Project
12D miR HR13_24
Navigation
Downloads
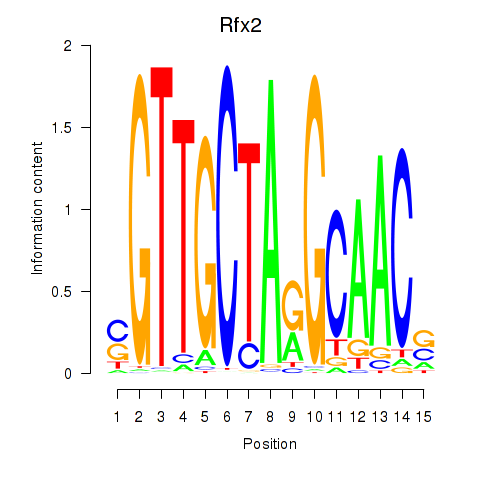
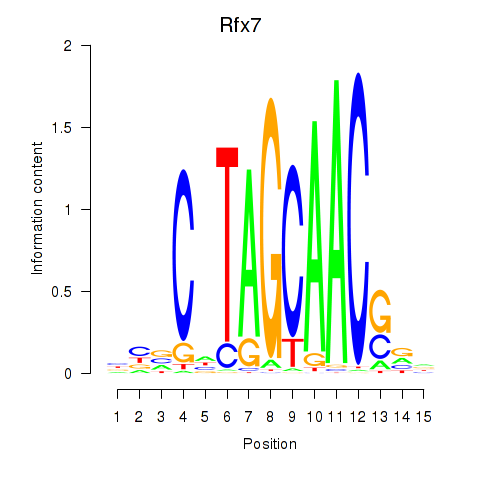
Results for Rfx2_Rfx7
Z-value: 1.59
Transcription factors associated with Rfx2_Rfx7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx2
|
ENSMUSG00000024206.8 | regulatory factor X, 2 (influences HLA class II expression) |
|
Rfx7
|
ENSMUSG00000037674.9 | regulatory factor X, 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx2 | mm10_v2_chr17_-_56830916_56831008 | 0.36 | 2.8e-01 | Click! |
| Rfx7 | mm10_v2_chr9_+_72532609_72532828 | 0.11 | 7.5e-01 | Click! |
Activity profile of Rfx2_Rfx7 motif
Sorted Z-values of Rfx2_Rfx7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_28466266 | 3.16 |
ENSMUST00000127683.1
ENSMUST00000086370.4 |
1700007K13Rik
|
RIKEN cDNA 1700007K13 gene |
| chr4_+_129960760 | 2.36 |
ENSMUST00000139884.1
|
1700003M07Rik
|
RIKEN cDNA 1700003M07 gene |
| chr2_-_152831665 | 1.63 |
ENSMUST00000156688.1
ENSMUST00000007803.5 |
Bcl2l1
|
BCL2-like 1 |
| chr4_-_126325641 | 1.42 |
ENSMUST00000131113.1
|
Tekt2
|
tektin 2 |
| chr11_+_117523526 | 1.36 |
ENSMUST00000132261.1
|
Gm11734
|
predicted gene 11734 |
| chr2_-_152831112 | 1.30 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr3_+_156562141 | 1.29 |
ENSMUST00000175773.1
|
Negr1
|
neuronal growth regulator 1 |
| chr2_-_181698433 | 1.20 |
ENSMUST00000052416.3
|
4930526D03Rik
|
RIKEN cDNA 4930526D03 gene |
| chr17_+_24696234 | 1.17 |
ENSMUST00000019464.7
|
Noxo1
|
NADPH oxidase organizer 1 |
| chrX_+_52791179 | 1.14 |
ENSMUST00000101588.1
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr12_+_33394854 | 1.04 |
ENSMUST00000020878.6
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr10_+_75037291 | 0.97 |
ENSMUST00000139384.1
|
Rab36
|
RAB36, member RAS oncogene family |
| chr7_+_44468051 | 0.97 |
ENSMUST00000118493.1
|
Josd2
|
Josephin domain containing 2 |
| chr15_-_89379246 | 0.92 |
ENSMUST00000049968.7
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr9_+_49102720 | 0.91 |
ENSMUST00000070390.5
ENSMUST00000167095.1 |
Tmprss5
|
transmembrane protease, serine 5 (spinesin) |
| chr2_-_3419019 | 0.88 |
ENSMUST00000115084.1
ENSMUST00000115083.1 |
Meig1
|
meiosis expressed gene 1 |
| chr4_-_126325672 | 0.88 |
ENSMUST00000102616.1
|
Tekt2
|
tektin 2 |
| chr17_-_29078953 | 0.84 |
ENSMUST00000133221.1
|
Trp53cor1
|
tumor protein p53 pathway corepressor 1 |
| chr7_+_75455534 | 0.83 |
ENSMUST00000147005.1
ENSMUST00000166315.1 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr4_+_148000722 | 0.81 |
ENSMUST00000103230.4
|
Nppa
|
natriuretic peptide type A |
| chr4_+_100478806 | 0.80 |
ENSMUST00000133493.2
ENSMUST00000092730.3 ENSMUST00000106979.3 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr1_-_10009098 | 0.79 |
ENSMUST00000176398.1
ENSMUST00000027049.3 |
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr10_+_33905015 | 0.78 |
ENSMUST00000169670.1
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr10_+_112163621 | 0.76 |
ENSMUST00000092176.1
|
Caps2
|
calcyphosphine 2 |
| chr2_-_32775584 | 0.69 |
ENSMUST00000161430.1
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr2_-_3419066 | 0.67 |
ENSMUST00000115082.3
|
Meig1
|
meiosis expressed gene 1 |
| chr11_-_53480178 | 0.66 |
ENSMUST00000104955.2
|
Sowaha
|
sosondowah ankyrin repeat domain family member A |
| chr1_-_119836999 | 0.64 |
ENSMUST00000163621.1
ENSMUST00000168303.1 |
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr2_-_32775330 | 0.63 |
ENSMUST00000161089.1
ENSMUST00000066478.2 ENSMUST00000161950.1 |
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr11_+_102881204 | 0.62 |
ENSMUST00000021307.3
ENSMUST00000159834.1 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr8_-_120177420 | 0.62 |
ENSMUST00000135567.2
|
Fam92b
|
family with sequence similarity 92, member B |
| chr15_+_89532816 | 0.61 |
ENSMUST00000167173.1
|
Shank3
|
SH3/ankyrin domain gene 3 |
| chr7_-_119896285 | 0.60 |
ENSMUST00000106519.1
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr2_+_32775769 | 0.56 |
ENSMUST00000066352.5
|
Ptrh1
|
peptidyl-tRNA hydrolase 1 homolog (S. cerevisiae) |
| chr11_-_105456674 | 0.56 |
ENSMUST00000049995.3
ENSMUST00000100332.3 |
March10
|
membrane-associated ring finger (C3HC4) 10 |
| chr4_+_138262189 | 0.54 |
ENSMUST00000030539.3
|
Kif17
|
kinesin family member 17 |
| chr11_-_30471792 | 0.54 |
ENSMUST00000041763.7
|
4930505A04Rik
|
RIKEN cDNA 4930505A04 gene |
| chr3_+_156561950 | 0.53 |
ENSMUST00000041425.5
ENSMUST00000106065.1 |
Negr1
|
neuronal growth regulator 1 |
| chr9_-_22002599 | 0.53 |
ENSMUST00000115336.2
ENSMUST00000044926.5 |
Ccdc151
|
coiled-coil domain containing 151 |
| chr15_-_79834261 | 0.53 |
ENSMUST00000148358.1
|
Cbx6
|
chromobox 6 |
| chr15_+_98708187 | 0.52 |
ENSMUST00000003444.4
|
Ccdc65
|
coiled-coil domain containing 65 |
| chr7_-_68749170 | 0.49 |
ENSMUST00000118110.1
ENSMUST00000048068.7 |
Arrdc4
|
arrestin domain containing 4 |
| chr12_+_112808914 | 0.49 |
ENSMUST00000037014.3
ENSMUST00000177808.1 |
BC022687
|
cDNA sequence BC022687 |
| chr12_+_111166413 | 0.49 |
ENSMUST00000021706.4
|
Traf3
|
TNF receptor-associated factor 3 |
| chr4_+_45012830 | 0.49 |
ENSMUST00000095105.1
|
1700055D18Rik
|
RIKEN cDNA 1700055D18 gene |
| chr10_-_100487267 | 0.48 |
ENSMUST00000128009.1
|
Tmtc3
|
transmembrane and tetratricopeptide repeat containing 3 |
| chr19_-_28967794 | 0.48 |
ENSMUST00000162110.1
|
4430402I18Rik
|
RIKEN cDNA 4430402I18 gene |
| chr9_+_110476985 | 0.48 |
ENSMUST00000084948.4
ENSMUST00000061155.6 ENSMUST00000140686.1 ENSMUST00000084952.5 |
Kif9
|
kinesin family member 9 |
| chr12_+_111166536 | 0.45 |
ENSMUST00000060274.6
|
Traf3
|
TNF receptor-associated factor 3 |
| chr4_-_117251879 | 0.45 |
ENSMUST00000165128.1
ENSMUST00000077500.4 |
Gm1661
|
predicted gene 1661 |
| chr5_-_128953303 | 0.45 |
ENSMUST00000111346.1
|
Rimbp2
|
RIMS binding protein 2 |
| chr1_-_75210732 | 0.45 |
ENSMUST00000113623.1
|
Glb1l
|
galactosidase, beta 1-like |
| chr7_-_4546567 | 0.44 |
ENSMUST00000065957.5
|
Syt5
|
synaptotagmin V |
| chr10_+_75037066 | 0.44 |
ENSMUST00000147802.1
ENSMUST00000020391.5 |
Rab36
|
RAB36, member RAS oncogene family |
| chr11_-_97041395 | 0.44 |
ENSMUST00000021251.6
|
Lrrc46
|
leucine rich repeat containing 46 |
| chr7_+_44896077 | 0.43 |
ENSMUST00000071207.7
ENSMUST00000166849.1 ENSMUST00000168712.1 ENSMUST00000168389.1 |
Fuz
|
fuzzy homolog (Drosophila) |
| chrX_-_37628612 | 0.43 |
ENSMUST00000119965.2
|
Rhox3g
|
reproductive homeobox 3G |
| chr13_+_21735055 | 0.43 |
ENSMUST00000087714.4
|
Hist1h4j
|
histone cluster 1, H4j |
| chrX_-_17319316 | 0.43 |
ENSMUST00000026014.7
|
Efhc2
|
EF-hand domain (C-terminal) containing 2 |
| chrX_+_37546975 | 0.43 |
ENSMUST00000081327.5
ENSMUST00000184210.1 ENSMUST00000184270.1 |
Rhox3e
|
reproductive homeobox 3E |
| chr8_+_45999297 | 0.43 |
ENSMUST00000110380.1
ENSMUST00000066451.3 |
Lrp2bp
|
Lrp2 binding protein |
| chr11_+_53480166 | 0.43 |
ENSMUST00000067523.1
|
Gm9945
|
predicted gene 9945 |
| chr18_-_3309858 | 0.42 |
ENSMUST00000144496.1
ENSMUST00000154715.1 |
Crem
|
cAMP responsive element modulator |
| chr12_-_83439910 | 0.42 |
ENSMUST00000177801.1
|
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr10_+_93589413 | 0.41 |
ENSMUST00000181835.1
|
4933408J17Rik
|
RIKEN cDNA 4933408J17 gene |
| chr12_+_111166485 | 0.41 |
ENSMUST00000139162.1
|
Traf3
|
TNF receptor-associated factor 3 |
| chr18_-_3309723 | 0.41 |
ENSMUST00000136961.1
ENSMUST00000152108.1 |
Crem
|
cAMP responsive element modulator |
| chr8_-_45382198 | 0.40 |
ENSMUST00000093526.6
|
Fam149a
|
family with sequence similarity 149, member A |
| chr8_-_70792392 | 0.40 |
ENSMUST00000166004.1
|
Mast3
|
microtubule associated serine/threonine kinase 3 |
| chr2_+_28840406 | 0.39 |
ENSMUST00000113853.2
|
Ddx31
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 31 |
| chr7_+_139214661 | 0.39 |
ENSMUST00000135509.1
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr5_+_30466044 | 0.38 |
ENSMUST00000031078.3
ENSMUST00000114743.1 |
1700001C02Rik
|
RIKEN cDNA 1700001C02 gene |
| chr5_-_34187670 | 0.38 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr1_-_37865040 | 0.38 |
ENSMUST00000041815.8
|
Tsga10
|
testis specific 10 |
| chr1_-_119837338 | 0.38 |
ENSMUST00000163435.1
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr2_+_102550012 | 0.37 |
ENSMUST00000028612.7
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr11_+_114727384 | 0.37 |
ENSMUST00000069325.7
|
Dnaic2
|
dynein, axonemal, intermediate chain 2 |
| chr12_+_73286779 | 0.37 |
ENSMUST00000140523.1
|
Slc38a6
|
solute carrier family 38, member 6 |
| chr13_-_92131494 | 0.37 |
ENSMUST00000099326.3
ENSMUST00000146492.1 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr14_-_32388355 | 0.36 |
ENSMUST00000100723.2
|
1700024G13Rik
|
RIKEN cDNA 1700024G13 gene |
| chr6_+_8259288 | 0.36 |
ENSMUST00000159335.1
|
Gm16039
|
predicted gene 16039 |
| chr11_-_60352869 | 0.35 |
ENSMUST00000095254.5
ENSMUST00000102683.4 ENSMUST00000093048.6 ENSMUST00000093046.6 ENSMUST00000064019.8 ENSMUST00000102682.4 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr11_-_66168505 | 0.35 |
ENSMUST00000080665.3
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chrX_+_37380388 | 0.34 |
ENSMUST00000096459.4
ENSMUST00000183543.1 ENSMUST00000183901.1 |
Gm21085
|
predicted gene, 21085 |
| chr7_+_119895836 | 0.34 |
ENSMUST00000106518.1
ENSMUST00000054440.3 |
Lyrm1
|
LYR motif containing 1 |
| chr1_+_46066738 | 0.34 |
ENSMUST00000069293.7
|
Dnah7b
|
dynein, axonemal, heavy chain 7B |
| chr16_+_52031549 | 0.33 |
ENSMUST00000114471.1
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr7_-_139683797 | 0.33 |
ENSMUST00000129990.1
ENSMUST00000130453.1 |
9330101J02Rik
|
RIKEN cDNA 9330101J02 gene |
| chr5_-_135778340 | 0.33 |
ENSMUST00000178796.1
ENSMUST00000178515.1 ENSMUST00000111164.2 |
Styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr2_+_168230597 | 0.33 |
ENSMUST00000099071.3
|
Mocs3
|
molybdenum cofactor synthesis 3 |
| chr11_-_53423123 | 0.32 |
ENSMUST00000036045.5
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr10_+_127195240 | 0.32 |
ENSMUST00000181578.1
|
F420014N23Rik
|
RIKEN cDNA F420014N23 gene |
| chr12_+_85288591 | 0.32 |
ENSMUST00000059341.4
|
Zc2hc1c
|
zinc finger, C2HC-type containing 1C |
| chrX_-_37661662 | 0.31 |
ENSMUST00000184824.1
ENSMUST00000185138.1 |
Rhox3h
|
reproductive homeobox 3H |
| chr5_+_27261916 | 0.31 |
ENSMUST00000101471.3
|
Dpp6
|
dipeptidylpeptidase 6 |
| chr12_+_113185877 | 0.31 |
ENSMUST00000058491.6
|
Tmem121
|
transmembrane protein 121 |
| chr7_+_90426312 | 0.31 |
ENSMUST00000061391.7
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr12_+_111166349 | 0.31 |
ENSMUST00000117269.1
|
Traf3
|
TNF receptor-associated factor 3 |
| chr3_-_89393294 | 0.30 |
ENSMUST00000142119.1
ENSMUST00000029677.8 ENSMUST00000148361.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr10_+_57645861 | 0.30 |
ENSMUST00000177473.1
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr11_+_60353324 | 0.30 |
ENSMUST00000070805.6
ENSMUST00000094140.2 ENSMUST00000108723.2 ENSMUST00000108722.4 |
Lrrc48
|
leucine rich repeat containing 48 |
| chr1_-_130715734 | 0.30 |
ENSMUST00000066863.6
ENSMUST00000050406.4 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr10_-_100487316 | 0.30 |
ENSMUST00000134477.1
ENSMUST00000099318.3 ENSMUST00000058154.8 |
Tmtc3
|
transmembrane and tetratricopeptide repeat containing 3 |
| chr7_-_141214080 | 0.30 |
ENSMUST00000026573.5
ENSMUST00000170841.1 |
1600016N20Rik
|
RIKEN cDNA 1600016N20 gene |
| chr15_-_79834323 | 0.29 |
ENSMUST00000177316.2
ENSMUST00000175858.2 |
Nptxr
|
neuronal pentraxin receptor |
| chr10_+_57645834 | 0.29 |
ENSMUST00000177325.1
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr9_+_22003035 | 0.29 |
ENSMUST00000115331.2
ENSMUST00000003493.7 |
Prkcsh
|
protein kinase C substrate 80K-H |
| chr11_-_66168444 | 0.28 |
ENSMUST00000108691.1
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chr7_-_105399991 | 0.28 |
ENSMUST00000118726.1
ENSMUST00000074686.7 ENSMUST00000122327.1 ENSMUST00000179474.1 ENSMUST00000048079.6 |
Fam160a2
|
family with sequence similarity 160, member A2 |
| chr4_-_129614254 | 0.28 |
ENSMUST00000106037.2
ENSMUST00000179209.1 |
Dcdc2b
|
doublecortin domain containing 2b |
| chr16_+_44394771 | 0.28 |
ENSMUST00000099742.2
|
Wdr52
|
WD repeat domain 52 |
| chr7_+_44468020 | 0.28 |
ENSMUST00000117324.1
ENSMUST00000120852.1 ENSMUST00000118628.1 |
Josd2
|
Josephin domain containing 2 |
| chr1_-_133801031 | 0.27 |
ENSMUST00000143567.1
|
Atp2b4
|
ATPase, Ca++ transporting, plasma membrane 4 |
| chr11_-_77725281 | 0.27 |
ENSMUST00000078623.4
|
Cryba1
|
crystallin, beta A1 |
| chr2_+_152962485 | 0.27 |
ENSMUST00000099197.2
ENSMUST00000103155.3 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr3_+_146404844 | 0.27 |
ENSMUST00000106149.1
|
Ssx2ip
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr13_-_62858364 | 0.26 |
ENSMUST00000021907.7
|
Fbp2
|
fructose bisphosphatase 2 |
| chr9_-_123717576 | 0.26 |
ENSMUST00000026274.7
|
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr5_+_30814571 | 0.26 |
ENSMUST00000031058.8
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr3_-_89393629 | 0.26 |
ENSMUST00000124783.1
ENSMUST00000126027.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr17_-_26244118 | 0.26 |
ENSMUST00000118487.1
|
Itfg3
|
integrin alpha FG-GAP repeat containing 3 |
| chr4_+_62525369 | 0.25 |
ENSMUST00000062145.1
|
4933430I17Rik
|
RIKEN cDNA 4933430I17 gene |
| chrX_+_95711641 | 0.25 |
ENSMUST00000150123.1
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr9_-_108578657 | 0.25 |
ENSMUST00000068700.5
|
Wdr6
|
WD repeat domain 6 |
| chr13_+_97241096 | 0.25 |
ENSMUST00000041623.7
|
Enc1
|
ectodermal-neural cortex 1 |
| chr5_-_5664196 | 0.25 |
ENSMUST00000061008.3
ENSMUST00000054865.6 |
A330021E22Rik
|
RIKEN cDNA A330021E22 gene |
| chr17_-_26244203 | 0.25 |
ENSMUST00000114988.1
|
Itfg3
|
integrin alpha FG-GAP repeat containing 3 |
| chr17_+_7945653 | 0.25 |
ENSMUST00000097423.2
|
Rsph3a
|
radial spoke 3A homolog (Chlamydomonas) |
| chr15_-_82338801 | 0.25 |
ENSMUST00000023088.7
|
Naga
|
N-acetyl galactosaminidase, alpha |
| chr11_+_119229092 | 0.25 |
ENSMUST00000053440.7
|
Ccdc40
|
coiled-coil domain containing 40 |
| chr17_-_6948283 | 0.25 |
ENSMUST00000024572.9
|
Rsph3b
|
radial spoke 3B homolog (Chlamydomonas) |
| chrX_+_37255007 | 0.24 |
ENSMUST00000185028.1
ENSMUST00000185050.1 |
Rhox3a
|
reproductive homeobox 3A |
| chr18_+_75820174 | 0.24 |
ENSMUST00000058997.7
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr15_-_79834224 | 0.24 |
ENSMUST00000109623.1
ENSMUST00000109625.1 ENSMUST00000023060.6 ENSMUST00000089299.5 |
Cbx6
Npcd
|
chromobox 6 neuronal pentraxin chromo domain |
| chrX_-_163761323 | 0.24 |
ENSMUST00000059320.2
|
Rnf138rt1
|
ring finger protein 138, retrogene 1 |
| chr15_+_85017138 | 0.24 |
ENSMUST00000023070.5
|
Upk3a
|
uroplakin 3A |
| chr2_-_151980135 | 0.23 |
ENSMUST00000062047.5
|
Fam110a
|
family with sequence similarity 110, member A |
| chr3_+_108537523 | 0.23 |
ENSMUST00000029485.5
|
1700013F07Rik
|
RIKEN cDNA 1700013F07 gene |
| chrX_+_37581352 | 0.23 |
ENSMUST00000115172.5
ENSMUST00000185008.1 ENSMUST00000183696.1 |
Rhox3f
|
reproductive homeobox 3F |
| chr9_+_46012810 | 0.23 |
ENSMUST00000126865.1
|
Sik3
|
SIK family kinase 3 |
| chr9_+_72985504 | 0.23 |
ENSMUST00000156879.1
|
Ccpg1
|
cell cycle progression 1 |
| chr18_+_31609512 | 0.23 |
ENSMUST00000164667.1
|
B930094E09Rik
|
RIKEN cDNA B930094E09 gene |
| chr7_-_13054665 | 0.23 |
ENSMUST00000182515.1
ENSMUST00000069289.8 |
Mzf1
|
myeloid zinc finger 1 |
| chr6_-_38875965 | 0.22 |
ENSMUST00000160360.1
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr5_+_110879788 | 0.22 |
ENSMUST00000156290.2
ENSMUST00000040111.9 |
Ttc28
|
tetratricopeptide repeat domain 28 |
| chr5_-_21645541 | 0.22 |
ENSMUST00000115234.1
ENSMUST00000051358.4 |
Fbxl13
|
F-box and leucine-rich repeat protein 13 |
| chr1_+_178405881 | 0.22 |
ENSMUST00000027775.7
|
Efcab2
|
EF-hand calcium binding domain 2 |
| chr5_+_30814722 | 0.22 |
ENSMUST00000114724.1
|
Mapre3
|
microtubule-associated protein, RP/EB family, member 3 |
| chr11_+_100545607 | 0.22 |
ENSMUST00000092684.5
ENSMUST00000006976.7 |
Ttc25
|
tetratricopeptide repeat domain 25 |
| chr18_-_7297901 | 0.22 |
ENSMUST00000081275.4
|
Armc4
|
armadillo repeat containing 4 |
| chr13_+_56703504 | 0.22 |
ENSMUST00000109874.1
|
Smad5
|
SMAD family member 5 |
| chr11_+_70030023 | 0.22 |
ENSMUST00000143920.2
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chrX_+_37380028 | 0.22 |
ENSMUST00000184565.1
|
Gm21085
|
predicted gene, 21085 |
| chr7_-_44375006 | 0.21 |
ENSMUST00000107933.1
|
1700008O03Rik
|
RIKEN cDNA 1700008O03 gene |
| chr7_-_31051431 | 0.21 |
ENSMUST00000073892.4
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr5_-_135778238 | 0.21 |
ENSMUST00000053906.4
ENSMUST00000177559.1 ENSMUST00000111161.2 ENSMUST00000111162.1 ENSMUST00000111163.2 |
Styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr12_-_111574384 | 0.21 |
ENSMUST00000180698.1
|
2810029C07Rik
|
RIKEN cDNA 2810029C07 gene |
| chr4_-_12087912 | 0.21 |
ENSMUST00000050686.3
|
Tmem67
|
transmembrane protein 67 |
| chr16_+_36875119 | 0.21 |
ENSMUST00000135406.1
ENSMUST00000114812.1 ENSMUST00000134616.1 ENSMUST00000023534.6 |
Golgb1
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr7_+_44896125 | 0.21 |
ENSMUST00000166552.1
ENSMUST00000168207.1 |
Fuz
|
fuzzy homolog (Drosophila) |
| chr9_+_65908967 | 0.21 |
ENSMUST00000034949.3
ENSMUST00000154589.1 |
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr3_+_146404978 | 0.21 |
ENSMUST00000129978.1
|
Ssx2ip
|
synovial sarcoma, X breakpoint 2 interacting protein |
| chr6_-_38875923 | 0.21 |
ENSMUST00000162359.1
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr17_+_34092340 | 0.21 |
ENSMUST00000025192.7
|
H2-Oa
|
histocompatibility 2, O region alpha locus |
| chr11_+_54314896 | 0.21 |
ENSMUST00000072178.4
ENSMUST00000101211.2 ENSMUST00000101213.2 |
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr17_-_10840285 | 0.20 |
ENSMUST00000041463.6
|
Pacrg
|
PARK2 co-regulated |
| chr7_-_13054514 | 0.20 |
ENSMUST00000182087.1
|
Mzf1
|
myeloid zinc finger 1 |
| chrX_+_37469868 | 0.20 |
ENSMUST00000115189.3
ENSMUST00000123851.1 ENSMUST00000151387.1 |
Rhox3c
|
reproductive homeobox 3C |
| chr1_+_91322075 | 0.20 |
ENSMUST00000088904.3
|
Espnl
|
espin-like |
| chr7_-_127588595 | 0.20 |
ENSMUST00000072155.3
|
Gm166
|
predicted gene 166 |
| chr17_+_15396240 | 0.20 |
ENSMUST00000055352.6
|
Fam120b
|
family with sequence similarity 120, member B |
| chr9_-_45828618 | 0.20 |
ENSMUST00000117194.1
|
Cep164
|
centrosomal protein 164 |
| chr10_+_79669410 | 0.20 |
ENSMUST00000020552.5
|
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr2_-_118703963 | 0.20 |
ENSMUST00000104937.1
|
Ankrd63
|
ankyrin repeat domain 63 |
| chr1_+_74661794 | 0.19 |
ENSMUST00000129890.1
|
Ttll4
|
tubulin tyrosine ligase-like family, member 4 |
| chr9_-_106685892 | 0.19 |
ENSMUST00000169068.1
ENSMUST00000046735.4 |
Tex264
|
testis expressed gene 264 |
| chr19_+_6046576 | 0.19 |
ENSMUST00000138532.1
ENSMUST00000129081.1 ENSMUST00000156550.1 |
Syvn1
|
synovial apoptosis inhibitor 1, synoviolin |
| chr7_-_102687252 | 0.19 |
ENSMUST00000082175.5
|
Trim68
|
tripartite motif-containing 68 |
| chr11_+_102665566 | 0.19 |
ENSMUST00000100378.3
|
Gm1564
|
predicted gene 1564 |
| chr1_+_91322117 | 0.18 |
ENSMUST00000176156.1
|
Espnl
|
espin-like |
| chr2_-_144527341 | 0.18 |
ENSMUST00000163701.1
ENSMUST00000081982.5 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr2_+_69789621 | 0.18 |
ENSMUST00000151298.1
ENSMUST00000028494.2 |
Phospho2
|
phosphatase, orphan 2 |
| chr4_-_106617232 | 0.18 |
ENSMUST00000106788.1
|
BC055111
|
cDNA sequence BC055111 |
| chr8_-_33843562 | 0.18 |
ENSMUST00000183062.1
|
Rbpms
|
RNA binding protein gene with multiple splicing |
| chr15_-_76009440 | 0.18 |
ENSMUST00000170153.1
|
Fam83h
|
family with sequence similarity 83, member H |
| chr10_-_93081596 | 0.18 |
ENSMUST00000168617.1
ENSMUST00000168110.1 ENSMUST00000020200.7 |
Gm872
|
RIKEN cDNA 4930485B16 gene |
| chr3_+_156561792 | 0.18 |
ENSMUST00000074015.4
|
Negr1
|
neuronal growth regulator 1 |
| chr9_+_46012822 | 0.18 |
ENSMUST00000120463.2
ENSMUST00000120247.1 |
Sik3
|
SIK family kinase 3 |
| chr7_+_19965365 | 0.17 |
ENSMUST00000094753.4
|
Ceacam20
|
carcinoembryonic antigen-related cell adhesion molecule 20 |
| chr9_-_106685653 | 0.17 |
ENSMUST00000163441.1
|
Tex264
|
testis expressed gene 264 |
| chr4_+_155086577 | 0.17 |
ENSMUST00000030915.4
ENSMUST00000155775.1 ENSMUST00000127457.1 |
Morn1
|
MORN repeat containing 1 |
| chr10_+_69213084 | 0.17 |
ENSMUST00000163497.1
ENSMUST00000164212.1 ENSMUST00000067908.7 |
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr4_-_132922487 | 0.17 |
ENSMUST00000097856.3
|
Fam76a
|
family with sequence similarity 76, member A |
| chr12_+_8973892 | 0.17 |
ENSMUST00000085745.6
ENSMUST00000111113.2 |
Wdr35
|
WD repeat domain 35 |
| chr12_+_105705970 | 0.17 |
ENSMUST00000040876.5
|
Ak7
|
adenylate kinase 7 |
| chrX_+_134686519 | 0.17 |
ENSMUST00000124226.2
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr1_-_155146755 | 0.17 |
ENSMUST00000027744.8
|
Mr1
|
major histocompatibility complex, class I-related |
| chr1_+_74601548 | 0.17 |
ENSMUST00000087186.4
|
Stk36
|
serine/threonine kinase 36 |
| chr3_-_53863764 | 0.17 |
ENSMUST00000122330.1
ENSMUST00000146598.1 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chrX_-_79517285 | 0.17 |
ENSMUST00000101410.2
|
Gm7173
|
predicted gene 7173 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx2_Rfx7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.9 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 3.5 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.2 | 0.6 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.2 | 0.8 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.2 | 0.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.2 | 0.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.2 | 1.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.6 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.1 | 1.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.1 | 0.6 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.1 | 0.5 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 1.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.1 | 0.3 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.1 | 0.3 | GO:0018307 | tRNA wobble position uridine thiolation(GO:0002143) enzyme active site formation(GO:0018307) |
| 0.1 | 0.3 | GO:0002587 | negative regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002587) |
| 0.1 | 0.3 | GO:1903750 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.1 | 0.3 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.3 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.2 | GO:2000256 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) positive regulation of male germ cell proliferation(GO:2000256) |
| 0.1 | 0.4 | GO:0010747 | positive regulation of plasma membrane long-chain fatty acid transport(GO:0010747) |
| 0.1 | 0.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) |
| 0.1 | 0.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 0.2 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.4 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.3 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.2 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.4 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.0 | 1.6 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.3 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.2 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.6 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.6 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.1 | GO:1900425 | positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.3 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 0.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.0 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.0 | GO:2000825 | positive regulation of androgen receptor activity(GO:2000825) |
| 0.0 | 0.1 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.9 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.3 | GO:1901620 | regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901620) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:0046909 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:0010826 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.2 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.0 | 0.1 | GO:1901580 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 0.1 | GO:1903978 | regulation of microglial cell activation(GO:1903978) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 0.1 | GO:0023021 | sphingomyelin catabolic process(GO:0006685) termination of signal transduction(GO:0023021) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.9 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.0 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 2.1 | GO:0007631 | feeding behavior(GO:0007631) |
| 0.0 | 0.1 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.0 | 0.1 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.2 | GO:0045060 | negative thymic T cell selection(GO:0045060) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.1 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.0 | 0.1 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.3 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.0 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.9 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 1.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 1.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.1 | 0.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.1 | 0.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.4 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.1 | 0.7 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.0 | 0.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.4 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.3 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 0.1 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.0 | 0.4 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 2.6 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.1 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.4 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.6 | GO:0060170 | ciliary membrane(GO:0060170) |
| 0.0 | 0.6 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.0 | 0.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.2 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.1 | GO:0032437 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.0 | 0.0 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 1.2 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) |
| 0.0 | 0.5 | GO:0002102 | podosome(GO:0002102) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 1.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.6 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.6 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.1 | 1.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 1.6 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.1 | 0.7 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.1 | 0.3 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.1 | 0.8 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.8 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.2 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.1 | 0.3 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.1 | 0.3 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.3 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.4 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.2 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.0 | 0.6 | GO:0050308 | sugar-phosphatase activity(GO:0050308) |
| 0.0 | 1.5 | GO:0045503 | dynein light chain binding(GO:0045503) |
| 0.0 | 0.2 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.8 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.3 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.1 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.6 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.9 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.0 | 0.2 | GO:0031811 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.0 | 0.1 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.4 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.1 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.5 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) 7S RNA binding(GO:0008312) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.0 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.0 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.0 | 0.0 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.1 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.0 | 0.1 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.3 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 2.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.8 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.6 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.8 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 0.6 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.7 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.2 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.5 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |