Project
12D miR HR13_24
Navigation
Downloads
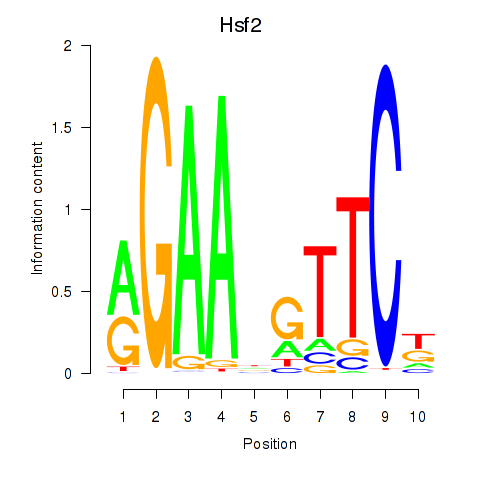
Results for Hsf2
Z-value: 1.29
Transcription factors associated with Hsf2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hsf2
|
ENSMUSG00000019878.7 | heat shock factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hsf2 | mm10_v2_chr10_+_57486354_57486414 | 0.40 | 2.2e-01 | Click! |
Activity profile of Hsf2 motif
Sorted Z-values of Hsf2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_+_4710119 | 3.88 |
ENSMUST00000105588.1
ENSMUST00000105589.1 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr13_+_51645232 | 1.45 |
ENSMUST00000075853.5
|
Cks2
|
CDC28 protein kinase regulatory subunit 2 |
| chr7_-_4752972 | 1.25 |
ENSMUST00000183971.1
ENSMUST00000182173.1 ENSMUST00000182738.1 ENSMUST00000184143.1 ENSMUST00000182111.1 ENSMUST00000182048.1 ENSMUST00000063324.7 |
Cox6b2
|
cytochrome c oxidase subunit VIb polypeptide 2 |
| chr15_+_62039216 | 1.21 |
ENSMUST00000183297.1
|
Pvt1
|
plasmacytoma variant translocation 1 |
| chr17_-_25797032 | 1.06 |
ENSMUST00000165838.1
ENSMUST00000002344.6 |
Metrn
|
meteorin, glial cell differentiation regulator |
| chr8_+_75214502 | 1.05 |
ENSMUST00000132133.1
|
Rasd2
|
RASD family, member 2 |
| chr2_-_181135103 | 1.02 |
ENSMUST00000149964.2
ENSMUST00000103050.3 ENSMUST00000081528.6 ENSMUST00000049792.8 ENSMUST00000103048.3 ENSMUST00000103047.3 ENSMUST00000129073.1 ENSMUST00000144592.1 ENSMUST00000139458.1 ENSMUST00000154164.1 ENSMUST00000123336.1 ENSMUST00000129361.1 ENSMUST00000103051.2 |
Kcnq2
|
potassium voltage-gated channel, subfamily Q, member 2 |
| chr5_+_76656512 | 0.97 |
ENSMUST00000086909.4
|
Gm10430
|
predicted gene 10430 |
| chr8_+_75213944 | 0.97 |
ENSMUST00000139848.1
|
Rasd2
|
RASD family, member 2 |
| chr5_-_77310049 | 0.96 |
ENSMUST00000047860.8
|
Noa1
|
nitric oxide associated 1 |
| chr12_+_81026800 | 0.94 |
ENSMUST00000110347.2
ENSMUST00000021564.4 ENSMUST00000129362.1 |
Smoc1
|
SPARC related modular calcium binding 1 |
| chr1_-_55088156 | 0.93 |
ENSMUST00000127861.1
ENSMUST00000144077.1 |
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr16_-_3909192 | 0.92 |
ENSMUST00000181699.1
|
1700016D08Rik
|
RIKEN cDNA 1700016D08 gene |
| chr10_-_80844025 | 0.91 |
ENSMUST00000053986.7
|
Lingo3
|
leucine rich repeat and Ig domain containing 3 |
| chr3_-_10208569 | 0.91 |
ENSMUST00000029041.4
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr4_-_42756543 | 0.91 |
ENSMUST00000102957.3
|
Ccl19
|
chemokine (C-C motif) ligand 19 |
| chr6_-_83536215 | 0.91 |
ENSMUST00000075161.5
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr1_+_136467958 | 0.90 |
ENSMUST00000047817.6
|
Kif14
|
kinesin family member 14 |
| chr11_+_116671658 | 0.88 |
ENSMUST00000106378.1
ENSMUST00000144049.1 |
1810032O08Rik
|
RIKEN cDNA 1810032O08 gene |
| chr9_-_110989611 | 0.86 |
ENSMUST00000084922.5
|
Rtp3
|
receptor transporter protein 3 |
| chr2_+_130277157 | 0.86 |
ENSMUST00000028890.8
ENSMUST00000159373.1 |
Nop56
|
NOP56 ribonucleoprotein |
| chr1_-_79858627 | 0.86 |
ENSMUST00000027467.4
|
Serpine2
|
serine (or cysteine) peptidase inhibitor, clade E, member 2 |
| chr2_+_30061754 | 0.85 |
ENSMUST00000149578.1
ENSMUST00000102866.3 |
Set
|
SET nuclear oncogene |
| chr17_-_34627148 | 0.82 |
ENSMUST00000171376.1
ENSMUST00000169287.1 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr17_-_34628005 | 0.82 |
ENSMUST00000166040.2
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr17_-_34627365 | 0.81 |
ENSMUST00000064953.8
ENSMUST00000170345.1 ENSMUST00000171121.2 ENSMUST00000168391.2 ENSMUST00000169067.2 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr17_-_50094277 | 0.80 |
ENSMUST00000113195.1
|
Rftn1
|
raftlin lipid raft linker 1 |
| chr14_+_66911170 | 0.80 |
ENSMUST00000089236.3
ENSMUST00000122431.2 |
Pnma2
|
paraneoplastic antigen MA2 |
| chr1_-_21961581 | 0.79 |
ENSMUST00000029667.6
ENSMUST00000173058.1 ENSMUST00000173404.1 |
Kcnq5
|
potassium voltage-gated channel, subfamily Q, member 5 |
| chr7_-_19280032 | 0.79 |
ENSMUST00000032560.4
|
Ppm1n
|
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr11_+_120949053 | 0.79 |
ENSMUST00000154187.1
ENSMUST00000100130.3 ENSMUST00000129473.1 ENSMUST00000168579.1 |
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr11_-_48946148 | 0.77 |
ENSMUST00000104958.1
|
Psme2b
|
protease (prosome, macropain) activator subunit 2B |
| chr2_+_25657944 | 0.75 |
ENSMUST00000100312.3
ENSMUST00000028306.6 ENSMUST00000100313.3 |
Lcn5
|
lipocalin 5 |
| chr6_+_128399766 | 0.75 |
ENSMUST00000001561.5
|
Nrip2
|
nuclear receptor interacting protein 2 |
| chr17_-_31129602 | 0.75 |
ENSMUST00000024827.4
|
Tff3
|
trefoil factor 3, intestinal |
| chr2_+_152847961 | 0.74 |
ENSMUST00000164120.1
ENSMUST00000178997.1 ENSMUST00000109816.1 |
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis) |
| chr5_-_23783700 | 0.74 |
ENSMUST00000119946.1
|
Pus7
|
pseudouridylate synthase 7 homolog (S. cerevisiae) |
| chr5_-_136244865 | 0.74 |
ENSMUST00000005188.9
|
Sh2b2
|
SH2B adaptor protein 2 |
| chrX_+_159459125 | 0.74 |
ENSMUST00000043151.5
ENSMUST00000112470.1 ENSMUST00000156172.1 |
Map7d2
|
MAP7 domain containing 2 |
| chr4_+_17853451 | 0.73 |
ENSMUST00000029881.3
|
Mmp16
|
matrix metallopeptidase 16 |
| chr2_-_28466266 | 0.69 |
ENSMUST00000127683.1
ENSMUST00000086370.4 |
1700007K13Rik
|
RIKEN cDNA 1700007K13 gene |
| chr9_-_107985863 | 0.68 |
ENSMUST00000048568.4
|
Fam212a
|
family with sequence similarity 212, member A |
| chr10_+_41810528 | 0.67 |
ENSMUST00000099931.3
|
Sesn1
|
sestrin 1 |
| chr10_+_127000709 | 0.67 |
ENSMUST00000026500.5
ENSMUST00000142698.1 |
Avil
|
advillin |
| chr6_+_134929089 | 0.67 |
ENSMUST00000183867.1
ENSMUST00000184991.1 ENSMUST00000183905.1 |
RP23-45G16.5
|
RP23-45G16.5 |
| chr2_-_181135220 | 0.66 |
ENSMUST00000016491.7
|
Kcnq2
|
potassium voltage-gated channel, subfamily Q, member 2 |
| chr2_+_152847993 | 0.65 |
ENSMUST00000028969.8
|
Tpx2
|
TPX2, microtubule-associated protein homolog (Xenopus laevis) |
| chr6_+_103510874 | 0.65 |
ENSMUST00000066905.6
|
Chl1
|
cell adhesion molecule with homology to L1CAM |
| chr18_+_61953048 | 0.65 |
ENSMUST00000051720.5
|
Sh3tc2
|
SH3 domain and tetratricopeptide repeats 2 |
| chr3_+_79591356 | 0.64 |
ENSMUST00000029382.7
|
Ppid
|
peptidylprolyl isomerase D (cyclophilin D) |
| chr1_-_55088024 | 0.63 |
ENSMUST00000027123.8
|
Hspd1
|
heat shock protein 1 (chaperonin) |
| chr12_-_17324703 | 0.62 |
ENSMUST00000020884.9
ENSMUST00000095820.5 ENSMUST00000127185.1 |
Atp6v1c2
|
ATPase, H+ transporting, lysosomal V1 subunit C2 |
| chr9_-_97018823 | 0.62 |
ENSMUST00000055433.4
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr15_+_78428564 | 0.61 |
ENSMUST00000166142.2
ENSMUST00000162517.1 ENSMUST00000089414.4 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr7_-_101933815 | 0.61 |
ENSMUST00000106963.1
ENSMUST00000106966.1 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr7_+_44112657 | 0.61 |
ENSMUST00000077528.6
|
Klk1b22
|
kallikrein 1-related peptidase b22 |
| chr2_-_144527341 | 0.60 |
ENSMUST00000163701.1
ENSMUST00000081982.5 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr9_+_8544196 | 0.60 |
ENSMUST00000050433.6
|
Trpc6
|
transient receptor potential cation channel, subfamily C, member 6 |
| chr3_-_32985076 | 0.60 |
ENSMUST00000108221.1
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr9_+_44134562 | 0.60 |
ENSMUST00000034650.8
ENSMUST00000098852.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr6_+_21986887 | 0.59 |
ENSMUST00000151315.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr17_-_26886175 | 0.59 |
ENSMUST00000108741.2
|
Gm17382
|
predicted gene, 17382 |
| chr14_-_7332395 | 0.58 |
ENSMUST00000100886.3
|
Gm5797
|
predicted gene 5797 |
| chr5_-_137858034 | 0.58 |
ENSMUST00000110978.2
|
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr10_-_67912620 | 0.57 |
ENSMUST00000064656.7
|
Zfp365
|
zinc finger protein 365 |
| chr4_-_117125618 | 0.56 |
ENSMUST00000183310.1
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr5_-_100820929 | 0.56 |
ENSMUST00000117364.1
ENSMUST00000055245.6 |
Fam175a
|
family with sequence similarity 175, member A |
| chr7_+_18987518 | 0.55 |
ENSMUST00000063563.7
|
Nanos2
|
nanos homolog 2 (Drosophila) |
| chr14_-_65833963 | 0.55 |
ENSMUST00000022613.9
|
Esco2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae) |
| chr1_+_66321708 | 0.54 |
ENSMUST00000114013.1
|
Map2
|
microtubule-associated protein 2 |
| chr8_-_70892752 | 0.54 |
ENSMUST00000000809.2
|
Slc5a5
|
solute carrier family 5 (sodium iodide symporter), member 5 |
| chrX_+_139800795 | 0.54 |
ENSMUST00000054889.3
|
Cldn2
|
claudin 2 |
| chr17_-_25727364 | 0.53 |
ENSMUST00000170070.1
ENSMUST00000048054.7 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr17_+_12119274 | 0.53 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr11_+_115564434 | 0.53 |
ENSMUST00000021085.4
|
Nup85
|
nucleoporin 85 |
| chr7_+_16781341 | 0.53 |
ENSMUST00000108496.2
|
Slc1a5
|
solute carrier family 1 (neutral amino acid transporter), member 5 |
| chr7_-_101933780 | 0.52 |
ENSMUST00000106964.1
ENSMUST00000078448.3 |
Lrrc51
|
leucine rich repeat containing 51 |
| chr2_+_31759993 | 0.50 |
ENSMUST00000124089.1
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr19_+_30232921 | 0.50 |
ENSMUST00000025797.5
|
Mbl2
|
mannose-binding lectin (protein C) 2 |
| chrX_-_75843063 | 0.50 |
ENSMUST00000114057.1
|
Pls3
|
plastin 3 (T-isoform) |
| chr5_-_124578992 | 0.50 |
ENSMUST00000128920.1
|
Eif2b1
|
eukaryotic translation initiation factor 2B, subunit 1 (alpha) |
| chrX_-_74246534 | 0.50 |
ENSMUST00000101454.2
ENSMUST00000033699.6 |
Flna
|
filamin, alpha |
| chr19_-_10203880 | 0.48 |
ENSMUST00000142241.1
ENSMUST00000116542.2 ENSMUST00000025651.5 ENSMUST00000156291.1 |
Fen1
|
flap structure specific endonuclease 1 |
| chr18_+_23415400 | 0.47 |
ENSMUST00000115832.2
ENSMUST00000047954.7 |
Dtna
|
dystrobrevin alpha |
| chr2_-_65364000 | 0.47 |
ENSMUST00000155962.1
ENSMUST00000112420.1 ENSMUST00000152324.1 |
Slc38a11
|
solute carrier family 38, member 11 |
| chr1_-_78488846 | 0.47 |
ENSMUST00000068333.7
ENSMUST00000170217.1 |
Farsb
|
phenylalanyl-tRNA synthetase, beta subunit |
| chrX_+_37255007 | 0.47 |
ENSMUST00000185028.1
ENSMUST00000185050.1 |
Rhox3a
|
reproductive homeobox 3A |
| chr17_+_34629533 | 0.47 |
ENSMUST00000015620.6
|
Prrt1
|
proline-rich transmembrane protein 1 |
| chr1_+_166130238 | 0.47 |
ENSMUST00000060833.7
ENSMUST00000166159.1 |
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr12_-_110696289 | 0.47 |
ENSMUST00000021698.6
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr10_+_7667503 | 0.46 |
ENSMUST00000040135.8
|
Nup43
|
nucleoporin 43 |
| chr4_+_150236685 | 0.46 |
ENSMUST00000150175.1
|
Eno1
|
enolase 1, alpha non-neuron |
| chr12_-_110696248 | 0.46 |
ENSMUST00000124156.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr7_-_105482197 | 0.46 |
ENSMUST00000047040.2
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr7_+_28350652 | 0.46 |
ENSMUST00000082134.4
|
Rps16
|
ribosomal protein S16 |
| chr2_+_25180737 | 0.46 |
ENSMUST00000104999.2
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr7_+_79743142 | 0.46 |
ENSMUST00000035622.7
|
Wdr93
|
WD repeat domain 93 |
| chr17_-_70849644 | 0.46 |
ENSMUST00000134654.1
ENSMUST00000172229.1 ENSMUST00000127719.1 |
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr2_+_43748802 | 0.46 |
ENSMUST00000112824.1
ENSMUST00000055776.7 |
Arhgap15
|
Rho GTPase activating protein 15 |
| chrX_-_157492280 | 0.45 |
ENSMUST00000112529.1
|
Sms
|
spermine synthase |
| chr4_+_95579463 | 0.45 |
ENSMUST00000150830.1
ENSMUST00000134012.2 |
Fggy
|
FGGY carbohydrate kinase domain containing |
| chrX_-_74246364 | 0.45 |
ENSMUST00000130007.1
|
Flna
|
filamin, alpha |
| chr6_+_21986438 | 0.44 |
ENSMUST00000115382.1
|
Cped1
|
cadherin-like and PC-esterase domain containing 1 |
| chr13_+_75839868 | 0.44 |
ENSMUST00000022082.7
|
Glrx
|
glutaredoxin |
| chr16_-_16869255 | 0.44 |
ENSMUST00000075017.4
|
Vpreb1
|
pre-B lymphocyte gene 1 |
| chr7_-_118995211 | 0.44 |
ENSMUST00000008878.8
|
Gprc5b
|
G protein-coupled receptor, family C, group 5, member B |
| chr6_-_102464667 | 0.44 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chr8_-_48555846 | 0.43 |
ENSMUST00000110345.1
ENSMUST00000110343.1 |
Tenm3
|
teneurin transmembrane protein 3 |
| chr3_+_79884496 | 0.43 |
ENSMUST00000118853.1
|
Fam198b
|
family with sequence similarity 198, member B |
| chr8_+_15011025 | 0.42 |
ENSMUST00000069399.6
|
Kbtbd11
|
kelch repeat and BTB (POZ) domain containing 11 |
| chr1_+_166130467 | 0.42 |
ENSMUST00000166860.1
|
Gpa33
|
glycoprotein A33 (transmembrane) |
| chr17_-_70851189 | 0.42 |
ENSMUST00000059775.8
|
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr10_+_121033960 | 0.42 |
ENSMUST00000020439.4
ENSMUST00000175867.1 |
Wif1
|
Wnt inhibitory factor 1 |
| chr13_-_110280103 | 0.41 |
ENSMUST00000167824.1
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr17_-_45686120 | 0.41 |
ENSMUST00000143907.1
ENSMUST00000127065.1 |
Tmem63b
|
transmembrane protein 63b |
| chr2_-_132029845 | 0.41 |
ENSMUST00000028814.8
|
Rassf2
|
Ras association (RalGDS/AF-6) domain family member 2 |
| chr7_+_105375053 | 0.41 |
ENSMUST00000106805.2
|
Gm5901
|
predicted gene 5901 |
| chr2_-_172370506 | 0.41 |
ENSMUST00000109139.1
ENSMUST00000028997.7 ENSMUST00000109140.3 |
Aurka
|
aurora kinase A |
| chr9_+_40269202 | 0.41 |
ENSMUST00000114956.3
ENSMUST00000049941.5 |
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr7_-_70366735 | 0.41 |
ENSMUST00000089565.5
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr13_+_41001002 | 0.40 |
ENSMUST00000046951.9
|
Pak1ip1
|
PAK1 interacting protein 1 |
| chr18_-_47368446 | 0.40 |
ENSMUST00000076043.6
ENSMUST00000135790.1 |
Sema6a
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr5_+_25246775 | 0.40 |
ENSMUST00000144971.1
|
Galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 |
| chr5_+_92603039 | 0.39 |
ENSMUST00000050952.3
|
Stbd1
|
starch binding domain 1 |
| chrX_-_48594373 | 0.39 |
ENSMUST00000088898.4
ENSMUST00000072292.5 |
Zfp280c
|
zinc finger protein 280C |
| chr11_-_106973090 | 0.39 |
ENSMUST00000150366.1
|
Gm11707
|
predicted gene 11707 |
| chr3_+_27984145 | 0.39 |
ENSMUST00000067757.4
|
Pld1
|
phospholipase D1 |
| chr12_-_110696332 | 0.38 |
ENSMUST00000094361.4
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr6_+_85431970 | 0.38 |
ENSMUST00000045693.7
|
Smyd5
|
SET and MYND domain containing 5 |
| chr3_-_101287897 | 0.37 |
ENSMUST00000029456.4
|
Cd2
|
CD2 antigen |
| chr3_-_108017806 | 0.37 |
ENSMUST00000126593.1
|
Gstm1
|
glutathione S-transferase, mu 1 |
| chr5_-_62766153 | 0.37 |
ENSMUST00000076623.4
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chrX_+_152781986 | 0.36 |
ENSMUST00000026383.3
|
Gpr143
|
G protein-coupled receptor 143 |
| chr9_+_40269273 | 0.36 |
ENSMUST00000176185.1
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr10_-_44458715 | 0.36 |
ENSMUST00000039174.4
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr5_+_92331828 | 0.36 |
ENSMUST00000125462.1
ENSMUST00000121096.1 ENSMUST00000113083.2 |
Art3
|
ADP-ribosyltransferase 3 |
| chr14_+_70077375 | 0.36 |
ENSMUST00000035908.1
|
Egr3
|
early growth response 3 |
| chr6_+_24528144 | 0.36 |
ENSMUST00000031696.3
|
Asb15
|
ankyrin repeat and SOCS box-containing 15 |
| chr17_+_55749978 | 0.36 |
ENSMUST00000025004.6
|
Emr4
|
EGF-like module containing, mucin-like, hormone receptor-like sequence 4 |
| chr10_-_117063764 | 0.36 |
ENSMUST00000047672.7
|
Cct2
|
chaperonin containing Tcp1, subunit 2 (beta) |
| chr7_+_102702331 | 0.36 |
ENSMUST00000094124.3
|
Olfr558
|
olfactory receptor 558 |
| chr7_-_140787826 | 0.36 |
ENSMUST00000026553.4
|
Syce1
|
synaptonemal complex central element protein 1 |
| chr9_-_107609215 | 0.35 |
ENSMUST00000102532.3
|
Sema3b
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B |
| chr2_-_151039363 | 0.35 |
ENSMUST00000128627.1
ENSMUST00000066640.4 |
Ninl
Nanp
|
ninein-like N-acetylneuraminic acid phosphatase |
| chr7_-_141443314 | 0.35 |
ENSMUST00000106005.2
|
Lrdd
|
leucine-rich and death domain containing |
| chr1_+_40515362 | 0.34 |
ENSMUST00000027237.5
|
Il18rap
|
interleukin 18 receptor accessory protein |
| chr3_-_121171678 | 0.34 |
ENSMUST00000170781.1
ENSMUST00000039761.5 ENSMUST00000106467.1 ENSMUST00000106466.3 ENSMUST00000164925.2 |
Rwdd3
|
RWD domain containing 3 |
| chr14_+_77904239 | 0.34 |
ENSMUST00000022591.8
|
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr12_-_28635914 | 0.33 |
ENSMUST00000074267.3
|
Rps7
|
ribosomal protein S7 |
| chr6_+_63255971 | 0.33 |
ENSMUST00000159561.1
ENSMUST00000095852.3 |
Grid2
|
glutamate receptor, ionotropic, delta 2 |
| chr13_+_31806627 | 0.33 |
ENSMUST00000062292.2
|
Foxc1
|
forkhead box C1 |
| chr6_-_34910563 | 0.33 |
ENSMUST00000152488.1
ENSMUST00000149448.1 ENSMUST00000133336.1 |
Wdr91
|
WD repeat domain 91 |
| chr1_-_72874877 | 0.33 |
ENSMUST00000027377.8
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr4_+_130047840 | 0.33 |
ENSMUST00000044565.8
ENSMUST00000132251.1 |
Col16a1
|
collagen, type XVI, alpha 1 |
| chr4_+_95579417 | 0.33 |
ENSMUST00000043335.4
|
Fggy
|
FGGY carbohydrate kinase domain containing |
| chr5_+_112255813 | 0.33 |
ENSMUST00000031286.6
ENSMUST00000131673.1 ENSMUST00000112375.1 |
Crybb1
|
crystallin, beta B1 |
| chr6_+_129397297 | 0.33 |
ENSMUST00000032262.7
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr6_+_113046225 | 0.33 |
ENSMUST00000032398.8
ENSMUST00000155378.1 |
Thumpd3
|
THUMP domain containing 3 |
| chr1_+_164796723 | 0.32 |
ENSMUST00000027861.4
|
Dpt
|
dermatopontin |
| chr2_+_162987330 | 0.32 |
ENSMUST00000018012.7
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr9_-_116175318 | 0.32 |
ENSMUST00000061101.4
ENSMUST00000035014.6 |
Tgfbr2
|
transforming growth factor, beta receptor II |
| chr7_-_141443989 | 0.32 |
ENSMUST00000026580.5
|
Lrdd
|
leucine-rich and death domain containing |
| chr10_-_78591945 | 0.31 |
ENSMUST00000040580.6
|
Syde1
|
synapse defective 1, Rho GTPase, homolog 1 (C. elegans) |
| chr6_+_56985295 | 0.31 |
ENSMUST00000164307.1
|
Vmn1r5
|
vomeronasal 1 receptor 5 |
| chr17_-_35838208 | 0.31 |
ENSMUST00000134978.2
|
Tubb5
|
tubulin, beta 5 class I |
| chr9_+_40269430 | 0.31 |
ENSMUST00000171835.2
|
Scn3b
|
sodium channel, voltage-gated, type III, beta |
| chr13_+_91461050 | 0.31 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr9_+_20868628 | 0.31 |
ENSMUST00000043911.7
|
A230050P20Rik
|
RIKEN cDNA A230050P20 gene |
| chr9_-_110624361 | 0.31 |
ENSMUST00000035069.9
|
Nradd
|
neurotrophin receptor associated death domain |
| chr16_+_36156801 | 0.30 |
ENSMUST00000079184.4
|
Stfa2l1
|
stefin A2 like 1 |
| chr12_-_84400929 | 0.30 |
ENSMUST00000122194.1
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr6_-_48766519 | 0.30 |
ENSMUST00000038811.8
|
Gimap3
|
GTPase, IMAP family member 3 |
| chrX_+_37546975 | 0.30 |
ENSMUST00000081327.5
ENSMUST00000184210.1 ENSMUST00000184270.1 |
Rhox3e
|
reproductive homeobox 3E |
| chr19_-_4839286 | 0.30 |
ENSMUST00000037246.5
|
Ccs
|
copper chaperone for superoxide dismutase |
| chr6_-_95718800 | 0.30 |
ENSMUST00000079847.5
|
Suclg2
|
succinate-Coenzyme A ligase, GDP-forming, beta subunit |
| chr12_+_29938036 | 0.30 |
ENSMUST00000122328.1
ENSMUST00000118321.1 |
Pxdn
|
peroxidasin homolog (Drosophila) |
| chr19_+_6057888 | 0.30 |
ENSMUST00000043074.5
ENSMUST00000178310.1 |
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr19_+_6057925 | 0.29 |
ENSMUST00000179142.1
|
Fau
|
Finkel-Biskis-Reilly murine sarcoma virus (FBR-MuSV) ubiquitously expressed (fox derived) |
| chr3_-_101287879 | 0.29 |
ENSMUST00000152321.1
|
Cd2
|
CD2 antigen |
| chr13_-_92794809 | 0.29 |
ENSMUST00000022213.7
|
Thbs4
|
thrombospondin 4 |
| chr10_+_80292453 | 0.29 |
ENSMUST00000068408.7
ENSMUST00000062674.6 |
Rps15
|
ribosomal protein S15 |
| chr12_+_37880700 | 0.29 |
ENSMUST00000040500.7
|
Dgkb
|
diacylglycerol kinase, beta |
| chr3_+_68468162 | 0.29 |
ENSMUST00000182532.1
|
Schip1
|
schwannomin interacting protein 1 |
| chr3_-_144760841 | 0.29 |
ENSMUST00000059091.5
|
Clca1
|
chloride channel calcium activated 1 |
| chr10_+_40629987 | 0.29 |
ENSMUST00000019977.7
|
Ddo
|
D-aspartate oxidase |
| chr7_+_25681158 | 0.28 |
ENSMUST00000108403.3
|
B9d2
|
B9 protein domain 2 |
| chr3_-_88772578 | 0.28 |
ENSMUST00000090945.4
|
Syt11
|
synaptotagmin XI |
| chr7_+_101663705 | 0.28 |
ENSMUST00000106998.1
|
Clpb
|
ClpB caseinolytic peptidase B |
| chr18_-_56562187 | 0.28 |
ENSMUST00000171844.2
|
Aldh7a1
|
aldehyde dehydrogenase family 7, member A1 |
| chr5_-_137871739 | 0.28 |
ENSMUST00000164886.1
|
Pilrb2
|
paired immunoglobin-like type 2 receptor beta 2 |
| chr5_+_151368683 | 0.28 |
ENSMUST00000181114.1
ENSMUST00000181555.1 |
1700028E10Rik
|
RIKEN cDNA 1700028E10 gene |
| chr11_+_32347800 | 0.28 |
ENSMUST00000038753.5
|
Sh3pxd2b
|
SH3 and PX domains 2B |
| chr7_-_89941084 | 0.28 |
ENSMUST00000075010.4
ENSMUST00000153470.1 |
l7Rn6
|
lethal, Chr 7, Rinchik 6 |
| chr2_+_163820832 | 0.28 |
ENSMUST00000029188.7
|
Wisp2
|
WNT1 inducible signaling pathway protein 2 |
| chr17_-_80373541 | 0.28 |
ENSMUST00000086549.1
|
Gm10190
|
predicted gene 10190 |
| chr10_+_84622365 | 0.28 |
ENSMUST00000077175.5
|
Polr3b
|
polymerase (RNA) III (DNA directed) polypeptide B |
| chr17_-_35838259 | 0.28 |
ENSMUST00000001566.8
|
Tubb5
|
tubulin, beta 5 class I |
| chr7_+_12922290 | 0.27 |
ENSMUST00000108539.1
ENSMUST00000004554.7 ENSMUST00000147435.1 ENSMUST00000137329.1 |
Rps5
|
ribosomal protein S5 |
| chr15_-_8710734 | 0.27 |
ENSMUST00000005493.7
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr12_-_84400851 | 0.27 |
ENSMUST00000117286.1
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hsf2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.9 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.5 | 1.6 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.3 | 0.9 | GO:1905000 | regulation of membrane repolarization during atrial cardiac muscle cell action potential(GO:1905000) |
| 0.3 | 0.9 | GO:2000547 | regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.3 | 0.9 | GO:0033624 | negative regulation of integrin activation(GO:0033624) |
| 0.3 | 1.5 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.2 | 0.6 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.2 | 0.9 | GO:0061110 | dense core granule biogenesis(GO:0061110) |
| 0.2 | 0.3 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.2 | 0.5 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.4 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.1 | 0.5 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.8 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 1.1 | GO:0060371 | regulation of atrial cardiac muscle cell membrane depolarization(GO:0060371) |
| 0.1 | 0.5 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.6 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.1 | 0.3 | GO:0019043 | establishment of viral latency(GO:0019043) |
| 0.1 | 0.3 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.1 | 0.2 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.1 | 0.3 | GO:0003275 | apoptotic process involved in outflow tract morphogenesis(GO:0003275) regulation of apoptotic process involved in outflow tract morphogenesis(GO:1902256) response to chemokine(GO:1990868) cellular response to chemokine(GO:1990869) |
| 0.1 | 0.4 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.7 | GO:0001922 | B-1 B cell homeostasis(GO:0001922) |
| 0.1 | 0.4 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) positive regulation of oocyte development(GO:0060282) |
| 0.1 | 0.3 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 1.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.1 | 0.3 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.1 | 0.3 | GO:1990927 | calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.3 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.1 | 1.0 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.1 | 0.1 | GO:2000724 | regulation of cardiac vascular smooth muscle cell differentiation(GO:2000722) positive regulation of cardiac vascular smooth muscle cell differentiation(GO:2000724) |
| 0.1 | 0.3 | GO:1900208 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.1 | 0.5 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.1 | 0.2 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 0.6 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.1 | 0.4 | GO:0060849 | regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.1 | 0.5 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.1 | 0.3 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.1 | 0.6 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.1 | 0.8 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 2.0 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.1 | 0.5 | GO:0015705 | iodide transport(GO:0015705) |
| 0.1 | 0.6 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.4 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.1 | 1.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.6 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.2 | GO:0046379 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.2 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 0.5 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.1 | 0.5 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.1 | 1.0 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.4 | GO:2000587 | negative regulation of platelet-derived growth factor receptor-beta signaling pathway(GO:2000587) |
| 0.1 | 0.5 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.1 | 0.2 | GO:0031554 | regulation of DNA-templated transcription, termination(GO:0031554) |
| 0.1 | 0.2 | GO:0009826 | unidimensional cell growth(GO:0009826) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.2 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.2 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.1 | 0.2 | GO:1903659 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.2 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) regulation of antimicrobial peptide production(GO:0002784) regulation of antibacterial peptide production(GO:0002786) positive regulation of antibacterial peptide production(GO:0002803) activation of MAPK activity involved in innate immune response(GO:0035419) |
| 0.1 | 0.5 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.1 | 0.2 | GO:1902913 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of neuroepithelial cell differentiation(GO:1902913) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 1.3 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
| 0.1 | 0.4 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.1 | 0.8 | GO:0019321 | pentose metabolic process(GO:0019321) |
| 0.1 | 0.2 | GO:2001160 | regulation of histone H3-K79 methylation(GO:2001160) |
| 0.1 | 0.7 | GO:1902715 | positive regulation of interferon-gamma secretion(GO:1902715) |
| 0.1 | 0.2 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.2 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.4 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.8 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.0 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.9 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.1 | GO:0070375 | ERK5 cascade(GO:0070375) |
| 0.0 | 0.5 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.2 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0060128 | corticotropin hormone secreting cell differentiation(GO:0060128) |
| 0.0 | 0.3 | GO:1904179 | positive regulation of adipose tissue development(GO:1904179) |
| 0.0 | 0.2 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.0 | 0.7 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.0 | GO:0061668 | mitochondrial ribosome assembly(GO:0061668) |
| 0.0 | 0.2 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.4 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.0 | 1.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.4 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.3 | GO:0003433 | chondrocyte development involved in endochondral bone morphogenesis(GO:0003433) |
| 0.0 | 0.0 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.0 | 0.2 | GO:0070125 | mitochondrial translational elongation(GO:0070125) |
| 0.0 | 0.7 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.2 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.9 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.2 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.0 | 0.5 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.5 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0090472 | dibasic protein processing(GO:0090472) |
| 0.0 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.6 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) regulation of gamma-delta T cell activation(GO:0046643) |
| 0.0 | 0.1 | GO:0033575 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.4 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.8 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.2 | GO:0044806 | G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.2 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.5 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.3 | GO:0006450 | regulation of translational fidelity(GO:0006450) |
| 0.0 | 0.4 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:2000812 | regulation of barbed-end actin filament capping(GO:2000812) |
| 0.0 | 0.6 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.7 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.2 | GO:1904338 | regulation of dopaminergic neuron differentiation(GO:1904338) |
| 0.0 | 0.3 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.3 | GO:1904861 | excitatory synapse assembly(GO:1904861) |
| 0.0 | 0.7 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.3 | GO:0051324 | meiotic prophase I(GO:0007128) prophase(GO:0051324) |
| 0.0 | 0.0 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.2 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.0 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.9 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0051194 | positive regulation of nucleotide catabolic process(GO:0030813) positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.2 | GO:0010991 | negative regulation of SMAD protein complex assembly(GO:0010991) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) |
| 0.0 | 0.2 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) cellular response to actinomycin D(GO:0072717) |
| 0.0 | 0.3 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.2 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.2 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.4 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.0 | GO:0036446 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.2 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.0 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.6 | GO:0000462 | maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000462) |
| 0.0 | 0.2 | GO:0030828 | positive regulation of cGMP biosynthetic process(GO:0030828) |
| 0.0 | 1.1 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.6 | GO:0006471 | protein ADP-ribosylation(GO:0006471) |
| 0.0 | 0.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:2000520 | regulation of immunological synapse formation(GO:2000520) negative regulation of immunological synapse formation(GO:2000521) regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001188) negative regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001189) |
| 0.0 | 0.7 | GO:0035640 | exploration behavior(GO:0035640) |
| 0.0 | 0.7 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.3 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.6 | GO:0045724 | positive regulation of cilium assembly(GO:0045724) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.6 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.3 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.6 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) regulation of microtubule depolymerization(GO:0031114) |
| 0.0 | 0.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0048246 | macrophage chemotaxis(GO:0048246) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.1 | GO:0021914 | negative regulation of smoothened signaling pathway involved in ventral spinal cord patterning(GO:0021914) |
| 0.0 | 0.3 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.5 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.5 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.0 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.5 | GO:0006801 | superoxide metabolic process(GO:0006801) |
| 0.0 | 0.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.1 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.2 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 3.9 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.3 | 1.5 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.2 | 1.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 0.9 | GO:0031523 | Myb complex(GO:0031523) |
| 0.2 | 1.4 | GO:0005818 | aster(GO:0005818) |
| 0.2 | 1.0 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.2 | 0.5 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.2 | 0.6 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.4 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.9 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.1 | 1.0 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.1 | 0.6 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 1.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.7 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.1 | 0.4 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 0.9 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.0 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.7 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.1 | 0.4 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.1 | 1.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.5 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.1 | 0.6 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 1.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.1 | 0.5 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.7 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 2.2 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.2 | GO:0097059 | CNTFR-CLCF1 complex(GO:0097059) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0097524 | sperm plasma membrane(GO:0097524) |
| 0.0 | 0.2 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.2 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.4 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.9 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.6 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.5 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.2 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0042272 | nucleocytoplasmic shuttling complex(GO:0031074) nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 1.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.8 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.2 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.0 | 0.2 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.4 | GO:0071339 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.7 | GO:0030118 | clathrin coat(GO:0030118) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0060091 | kinocilium(GO:0060091) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 3.9 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.4 | 1.5 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.3 | 1.7 | GO:0002135 | CTP binding(GO:0002135) |
| 0.3 | 0.8 | GO:0019150 | D-ribulokinase activity(GO:0019150) |
| 0.2 | 2.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.2 | 0.9 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.2 | 1.6 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 0.9 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.2 | 0.9 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 1.4 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.2 | 1.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.1 | 0.4 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.1 | 0.5 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.9 | GO:0016018 | cyclosporin A binding(GO:0016018) |
| 0.1 | 0.5 | GO:0008802 | betaine-aldehyde dehydrogenase activity(GO:0008802) |
| 0.1 | 0.6 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.6 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 0.8 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.4 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.1 | 0.5 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.1 | 0.6 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.3 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 0.4 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.1 | 0.5 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.1 | 0.5 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 2.0 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.1 | 0.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.5 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.3 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.3 | GO:0004952 | dopamine neurotransmitter receptor activity(GO:0004952) |
| 0.1 | 0.3 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.5 | GO:0005550 | pheromone binding(GO:0005550) |
| 0.1 | 0.4 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.4 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.1 | 1.0 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 0.6 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0003884 | D-amino-acid oxidase activity(GO:0003884) |
| 0.1 | 0.4 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.4 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.5 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 0.3 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 1.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 0.2 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.1 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.1 | 0.2 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.4 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.1 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.2 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.1 | 0.2 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.9 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 1.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.5 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 1.7 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.6 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.5 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.0 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.1 | GO:0070410 | JUN kinase binding(GO:0008432) co-SMAD binding(GO:0070410) |
| 0.0 | 0.2 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.0 | 0.3 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.5 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.5 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.3 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.3 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.4 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0008260 | 3-oxoacid CoA-transferase activity(GO:0008260) |
| 0.0 | 0.7 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.2 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.9 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.6 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.3 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0016888 | endodeoxyribonuclease activity, producing 5'-phosphomonoesters(GO:0016888) |
| 0.0 | 0.3 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.6 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) |
| 0.0 | 0.1 | GO:0036402 | proteasome-activating ATPase activity(GO:0036402) |
| 0.0 | 0.4 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.2 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.1 | GO:0004028 | retinal dehydrogenase activity(GO:0001758) 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.7 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.3 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.6 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.8 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.3 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0070883 | pre-miRNA binding(GO:0070883) |
| 0.0 | 0.2 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.5 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.7 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.7 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.8 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.1 | 1.7 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.1 | 2.1 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.1 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.1 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.0 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.8 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 1.2 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.4 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 1.5 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.9 | PID INSULIN PATHWAY | Insulin Pathway |
| 0.0 | 2.4 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.4 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.8 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.6 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.1 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.1 | 0.8 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 3.1 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.1 | 2.1 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.5 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 2.5 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 1.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 1.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.8 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 1.2 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.0 | 1.0 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.5 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.7 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.7 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 1.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.9 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.5 | REACTOME TRNA AMINOACYLATION | Genes involved in tRNA Aminoacylation |
| 0.0 | 0.4 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.5 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.5 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.6 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |