine stimulus(GO:0071872) |
| 0.0 | 0.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 1.1 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.4 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 1.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.8 | GO:0036342 | post-anal tail morphogenesis(GO:0036342) |
| 0.0 | 0.3 | GO:0060576 | intestinal epithelial cell development(GO:0060576) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.3 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 4.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 2.4 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.9 | GO:0045624 | positive regulation of T-helper cell differentiation(GO:0045624) |
| 0.0 | 0.6 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.7 | GO:0034030 | coenzyme A biosynthetic process(GO:0015937) nucleoside bisphosphate biosynthetic process(GO:0033866) ribonucleoside bisphosphate biosynthetic process(GO:0034030) purine nucleoside bisphosphate biosynthetic process(GO:0034033) |
| 0.0 | 0.1 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.4 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.6 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.5 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) postsynaptic signal transduction(GO:0098926) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.2 | GO:0030070 | insulin processing(GO:0030070) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.2 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 0.6 | GO:0030497 | fatty acid elongation(GO:0030497) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) negative regulation of SMAD protein complex assembly(GO:0010991) cardiac vascular smooth muscle cell development(GO:0060948) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone metabolic process(GO:0032352) positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.4 | GO:0060252 | positive regulation of glial cell proliferation(GO:0060252) |
| 0.0 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.3 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) paramesonephric duct development(GO:0061205) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.0 | 0.3 | GO:0006449 | regulation of translational termination(GO:0006449) |
| 0.0 | 0.2 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.7 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.4 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.6 | GO:0002230 | positive regulation of defense response to virus by host(GO:0002230) |
| 0.0 | 1.8 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:2000410 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) thymocyte migration(GO:0072679) regulation of thymocyte migration(GO:2000410) positive regulation of thymocyte migration(GO:2000412) regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.1 | GO:0070094 | positive regulation of glucagon secretion(GO:0070094) |
| 0.0 | 0.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.0 | 0.2 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.0 | 0.4 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.4 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.7 | GO:0050891 | multicellular organismal water homeostasis(GO:0050891) |
| 0.0 | 0.2 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.2 | GO:0061368 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.0 | 0.1 | GO:0071600 | otic vesicle morphogenesis(GO:0071600) |
| 0.0 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0046543 | development of secondary sexual characteristics(GO:0045136) development of secondary female sexual characteristics(GO:0046543) |
| 0.0 | 0.3 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.1 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.0 | 0.8 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.2 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 2.1 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.5 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.9 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.3 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.4 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.5 | GO:0045047 | protein targeting to ER(GO:0045047) |
| 0.0 | 0.1 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.0 | 0.3 | GO:0021548 | pons development(GO:0021548) |
| 0.0 | 1.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.4 | GO:0006613 | cotranslational protein targeting to membrane(GO:0006613) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.4 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0042136 | neurotransmitter biosynthetic process(GO:0042136) |
| 0.0 | 2.5 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.2 | GO:0035608 | protein deglutamylation(GO:0035608) |
| 0.0 | 0.2 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.7 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.3 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.4 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.4 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 1.3 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.2 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.6 | GO:0040023 | nuclear migration(GO:0007097) establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.5 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 1.6 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.3 | GO:0098877 | neurotransmitter receptor transport to plasma membrane(GO:0098877) neurotransmitter receptor transport to postsynaptic membrane(GO:0098969) establishment of protein localization to postsynaptic membrane(GO:1903540) |
| 0.0 | 0.5 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.1 | GO:0060591 | chondroblast differentiation(GO:0060591) |
| 0.0 | 0.3 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.1 | GO:1901673 | regulation of mitotic spindle assembly(GO:1901673) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.3 | GO:0042104 | positive regulation of activated T cell proliferation(GO:0042104) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 2.1 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.3 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.8 | GO:0071346 | cellular response to interferon-gamma(GO:0071346) |
| 0.0 | 3.2 | GO:0008360 | regulation of cell shape(GO:0008360) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
| 0.0 | 0.2 | GO:0007194 | negative regulation of adenylate cyclase activity(GO:0007194) |
| 0.0 | 0.4 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.4 | GO:0051930 | regulation of sensory perception of pain(GO:0051930) regulation of sensory perception(GO:0051931) |
| 0.0 | 0.1 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0060012 | synaptic transmission, glycinergic(GO:0060012) |
| 0.0 | 0.4 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.5 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0060979 | vasculogenesis involved in coronary vascular morphogenesis(GO:0060979) |
| 0.0 | 0.4 | GO:0006779 | porphyrin-containing compound biosynthetic process(GO:0006779) |
| 0.0 | 0.6 | GO:0006919 | activation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0006919) |
| 0.0 | 1.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 1.5 | GO:0010466 | negative regulation of peptidase activity(GO:0010466) |
| 0.0 | 0.5 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0031987 | locomotion involved in locomotory behavior(GO:0031987) |
| 0.0 | 0.1 | GO:0006501 | C-terminal protein lipidation(GO:0006501) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.4 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 0.8 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.2 | GO:0046174 | polyol catabolic process(GO:0046174) |
| 0.0 | 0.1 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.1 | GO:0072600 | establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.0 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.0 | 0.6 | GO:0030316 | osteoclast differentiation(GO:0030316) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0043651 | linoleic acid metabolic process(GO:0043651) |
| 0.0 | 0.0 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.0 | 0.3 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.2 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 6.5 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.4 | 1.2 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.4 | 2.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.4 | 6.2 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.3 | 3.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.3 | 7.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.3 | 5.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.3 | 0.8 | GO:0044299 | C-fiber(GO:0044299) |
| 0.3 | 1.3 | GO:0002079 | inner acrosomal membrane(GO:0002079) |
| 0.2 | 1.0 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.2 | 0.6 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.2 | 3.8 | GO:0031045 | dense core granule(GO:0031045) |
| 0.2 | 1.0 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.2 | 6.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.2 | 0.9 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 1.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.2 | 0.9 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 1.8 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 6.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.1 | 0.9 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 2.0 | GO:1902711 | GABA-A receptor complex(GO:1902711) |
| 0.1 | 1.6 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.1 | 1.8 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 2.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 0.4 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.3 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.1 | 4.7 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 0.6 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 0.7 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.5 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 0.2 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 0.5 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 7.0 | GO:0001750 | photoreceptor outer segment(GO:0001750) |
| 0.1 | 1.9 | GO:0097038 | perinuclear endoplasmic reticulum(GO:0097038) |
| 0.1 | 0.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.1 | 0.5 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 2.3 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.1 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 0.6 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.1 | 0.4 | GO:0045293 | mRNA editing complex(GO:0045293) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.3 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.1 | 0.3 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.5 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 1.4 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.7 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.1 | 1.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 0.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 1.2 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.2 | GO:0043540 | 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase complex(GO:0043540) |
| 0.1 | 0.4 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.5 | GO:0044217 | other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.1 | 0.6 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 2.9 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.1 | 1.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.5 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 1.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.1 | GO:0005595 | collagen type XII trimer(GO:0005595) anchoring collagen complex(GO:0030934) |
| 0.0 | 0.7 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 7.9 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.4 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 1.8 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 1.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.0 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 0.3 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.6 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0098642 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 3.0 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.5 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0030427 | site of polarized growth(GO:0030427) |
| 0.0 | 0.3 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.6 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.3 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 1.3 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.3 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.8 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 1.7 | GO:0016529 | sarcoplasmic reticulum(GO:0016529) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.6 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:1990131 | Iml1 complex(GO:1990130) Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 2.2 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 3.9 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 1.1 | GO:0000795 | synaptonemal complex(GO:0000795) |
| 0.0 | 2.6 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 6.4 | GO:0097060 | synaptic membrane(GO:0097060) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.3 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 2.0 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.5 | GO:0031306 | intrinsic component of mitochondrial outer membrane(GO:0031306) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 1.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 2.3 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 0.2 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 0.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.5 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.4 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 7.3 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.9 | 2.6 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.7 | 0.7 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.5 | 1.6 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.5 | 1.9 | GO:0004510 | tryptophan 5-monooxygenase activity(GO:0004510) |
| 0.4 | 1.7 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.4 | 3.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.4 | 2.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.4 | 2.7 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.4 | 3.0 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) |
| 0.4 | 1.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.3 | 2.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 1.4 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.3 | 0.9 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.3 | 1.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.3 | 0.8 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.2 | 5.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.2 | 1.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 0.7 | GO:0030977 | taurine binding(GO:0030977) |
| 0.2 | 0.9 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.2 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.2 | 1.7 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.2 | 0.8 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.2 | 0.2 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.2 | 1.7 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.2 | 0.6 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.2 | 1.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 2.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.2 | 1.0 | GO:0015100 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.2 | 0.8 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.2 | 1.4 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.2 | 0.9 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 6.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.2 | 1.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.2 | 0.6 | GO:0004909 | interleukin-1, Type I, activating receptor activity(GO:0004909) |
| 0.2 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.2 | 0.9 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.2 | 1.9 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.2 | 0.5 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.2 | 1.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.2 | 0.5 | GO:0002113 | interleukin-33 binding(GO:0002113) |
| 0.2 | 1.3 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) |
| 0.2 | 0.8 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.2 | 5.4 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.2 | 0.5 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.2 | 0.6 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 3.6 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.1 | 1.5 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.3 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.1 | 3.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 0.9 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.4 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.5 | GO:0070697 | activin receptor binding(GO:0070697) |
| 0.1 | 0.7 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.1 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.5 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 1.3 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.1 | 4.7 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 0.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.1 | 0.9 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.5 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.1 | 0.5 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.4 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.1 | 1.3 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.3 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 1.8 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.1 | 0.4 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.3 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.1 | 0.4 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 2.0 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 3.5 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.1 | 0.2 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.1 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.0 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 1.1 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 1.4 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.8 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.7 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.1 | 1.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.5 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.1 | 0.7 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.4 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.5 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 1.6 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 1.4 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.1 | 1.6 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.1 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 0.3 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.1 | 0.4 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.1 | 0.5 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.6 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.1 | 0.3 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.1 | 1.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 2.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.1 | 0.3 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.1 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.1 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 4.6 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.1 | 0.6 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.4 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.1 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.7 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.5 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.1 | 1.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 0.3 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 0.8 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.1 | 1.3 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.1 | 0.3 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.1 | 0.6 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 3.0 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.1 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.2 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.1 | 0.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.1 | 0.5 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.1 | 0.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 3.2 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.1 | 0.7 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0015198 | oligopeptide transporter activity(GO:0015198) |
| 0.1 | 0.2 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.7 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.1 | 0.3 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.1 | 0.2 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.1 | 0.6 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.1 | 0.4 | GO:0032564 | dATP binding(GO:0032564) |
| 0.1 | 0.4 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.1 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.7 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.2 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.8 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 4.7 | GO:0005262 | calcium channel activity(GO:0005262) |
| 0.0 | 0.3 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.3 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 4.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.3 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 3.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.5 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.9 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.2 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 1.5 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0005452 | inorganic anion exchanger activity(GO:0005452) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 3.8 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.0 | 0.2 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.1 | GO:0031699 | beta-3 adrenergic receptor binding(GO:0031699) |
| 0.0 | 3.3 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 6.1 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.4 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 2.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 3.7 | GO:0004860 | protein kinase inhibitor activity(GO:0004860) |
| 0.0 | 0.4 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0048495 | GTPase inhibitor activity(GO:0005095) laminin-1 binding(GO:0043237) Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.3 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.4 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.3 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.3 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0005375 | copper ion transmembrane transporter activity(GO:0005375) |
| 0.0 | 0.1 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.6 | GO:0016701 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.4 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.2 | GO:0030547 | receptor inhibitor activity(GO:0030547) |
| 0.0 | 4.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.3 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.2 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.8 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
| 0.0 | 2.2 | GO:0005057 | receptor signaling protein activity(GO:0005057) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.2 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.4 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.2 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0005537 | mannose binding(GO:0005537) |
| 0.0 | 0.3 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.4 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.3 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.5 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.7 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.3 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.3 | GO:0051721 | protein phosphatase 2A binding(GO:0051721) |
| 0.0 | 0.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 5.1 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.1 | 4.2 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 3.8 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.1 | 3.3 | PID FGF PATHWAY | FGF signaling pathway |
| 0.1 | 5.0 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.1 | 5.4 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.1 | 0.6 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.1 | 0.8 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 1.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.1 | 7.3 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.1 | 1.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.1 | 0.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.1 | 1.7 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.8 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 1.6 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.4 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 1.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.7 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.7 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.9 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.9 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 1.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.2 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.3 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 1.1 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 4.1 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.2 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.7 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.7 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.2 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.0 | 0.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.5 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.3 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.6 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.9 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.2 | 4.8 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.2 | 6.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 2.9 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.2 | 3.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.2 | 7.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.2 | 2.9 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.2 | 1.7 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.2 | 2.2 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.6 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.1 | 4.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 0.8 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.1 | 1.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 1.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.1 | 1.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 1.4 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 0.6 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.1 | 3.7 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.1 | 1.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 1.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.0 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.1 | 0.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 2.6 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.1 | 0.6 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 0.5 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 0.7 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.1 | 1.3 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.1 | 0.9 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.6 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.7 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 1.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.8 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.4 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.2 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 1.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.5 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 1.1 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 1.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.6 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 1.8 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 1.5 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.5 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |
| 0.0 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | Genes involved in Apoptotic cleavage of cellular proteins |
| 0.0 | 1.7 | REACTOME POTASSIUM CHANNELS | Genes involved in Potassium Channels |
| 0.0 | 0.1 | REACTOME NEF MEDIATES DOWN MODULATION OF CELL SURFACE RECEPTORS BY RECRUITING THEM TO CLATHRIN ADAPTERS | Genes involved in Nef-mediates down modulation of cell surface receptors by recruiting them to clathrin adapters |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME NCAM SIGNALING FOR NEURITE OUT GROWTH | Genes involved in NCAM signaling for neurite out-growth |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.5 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.0 | REACTOME SIGNAL TRANSDUCTION BY L1 | Genes involved in Signal transduction by L1 |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 3.1 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.4 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.6 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.6 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.5 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.6 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.4 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.6 | REACTOME ASPARAGINE N LINKED GLYCOSYLATION | Genes involved in Asparagine N-linked glycosylation |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.6 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.7 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 0.2 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.8 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.3 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.3 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |