Project
12D miR HR13_24
Navigation
Downloads
Results for Irf2_Irf1_Irf8_Irf9_Irf7
Z-value: 2.09
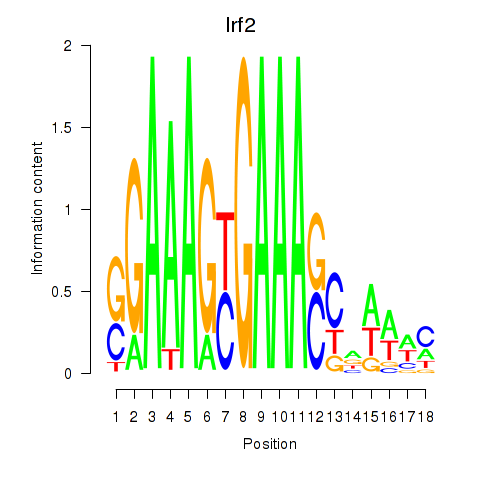
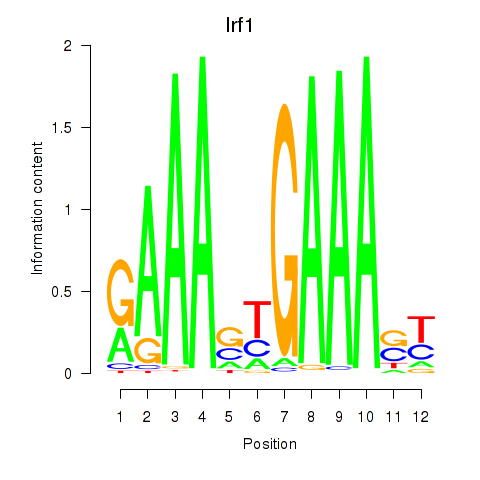
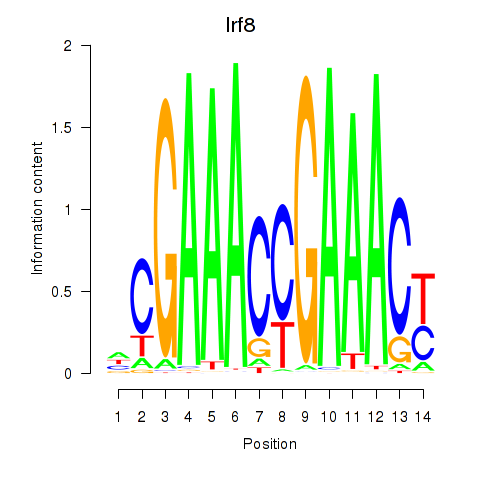
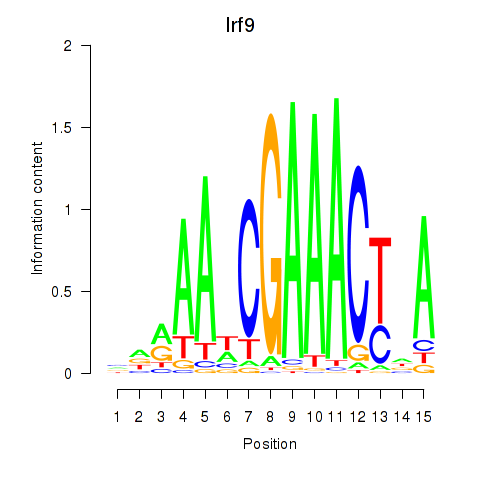
Transcription factors associated with Irf2_Irf1_Irf8_Irf9_Irf7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Irf2
|
ENSMUSG00000031627.7 | interferon regulatory factor 2 |
|
Irf1
|
ENSMUSG00000018899.10 | interferon regulatory factor 1 |
|
Irf8
|
ENSMUSG00000041515.3 | interferon regulatory factor 8 |
|
Irf9
|
ENSMUSG00000002325.8 | interferon regulatory factor 9 |
|
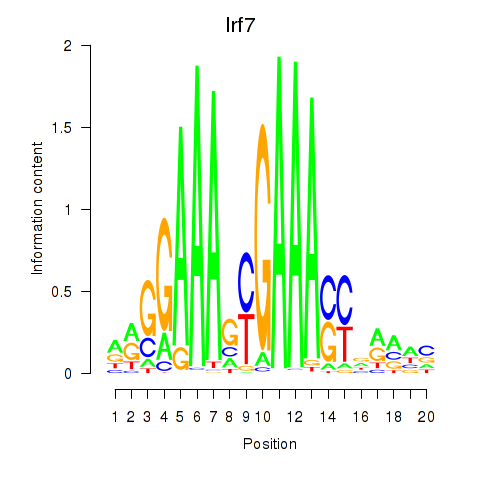
Irf7
|
ENSMUSG00000025498.8 | interferon regulatory factor 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Irf2 | mm10_v2_chr8_+_46739745_46739791 | -0.80 | 2.8e-03 | Click! |
| Irf8 | mm10_v2_chr8_+_120736352_120736385 | -0.52 | 1.0e-01 | Click! |
| Irf1 | mm10_v2_chr11_+_53770458_53770509 | -0.46 | 1.5e-01 | Click! |
| Irf9 | mm10_v2_chr14_+_55604550_55604579 | -0.34 | 3.0e-01 | Click! |
| Irf7 | mm10_v2_chr7_-_141266415_141266481 | -0.00 | 9.9e-01 | Click! |
Activity profile of Irf2_Irf1_Irf8_Irf9_Irf7 motif
Sorted Z-values of Irf2_Irf1_Irf8_Irf9_Irf7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_61928081 | 5.73 |
ENSMUST00000154398.1
ENSMUST00000093485.2 ENSMUST00000156980.1 ENSMUST00000070631.7 |
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr3_-_113574242 | 4.62 |
ENSMUST00000142505.2
|
Amy1
|
amylase 1, salivary |
| chr3_-_113574758 | 4.12 |
ENSMUST00000106540.1
|
Amy1
|
amylase 1, salivary |
| chr7_+_104244496 | 3.79 |
ENSMUST00000106854.1
ENSMUST00000143414.1 |
Trim34a
|
tripartite motif-containing 34A |
| chr7_+_104244449 | 3.78 |
ENSMUST00000106849.2
ENSMUST00000060315.5 |
Trim34a
|
tripartite motif-containing 34A |
| chr7_+_104244465 | 3.61 |
ENSMUST00000106848.1
|
Trim34a
|
tripartite motif-containing 34A |
| chr19_+_34640871 | 3.47 |
ENSMUST00000102824.3
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr12_-_79007276 | 3.45 |
ENSMUST00000056660.6
ENSMUST00000174721.1 |
Tmem229b
|
transmembrane protein 229B |
| chr5_-_92348871 | 2.94 |
ENSMUST00000038816.6
ENSMUST00000118006.1 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr8_-_105938384 | 2.75 |
ENSMUST00000034369.8
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10 |
| chr5_-_105239533 | 2.57 |
ENSMUST00000065588.6
|
Gbp10
|
guanylate-binding protein 10 |
| chr4_-_40239779 | 2.18 |
ENSMUST00000037907.6
|
Ddx58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr7_+_78913765 | 2.07 |
ENSMUST00000038142.8
|
Isg20
|
interferon-stimulated protein |
| chr12_-_78980758 | 2.03 |
ENSMUST00000174072.1
|
Tmem229b
|
transmembrane protein 229B |
| chr1_-_156674290 | 1.98 |
ENSMUST00000079625.4
|
Tor3a
|
torsin family 3, member A |
| chr4_-_40239700 | 1.97 |
ENSMUST00000142055.1
|
Ddx58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr19_+_34607927 | 1.93 |
ENSMUST00000076249.5
|
I830012O16Rik
|
RIKEN cDNA I830012O16 gene |
| chr17_+_33919332 | 1.90 |
ENSMUST00000025161.7
|
Tapbp
|
TAP binding protein |
| chr19_+_29367447 | 1.88 |
ENSMUST00000016640.7
|
Cd274
|
CD274 antigen |
| chr3_+_142620596 | 1.87 |
ENSMUST00000165774.1
|
Gbp2
|
guanylate binding protein 2 |
| chr10_-_75797728 | 1.82 |
ENSMUST00000139724.1
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr15_+_9436028 | 1.80 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chr19_+_55741810 | 1.78 |
ENSMUST00000111657.3
ENSMUST00000061496.9 ENSMUST00000041717.7 ENSMUST00000111662.4 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr12_+_26469204 | 1.78 |
ENSMUST00000020969.3
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr8_-_111338152 | 1.73 |
ENSMUST00000056157.7
ENSMUST00000120432.1 |
Mlkl
|
mixed lineage kinase domain-like |
| chr5_+_117357274 | 1.73 |
ENSMUST00000031309.9
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr2_-_103283760 | 1.70 |
ENSMUST00000111174.1
|
Ehf
|
ets homologous factor |
| chr9_+_5298517 | 1.65 |
ENSMUST00000027015.5
|
Casp1
|
caspase 1 |
| chr17_-_36042690 | 1.64 |
ENSMUST00000058801.8
ENSMUST00000080015.5 ENSMUST00000077960.6 |
H2-T22
|
histocompatibility 2, T region locus 22 |
| chr4_-_42773993 | 1.63 |
ENSMUST00000095114.4
|
Ccl21a
|
chemokine (C-C motif) ligand 21A (serine) |
| chr11_+_29718563 | 1.62 |
ENSMUST00000060992.5
|
Rtn4
|
reticulon 4 |
| chr4_+_42255767 | 1.59 |
ENSMUST00000178864.1
|
Ccl21b
|
chemokine (C-C motif) ligand 21B (leucine) |
| chr5_-_105293699 | 1.59 |
ENSMUST00000050011.8
|
Gbp6
|
guanylate binding protein 6 |
| chr2_-_51972990 | 1.58 |
ENSMUST00000145481.1
ENSMUST00000112705.2 |
Nmi
|
N-myc (and STAT) interactor |
| chr8_+_64947177 | 1.58 |
ENSMUST00000079896.7
ENSMUST00000026595.5 |
Tmem192
|
transmembrane protein 192 |
| chr12_-_26456423 | 1.54 |
ENSMUST00000020970.7
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr15_-_98728120 | 1.53 |
ENSMUST00000003445.6
|
Fkbp11
|
FK506 binding protein 11 |
| chr6_+_39381175 | 1.50 |
ENSMUST00000031986.4
|
Rab19
|
RAB19, member RAS oncogene family |
| chr6_-_125231772 | 1.50 |
ENSMUST00000043422.7
|
Tapbpl
|
TAP binding protein-like |
| chr1_+_52119438 | 1.49 |
ENSMUST00000070968.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr4_+_45972233 | 1.48 |
ENSMUST00000102929.1
|
Tdrd7
|
tudor domain containing 7 |
| chr6_-_39118211 | 1.45 |
ENSMUST00000038398.6
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr17_-_34187219 | 1.42 |
ENSMUST00000173831.1
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr19_+_22692613 | 1.42 |
ENSMUST00000099564.2
ENSMUST00000099566.3 |
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr4_-_145246855 | 1.38 |
ENSMUST00000030336.4
|
Tnfrsf1b
|
tumor necrosis factor receptor superfamily, member 1b |
| chr14_+_41105359 | 1.35 |
ENSMUST00000047286.6
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr16_-_24393588 | 1.35 |
ENSMUST00000181640.1
|
1110054M08Rik
|
RIKEN cDNA 1110054M08 gene |
| chr3_+_142560351 | 1.34 |
ENSMUST00000106221.1
|
Gbp3
|
guanylate binding protein 3 |
| chr9_+_107975529 | 1.34 |
ENSMUST00000035216.4
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr1_+_16688405 | 1.32 |
ENSMUST00000026881.4
|
Ly96
|
lymphocyte antigen 96 |
| chr3_+_142560108 | 1.31 |
ENSMUST00000128609.1
ENSMUST00000029935.7 |
Gbp3
|
guanylate binding protein 3 |
| chr7_-_102565425 | 1.31 |
ENSMUST00000106913.1
ENSMUST00000033264.4 |
Trim21
|
tripartite motif-containing 21 |
| chr7_-_141266415 | 1.29 |
ENSMUST00000106023.1
ENSMUST00000097952.2 ENSMUST00000026571.4 |
Irf7
|
interferon regulatory factor 7 |
| chr15_+_99392882 | 1.29 |
ENSMUST00000023749.8
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr1_+_153751859 | 1.26 |
ENSMUST00000182538.1
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr16_+_23609895 | 1.25 |
ENSMUST00000038423.5
|
Rtp4
|
receptor transporter protein 4 |
| chr7_-_104353328 | 1.24 |
ENSMUST00000130139.1
ENSMUST00000059037.8 |
Trim12c
|
tripartite motif-containing 12C |
| chr7_+_78914216 | 1.19 |
ENSMUST00000120331.2
|
Isg20
|
interferon-stimulated protein |
| chr10_+_128270546 | 1.18 |
ENSMUST00000105238.3
ENSMUST00000085708.2 |
Stat2
|
signal transducer and activator of transcription 2 |
| chr1_+_130826676 | 1.17 |
ENSMUST00000027675.7
|
Pigr
|
polymeric immunoglobulin receptor |
| chr11_+_101582236 | 1.16 |
ENSMUST00000039581.7
ENSMUST00000100403.2 ENSMUST00000107194.1 ENSMUST00000128614.1 |
Tmem106a
|
transmembrane protein 106A |
| chr5_-_120749848 | 1.14 |
ENSMUST00000053909.6
ENSMUST00000081491.6 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr2_+_24385313 | 1.13 |
ENSMUST00000056641.8
|
Psd4
|
pleckstrin and Sec7 domain containing 4 |
| chr5_+_117319292 | 1.12 |
ENSMUST00000086464.4
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr1_-_155146755 | 1.12 |
ENSMUST00000027744.8
|
Mr1
|
major histocompatibility complex, class I-related |
| chr3_+_142560052 | 1.11 |
ENSMUST00000106222.2
|
Gbp3
|
guanylate binding protein 3 |
| chr17_-_34862473 | 1.07 |
ENSMUST00000025229.4
ENSMUST00000176203.2 ENSMUST00000128767.1 |
Cfb
|
complement factor B |
| chr9_+_38718263 | 1.06 |
ENSMUST00000001544.5
ENSMUST00000118144.1 |
Vwa5a
|
von Willebrand factor A domain containing 5A |
| chr13_+_23738804 | 1.05 |
ENSMUST00000040914.1
|
Hist1h1c
|
histone cluster 1, H1c |
| chr11_+_69964758 | 1.04 |
ENSMUST00000108597.1
ENSMUST00000060651.5 ENSMUST00000108596.1 |
Cldn7
|
claudin 7 |
| chr2_-_62646146 | 1.03 |
ENSMUST00000112459.3
ENSMUST00000028259.5 |
Ifih1
|
interferon induced with helicase C domain 1 |
| chr2_-_51973219 | 1.01 |
ENSMUST00000028314.2
|
Nmi
|
N-myc (and STAT) interactor |
| chr6_+_96113146 | 0.99 |
ENSMUST00000122120.1
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chr11_+_114851142 | 0.99 |
ENSMUST00000133245.1
ENSMUST00000122967.2 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_-_48871344 | 0.99 |
ENSMUST00000049519.3
|
Irgm1
|
immunity-related GTPase family M member 1 |
| chr1_+_130826762 | 0.99 |
ENSMUST00000133792.1
|
Pigr
|
polymeric immunoglobulin receptor |
| chr4_-_46536134 | 0.98 |
ENSMUST00000046897.6
|
Trim14
|
tripartite motif-containing 14 |
| chr2_-_51934644 | 0.98 |
ENSMUST00000165313.1
|
Rbm43
|
RNA binding motif protein 43 |
| chr2_-_51934943 | 0.97 |
ENSMUST00000102767.1
ENSMUST00000102768.1 |
Rbm43
|
RNA binding motif protein 43 |
| chr13_+_56609516 | 0.97 |
ENSMUST00000045173.8
|
Tgfbi
|
transforming growth factor, beta induced |
| chr5_-_117319242 | 0.97 |
ENSMUST00000100834.1
|
Gm10399
|
predicted gene 10399 |
| chr3_+_27317028 | 0.97 |
ENSMUST00000046383.5
ENSMUST00000174840.1 |
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr5_+_114896936 | 0.96 |
ENSMUST00000031542.9
ENSMUST00000146072.1 ENSMUST00000150361.1 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr2_+_122147680 | 0.96 |
ENSMUST00000102476.4
|
B2m
|
beta-2 microglobulin |
| chr7_+_97453204 | 0.95 |
ENSMUST00000050732.7
ENSMUST00000121987.1 |
Kctd14
|
potassium channel tetramerisation domain containing 14 |
| chr17_-_34862122 | 0.95 |
ENSMUST00000154526.1
|
Cfb
|
complement factor B |
| chr4_+_41903610 | 0.95 |
ENSMUST00000098128.3
|
Gm21541
|
predicted gene, 21541 |
| chr6_-_54972603 | 0.95 |
ENSMUST00000060655.8
|
Nod1
|
nucleotide-binding oligomerization domain containing 1 |
| chr13_+_33004528 | 0.94 |
ENSMUST00000006391.4
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr4_+_42114817 | 0.93 |
ENSMUST00000098123.3
|
Gm13304
|
predicted gene 13304 |
| chr2_+_58755177 | 0.92 |
ENSMUST00000102755.3
|
Upp2
|
uridine phosphorylase 2 |
| chr5_+_117319258 | 0.92 |
ENSMUST00000111967.1
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr5_-_134229581 | 0.92 |
ENSMUST00000111275.1
ENSMUST00000016094.6 ENSMUST00000144086.1 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr5_+_147269959 | 0.91 |
ENSMUST00000085591.5
|
Pdx1
|
pancreatic and duodenal homeobox 1 |
| chr11_+_58199556 | 0.90 |
ENSMUST00000035266.4
ENSMUST00000094169.4 ENSMUST00000168280.1 ENSMUST00000058704.8 |
Igtp
Irgm2
|
interferon gamma induced GTPase immunity-related GTPase family M member 2 |
| chr7_+_51878967 | 0.89 |
ENSMUST00000051912.6
|
Gas2
|
growth arrest specific 2 |
| chr1_+_58711488 | 0.89 |
ENSMUST00000097722.2
ENSMUST00000114313.1 |
Cflar
|
CASP8 and FADD-like apoptosis regulator |
| chr8_+_127064107 | 0.89 |
ENSMUST00000162536.1
ENSMUST00000026921.6 ENSMUST00000162665.1 ENSMUST00000160766.1 ENSMUST00000162602.1 ENSMUST00000162531.1 ENSMUST00000160581.1 ENSMUST00000161355.1 ENSMUST00000159537.1 |
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr3_-_137981523 | 0.89 |
ENSMUST00000136613.1
ENSMUST00000029806.6 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chr3_+_68691424 | 0.89 |
ENSMUST00000107816.2
|
Il12a
|
interleukin 12a |
| chr18_-_61536522 | 0.87 |
ENSMUST00000171629.1
|
Arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr11_-_49064202 | 0.87 |
ENSMUST00000046745.6
|
Tgtp2
|
T cell specific GTPase 2 |
| chr19_-_11050500 | 0.86 |
ENSMUST00000099676.4
|
AW112010
|
expressed sequence AW112010 |
| chr16_-_97462903 | 0.85 |
ENSMUST00000142883.1
ENSMUST00000113768.1 ENSMUST00000023655.6 |
Mx1
|
myxovirus (influenza virus) resistance 1 |
| chr4_+_102589687 | 0.84 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr16_+_35938470 | 0.84 |
ENSMUST00000114878.1
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr1_+_153751946 | 0.83 |
ENSMUST00000183241.1
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr9_-_45204083 | 0.83 |
ENSMUST00000034599.8
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr7_-_45238794 | 0.83 |
ENSMUST00000098461.1
ENSMUST00000107797.1 |
Cd37
|
CD37 antigen |
| chr7_+_51879041 | 0.83 |
ENSMUST00000107591.2
|
Gas2
|
growth arrest specific 2 |
| chr13_+_51846673 | 0.82 |
ENSMUST00000021903.2
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr11_-_100704217 | 0.81 |
ENSMUST00000017974.6
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr11_-_48992226 | 0.80 |
ENSMUST00000059930.2
ENSMUST00000068063.3 |
Gm12185
Tgtp1
|
predicted gene 12185 T cell specific GTPase 1 |
| chr11_-_48871408 | 0.79 |
ENSMUST00000097271.2
|
Irgm1
|
immunity-related GTPase family M member 1 |
| chr2_-_24049389 | 0.78 |
ENSMUST00000051416.5
|
Hnmt
|
histamine N-methyltransferase |
| chr9_-_111057235 | 0.78 |
ENSMUST00000111888.1
|
Ccrl2
|
chemokine (C-C motif) receptor-like 2 |
| chr4_+_111719975 | 0.77 |
ENSMUST00000038868.7
ENSMUST00000070513.6 ENSMUST00000153746.1 |
Spata6
|
spermatogenesis associated 6 |
| chr5_-_120907510 | 0.77 |
ENSMUST00000080322.7
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr4_+_111720187 | 0.76 |
ENSMUST00000084354.3
|
Spata6
|
spermatogenesis associated 6 |
| chr2_+_58754910 | 0.76 |
ENSMUST00000059102.6
|
Upp2
|
uridine phosphorylase 2 |
| chr19_+_6164433 | 0.75 |
ENSMUST00000045042.7
|
Batf2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr3_+_60081861 | 0.75 |
ENSMUST00000029326.5
|
Sucnr1
|
succinate receptor 1 |
| chr3_+_106482427 | 0.75 |
ENSMUST00000029508.4
|
Dennd2d
|
DENN/MADD domain containing 2D |
| chr11_+_58215028 | 0.74 |
ENSMUST00000108836.1
|
Irgm2
|
immunity-related GTPase family M member 2 |
| chr16_+_97536079 | 0.73 |
ENSMUST00000024112.7
|
Mx2
|
myxovirus (influenza virus) resistance 2 |
| chr12_+_52699297 | 0.73 |
ENSMUST00000095737.3
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr12_+_103434211 | 0.73 |
ENSMUST00000079294.5
ENSMUST00000076788.5 ENSMUST00000076702.5 ENSMUST00000066701.6 ENSMUST00000085065.5 ENSMUST00000140838.1 |
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr6_-_34955903 | 0.72 |
ENSMUST00000147169.1
|
2010107G12Rik
|
RIKEN cDNA 2010107G12 gene |
| chr9_+_118040475 | 0.72 |
ENSMUST00000044454.5
|
Azi2
|
5-azacytidine induced gene 2 |
| chr5_+_114923234 | 0.72 |
ENSMUST00000031540.4
ENSMUST00000112143.3 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr1_-_174031712 | 0.71 |
ENSMUST00000059226.6
|
Ifi205
|
interferon activated gene 205 |
| chr14_-_26066961 | 0.71 |
ENSMUST00000100818.5
|
Tmem254c
|
transmembrane protein 254c |
| chr10_+_116177351 | 0.70 |
ENSMUST00000155606.1
ENSMUST00000128399.1 |
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr1_-_156036473 | 0.70 |
ENSMUST00000097527.3
ENSMUST00000027738.7 |
Tor1aip1
|
torsin A interacting protein 1 |
| chr5_+_35056813 | 0.70 |
ENSMUST00000101298.2
ENSMUST00000114270.1 ENSMUST00000133381.1 |
Dok7
|
docking protein 7 |
| chr4_+_54947976 | 0.70 |
ENSMUST00000098070.3
|
Zfp462
|
zinc finger protein 462 |
| chr10_+_116177217 | 0.69 |
ENSMUST00000148731.1
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr16_+_35938972 | 0.68 |
ENSMUST00000023622.6
ENSMUST00000114877.1 |
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr14_+_30716377 | 0.68 |
ENSMUST00000112177.1
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr6_+_34863130 | 0.68 |
ENSMUST00000074949.3
|
Tmem140
|
transmembrane protein 140 |
| chr11_+_61065798 | 0.67 |
ENSMUST00000041944.2
ENSMUST00000108717.2 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr13_+_42866247 | 0.67 |
ENSMUST00000131942.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chrX_-_9469288 | 0.67 |
ENSMUST00000015484.3
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr18_+_37264998 | 0.67 |
ENSMUST00000052366.3
|
Pcdhb1
|
protocadherin beta 1 |
| chr13_-_23710714 | 0.67 |
ENSMUST00000091707.6
ENSMUST00000006787.7 ENSMUST00000091706.6 |
Hfe
|
hemochromatosis |
| chr18_-_60273267 | 0.67 |
ENSMUST00000090260.4
|
Gm4841
|
predicted gene 4841 |
| chr5_-_65492984 | 0.66 |
ENSMUST00000139122.1
|
Smim14
|
small integral membrane protein 14 |
| chr19_+_36409719 | 0.66 |
ENSMUST00000062389.5
|
Pcgf5
|
polycomb group ring finger 5 |
| chr11_-_47379405 | 0.66 |
ENSMUST00000077221.5
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr1_+_61638819 | 0.65 |
ENSMUST00000138768.1
ENSMUST00000075374.3 |
Pard3b
|
par-3 partitioning defective 3 homolog B (C. elegans) |
| chr9_+_118040509 | 0.65 |
ENSMUST00000133580.1
|
Azi2
|
5-azacytidine induced gene 2 |
| chr1_-_69685937 | 0.64 |
ENSMUST00000027146.2
|
Ikzf2
|
IKAROS family zinc finger 2 |
| chr18_-_3299537 | 0.63 |
ENSMUST00000129435.1
ENSMUST00000122958.1 |
Crem
|
cAMP responsive element modulator |
| chr15_+_83563571 | 0.62 |
ENSMUST00000047419.6
|
Tspo
|
translocator protein |
| chr15_+_57694651 | 0.62 |
ENSMUST00000096430.4
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr11_+_3989924 | 0.62 |
ENSMUST00000109981.1
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr10_-_88356990 | 0.61 |
ENSMUST00000020249.1
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr14_+_77904365 | 0.61 |
ENSMUST00000169978.1
|
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr18_-_3299452 | 0.61 |
ENSMUST00000126578.1
|
Crem
|
cAMP responsive element modulator |
| chr6_-_38354243 | 0.61 |
ENSMUST00000114900.1
|
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr18_+_60376029 | 0.61 |
ENSMUST00000066912.5
ENSMUST00000032473.6 |
Iigp1
|
interferon inducible GTPase 1 |
| chr11_-_102296618 | 0.60 |
ENSMUST00000107132.2
ENSMUST00000073234.2 |
Atxn7l3
|
ataxin 7-like 3 |
| chr4_+_138972885 | 0.60 |
ENSMUST00000123636.1
ENSMUST00000043042.3 ENSMUST00000050949.2 |
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chrX_+_144688907 | 0.60 |
ENSMUST00000112843.1
|
Zcchc16
|
zinc finger, CCHC domain containing 16 |
| chr10_+_78069351 | 0.60 |
ENSMUST00000105393.1
|
Icosl
|
icos ligand |
| chr6_-_3399545 | 0.60 |
ENSMUST00000120087.3
|
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr17_+_30901811 | 0.59 |
ENSMUST00000114574.1
|
Glp1r
|
glucagon-like peptide 1 receptor |
| chr5_-_52471534 | 0.59 |
ENSMUST00000059428.5
|
Ccdc149
|
coiled-coil domain containing 149 |
| chr7_-_45239108 | 0.59 |
ENSMUST00000033063.6
|
Cd37
|
CD37 antigen |
| chr16_+_24393350 | 0.58 |
ENSMUST00000038053.6
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr8_+_83165348 | 0.58 |
ENSMUST00000034145.4
|
Tbc1d9
|
TBC1 domain family, member 9 |
| chr3_+_142530329 | 0.57 |
ENSMUST00000171263.1
ENSMUST00000045097.9 |
Gbp7
|
guanylate binding protein 7 |
| chr15_+_99392948 | 0.57 |
ENSMUST00000161250.1
ENSMUST00000160635.1 ENSMUST00000161778.1 |
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chrX_-_104857228 | 0.57 |
ENSMUST00000033575.5
|
Magee2
|
melanoma antigen, family E, 2 |
| chr9_-_14381242 | 0.56 |
ENSMUST00000167549.1
|
Endod1
|
endonuclease domain containing 1 |
| chr10_+_39612934 | 0.55 |
ENSMUST00000019987.6
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr6_+_41521782 | 0.55 |
ENSMUST00000070380.4
|
Prss2
|
protease, serine, 2 |
| chr3_+_95526777 | 0.54 |
ENSMUST00000015667.2
ENSMUST00000116304.2 |
Ctss
|
cathepsin S |
| chr8_+_35375719 | 0.54 |
ENSMUST00000070481.6
|
Ppp1r3b
|
protein phosphatase 1, regulatory (inhibitor) subunit 3B |
| chr18_+_37447641 | 0.54 |
ENSMUST00000052387.3
|
Pcdhb14
|
protocadherin beta 14 |
| chr9_-_15045378 | 0.54 |
ENSMUST00000164273.1
|
Panx1
|
pannexin 1 |
| chr3_-_88372740 | 0.54 |
ENSMUST00000107543.1
ENSMUST00000107542.1 |
Bglap3
|
bone gamma-carboxyglutamate protein 3 |
| chr11_-_58534825 | 0.53 |
ENSMUST00000170009.1
|
Olfr330
|
olfactory receptor 330 |
| chr2_-_167062981 | 0.53 |
ENSMUST00000048988.7
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr17_+_36042956 | 0.52 |
ENSMUST00000097331.1
|
Gm6034
|
predicted gene 6034 |
| chr11_-_109298066 | 0.52 |
ENSMUST00000106706.1
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr7_+_78913436 | 0.52 |
ENSMUST00000121645.1
|
Isg20
|
interferon-stimulated protein |
| chr10_-_75797528 | 0.52 |
ENSMUST00000120177.1
|
Gstt1
|
glutathione S-transferase, theta 1 |
| chr14_+_103513328 | 0.51 |
ENSMUST00000095576.3
|
Scel
|
sciellin |
| chr11_-_3931789 | 0.51 |
ENSMUST00000109992.1
ENSMUST00000109988.1 |
Tcn2
|
transcobalamin 2 |
| chr11_+_49087022 | 0.51 |
ENSMUST00000046704.6
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr3_+_127791374 | 0.51 |
ENSMUST00000171621.1
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr5_-_105139539 | 0.51 |
ENSMUST00000100961.4
ENSMUST00000031235.6 ENSMUST00000100962.3 |
Gbp9
Gbp8
Gbp4
|
guanylate-binding protein 9 guanylate-binding protein 8 guanylate binding protein 4 |
| chr17_-_31144271 | 0.51 |
ENSMUST00000024826.7
|
Tff2
|
trefoil factor 2 (spasmolytic protein 1) |
| chr2_+_104095796 | 0.51 |
ENSMUST00000040423.5
ENSMUST00000168176.1 |
Cd59a
|
CD59a antigen |
| chr11_-_61267177 | 0.51 |
ENSMUST00000066277.3
ENSMUST00000074127.7 ENSMUST00000108715.2 |
Aldh3a2
|
aldehyde dehydrogenase family 3, subfamily A2 |
| chr6_+_139843648 | 0.50 |
ENSMUST00000087657.6
|
Pik3c2g
|
phosphatidylinositol 3-kinase, C2 domain containing, gamma polypeptide |
| chr1_+_58802492 | 0.50 |
ENSMUST00000165549.1
|
Casp8
|
caspase 8 |
| chrX_-_134161928 | 0.50 |
ENSMUST00000033611.4
|
Xkrx
|
X Kell blood group precursor related X linked |
| chr5_-_116024475 | 0.50 |
ENSMUST00000111999.1
|
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Irf2_Irf1_Irf8_Irf9_Irf7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.5 | 1.5 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 1.0 | 3.8 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.9 | 2.6 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.8 | 5.0 | GO:0035546 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.8 | 2.3 | GO:0042196 | dichloromethane metabolic process(GO:0018900) chlorinated hydrocarbon metabolic process(GO:0042196) halogenated hydrocarbon metabolic process(GO:0042197) |
| 0.8 | 1.5 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.8 | 3.8 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.7 | 2.2 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.6 | 1.9 | GO:0006227 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.5 | 2.2 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.5 | 2.5 | GO:1904720 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.5 | 5.5 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.4 | 17.0 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.4 | 0.4 | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.4 | 1.7 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.4 | 1.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.4 | 1.8 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.3 | 1.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.3 | 0.9 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.3 | 0.6 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.3 | 2.1 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.3 | 0.8 | GO:0002014 | vasoconstriction of artery involved in ischemic response to lowering of systemic arterial blood pressure(GO:0002014) |
| 0.3 | 2.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.3 | 3.7 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.3 | 1.3 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.3 | 1.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.6 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.2 | 1.4 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.2 | 0.2 | GO:1990009 | retinal cell apoptotic process(GO:1990009) |
| 0.2 | 1.3 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.2 | 1.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.2 | 0.9 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.2 | 2.7 | GO:2000675 | negative regulation of type B pancreatic cell apoptotic process(GO:2000675) |
| 0.2 | 1.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 0.8 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.2 | 0.8 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 1.3 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.2 | 0.5 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.2 | 0.4 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.2 | 0.4 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.2 | 0.5 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.2 | 1.1 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.2 | 0.5 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.1 | 0.4 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.1 | 0.7 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.1 | 0.4 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.3 | GO:0039533 | regulation of MDA-5 signaling pathway(GO:0039533) |
| 0.1 | 0.6 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.7 | GO:0001969 | activation of membrane attack complex(GO:0001905) regulation of activation of membrane attack complex(GO:0001969) |
| 0.1 | 2.0 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 0.1 | GO:0045626 | negative regulation of T-helper 1 cell differentiation(GO:0045626) |
| 0.1 | 0.4 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.4 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.4 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) cellular response to fructose stimulus(GO:0071332) |
| 0.1 | 2.5 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 0.5 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.1 | 0.5 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.5 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.4 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.4 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 1.1 | GO:0098532 | histone H3-K27 trimethylation(GO:0098532) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.6 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.1 | 0.4 | GO:1904565 | response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904565) cellular response to 1-oleoyl-sn-glycerol 3-phosphate(GO:1904566) |
| 0.1 | 0.3 | GO:0002730 | regulation of dendritic cell cytokine production(GO:0002730) |
| 0.1 | 0.3 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 0.7 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.1 | 0.3 | GO:0048239 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.1 | 0.4 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.1 | 0.3 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.1 | 0.8 | GO:0070317 | negative regulation of G0 to G1 transition(GO:0070317) |
| 0.1 | 0.7 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.3 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.1 | 0.3 | GO:0007161 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.1 | 0.8 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:1900020 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.4 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.1 | 1.2 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.1 | 0.4 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.1 | 0.3 | GO:2000338 | positive regulation of interleukin-23 production(GO:0032747) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) |
| 0.1 | 0.3 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.1 | 0.5 | GO:0060455 | negative regulation of gastric acid secretion(GO:0060455) |
| 0.1 | 0.9 | GO:0003374 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.1 | 0.6 | GO:0036462 | TRAIL-activated apoptotic signaling pathway(GO:0036462) |
| 0.1 | 2.3 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.1 | 0.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.1 | 0.5 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.1 | 1.0 | GO:0090435 | protein localization to nuclear envelope(GO:0090435) |
| 0.1 | 0.4 | GO:0015692 | vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) |
| 0.1 | 0.5 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.2 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.4 | GO:0097106 | postsynaptic density organization(GO:0097106) |
| 0.1 | 0.3 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.3 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.1 | 0.7 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 1.6 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.1 | 0.4 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.1 | 0.6 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.1 | GO:1902938 | regulation of intracellular calcium activated chloride channel activity(GO:1902938) |
| 0.1 | 0.7 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.1 | 0.4 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.5 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.9 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.1 | 0.1 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.1 | 1.5 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.2 | GO:0061518 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) |
| 0.1 | 1.4 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 0.3 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.1 | 0.4 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 1.0 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.8 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.2 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.1 | 0.8 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.1 | 0.4 | GO:1900227 | positive regulation of NLRP3 inflammasome complex assembly(GO:1900227) |
| 0.1 | 0.4 | GO:0031642 | negative regulation of myelination(GO:0031642) |
| 0.1 | 0.4 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.3 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.1 | 9.4 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.1 | 0.2 | GO:0009814 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 1.0 | GO:0030238 | male sex determination(GO:0030238) |
| 0.1 | 0.3 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 0.2 | GO:1905204 | negative regulation of connective tissue replacement(GO:1905204) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 8.5 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.1 | 0.4 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.1 | 2.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.0 | 0.8 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0006583 | melanin biosynthetic process from tyrosine(GO:0006583) |
| 0.0 | 0.1 | GO:0045590 | negative regulation of regulatory T cell differentiation(GO:0045590) |
| 0.0 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:1901026 | ripoptosome assembly(GO:0097343) ripoptosome assembly involved in necroptotic process(GO:1901026) |
| 0.0 | 0.3 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.8 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.4 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.0 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.3 | GO:0031584 | activation of phospholipase D activity(GO:0031584) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 1.0 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.7 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.3 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.2 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.0 | 0.2 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0070425 | regulation of granuloma formation(GO:0002631) negative regulation of granuloma formation(GO:0002632) regulation of toll-like receptor 5 signaling pathway(GO:0034147) negative regulation of toll-like receptor 5 signaling pathway(GO:0034148) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070429) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) tolerance induction to lipopolysaccharide(GO:0072573) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.4 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.2 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.2 | GO:0035608 | protein deglutamylation(GO:0035608) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.4 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.4 | GO:0034626 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 1.0 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.2 | GO:1902856 | negative regulation of nonmotile primary cilium assembly(GO:1902856) |
| 0.0 | 0.8 | GO:0061462 | protein localization to lysosome(GO:0061462) |
| 0.0 | 0.4 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 1.4 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.4 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.2 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.3 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.3 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.0 | 0.1 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 1.2 | GO:0070306 | lens fiber cell differentiation(GO:0070306) |
| 0.0 | 0.1 | GO:0060809 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.3 | GO:0010529 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.1 | GO:0060022 | hard palate development(GO:0060022) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.1 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:1904959 | regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.0 | 0.2 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.5 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 1.6 | GO:0009166 | nucleotide catabolic process(GO:0009166) |
| 0.0 | 0.0 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.2 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.7 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:1904220 | regulation of serine C-palmitoyltransferase activity(GO:1904220) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.7 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:1905051 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.0 | 0.2 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0051771 | negative regulation of nitric-oxide synthase biosynthetic process(GO:0051771) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 1.0 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.7 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.2 | GO:1904424 | regulation of GTP binding(GO:1904424) |
| 0.0 | 0.1 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.3 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.3 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0031937 | positive regulation of chromatin silencing(GO:0031937) |
| 0.0 | 0.1 | GO:0002154 | thyroid hormone mediated signaling pathway(GO:0002154) regulation of thyroid hormone mediated signaling pathway(GO:0002155) |
| 0.0 | 0.6 | GO:0045823 | positive regulation of heart contraction(GO:0045823) |
| 0.0 | 2.4 | GO:0007601 | visual perception(GO:0007601) |
| 0.0 | 0.1 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) |
| 0.0 | 0.0 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.3 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.2 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.0 | GO:2000821 | regulation of grooming behavior(GO:2000821) |
| 0.0 | 0.5 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.4 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.5 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.2 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.6 | GO:0009250 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.2 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.0 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0098795 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.3 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 0.3 | GO:0036344 | platelet formation(GO:0030220) platelet morphogenesis(GO:0036344) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.1 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.3 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.0 | 0.2 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) |
| 0.0 | 0.1 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.6 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.0 | 0.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.0 | GO:1904109 | positive regulation of intracellular lipid transport(GO:0032379) positive regulation of intracellular sterol transport(GO:0032382) positive regulation of intracellular cholesterol transport(GO:0032385) lipid hydroperoxide transport(GO:1901373) positive regulation of cholesterol import(GO:1904109) positive regulation of sterol import(GO:2000911) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 4.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.8 | 11.1 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.4 | 3.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.3 | 1.2 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.3 | 3.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.2 | 1.4 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.2 | 1.6 | GO:1990462 | omegasome(GO:1990462) |
| 0.2 | 1.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.2 | 1.8 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.3 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.2 | 1.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 1.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 2.1 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 0.4 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
| 0.1 | 2.0 | GO:0043186 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 0.4 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.1 | 0.4 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.1 | 0.5 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.1 | 0.3 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.1 | 0.6 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.8 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 0.3 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.1 | 1.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.1 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.1 | 0.4 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.1 | 0.2 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.1 | 0.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 0.7 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.1 | 2.2 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 1.3 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.2 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.5 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 2.4 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.7 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 0.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 5.3 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 3.8 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.5 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.0 | 1.4 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 4.9 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.9 | GO:0048770 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.5 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.3 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 1.3 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.4 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.3 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.8 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 1.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.8 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 1.1 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 1.5 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.4 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.7 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.1 | GO:0044307 | dendritic branch(GO:0044307) |
| 0.0 | 0.6 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 1.4 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 1.4 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.7 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.9 | 3.8 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.8 | 2.3 | GO:0019120 | hydrolase activity, acting on acid halide bonds(GO:0016824) hydrolase activity, acting on acid halide bonds, in C-halide compounds(GO:0019120) alkylhalidase activity(GO:0047651) |
| 0.6 | 1.9 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.6 | 2.9 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.5 | 4.9 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.4 | 3.0 | GO:0046978 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.4 | 2.5 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.3 | 1.7 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.3 | 1.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 1.0 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.3 | 4.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.3 | 1.3 | GO:0001875 | lipopolysaccharide receptor activity(GO:0001875) |
| 0.2 | 1.2 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.2 | 2.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.2 | 1.5 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.2 | 0.6 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.2 | 1.6 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.2 | 0.5 | GO:0016899 | oxidoreductase activity, acting on the CH-OH group of donors, oxygen as acceptor(GO:0016899) |
| 0.2 | 0.7 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.2 | 1.7 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.2 | 1.9 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.2 | 0.6 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.1 | 0.1 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.1 | 0.4 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 1.6 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.1 | 0.5 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.1 | 2.9 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 2.0 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.1 | 0.5 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.1 | 0.3 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.1 | 0.4 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.1 | 0.4 | GO:0003844 | 1,4-alpha-glucan branching enzyme activity(GO:0003844) |
| 0.1 | 0.8 | GO:0015288 | porin activity(GO:0015288) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.3 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.3 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 8.0 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 0.4 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.1 | 2.3 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.1 | 0.3 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.1 | 0.3 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.4 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 1.8 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.2 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 0.2 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.1 | 0.2 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.1 | 0.4 | GO:0045340 | mercury ion binding(GO:0045340) |
| 0.1 | 1.6 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 0.2 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.4 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.1 | 1.2 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.7 | GO:0043495 | protein anchor(GO:0043495) |
| 0.1 | 1.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.5 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.8 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.4 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.3 | GO:0089720 | caspase binding(GO:0089720) |
| 0.1 | 0.2 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.1 | 2.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.2 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0004658 | propionyl-CoA carboxylase activity(GO:0004658) |
| 0.0 | 0.2 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.7 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 2.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 1.1 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.7 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.3 | GO:0022833 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.5 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.5 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 1.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.2 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 9.4 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 1.3 | GO:0043531 | ADP binding(GO:0043531) |
| 0.0 | 5.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.1 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 2.0 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.7 | GO:0001848 | complement binding(GO:0001848) |
| 0.0 | 0.3 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.3 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 1.5 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0003958 | NADPH-hemoprotein reductase activity(GO:0003958) |
| 0.0 | 1.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.7 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.3 | GO:0098505 | G-rich strand telomeric DNA binding(GO:0098505) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.8 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.4 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.2 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.8 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 1.7 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.2 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.8 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 1.5 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.0 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.0 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.0 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 0.1 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 1.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.4 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.1 | 2.9 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.2 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.1 | 1.6 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.1 | 1.7 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 6.4 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 0.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 0.8 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 2.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.9 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.1 | 1.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 2.8 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.9 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.2 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.6 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 1.9 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.6 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.6 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 2.6 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.9 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.5 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.3 | 5.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.3 | 14.5 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.2 | 2.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 1.6 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 2.3 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.2 | 2.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.2 | 2.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.1 | 1.8 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.1 | 3.3 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.1 | 2.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.0 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.1 | 1.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.1 | 0.5 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 1.0 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.1 | 0.8 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 2.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.3 | REACTOME P75NTR SIGNALS VIA NFKB | Genes involved in p75NTR signals via NF-kB |
| 0.1 | 1.3 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.3 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.4 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.0 | 1.3 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 1.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.2 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.7 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.4 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.8 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.5 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 1.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 1.1 | REACTOME GLUCOSE METABOLISM | Genes involved in Glucose metabolism |
| 0.0 | 0.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 1.5 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.2 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |