Project
12D miR HR13_24
Navigation
Downloads
Results for Rfx3_Rfx1_Rfx4
Z-value: 2.36
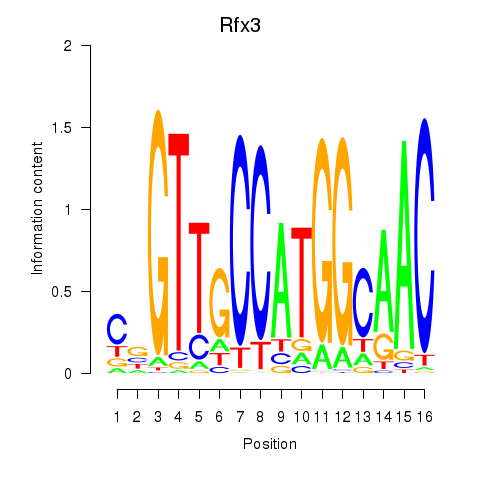
Transcription factors associated with Rfx3_Rfx1_Rfx4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx3
|
ENSMUSG00000040929.10 | regulatory factor X, 3 (influences HLA class II expression) |
|
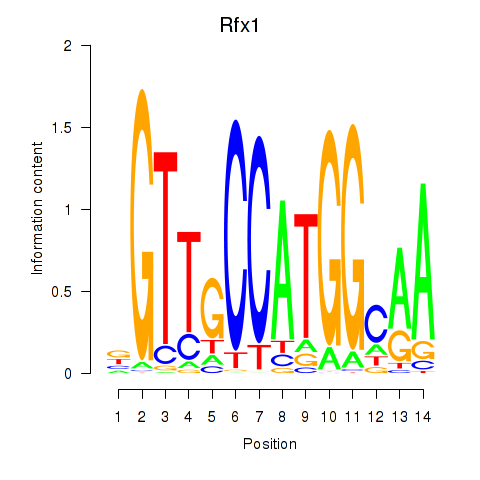
Rfx1
|
ENSMUSG00000031706.6 | regulatory factor X, 1 (influences HLA class II expression) |
|
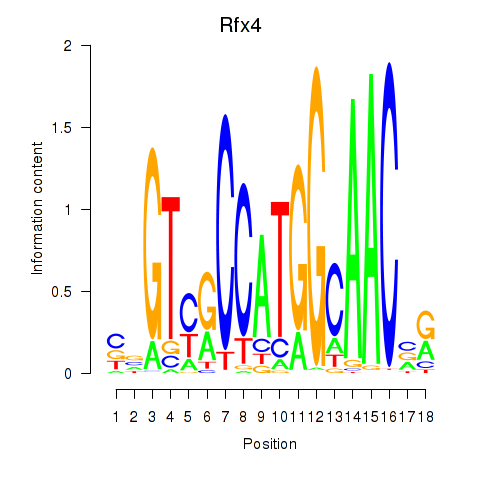
Rfx4
|
ENSMUSG00000020037.9 | regulatory factor X, 4 (influences HLA class II expression) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx1 | mm10_v2_chr8_+_84066824_84066882 | 0.90 | 1.9e-04 | Click! |
| Rfx3 | mm10_v2_chr19_-_28011138_28011181 | 0.63 | 3.9e-02 | Click! |
| Rfx4 | mm10_v2_chr10_+_84838143_84838153 | 0.28 | 4.0e-01 | Click! |
Activity profile of Rfx3_Rfx1_Rfx4 motif
Sorted Z-values of Rfx3_Rfx1_Rfx4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_60925612 | 7.59 |
ENSMUST00000102888.3
ENSMUST00000025519.4 |
Camk2a
|
calcium/calmodulin-dependent protein kinase II alpha |
| chr11_-_6606053 | 3.21 |
ENSMUST00000045713.3
|
Nacad
|
NAC alpha domain containing |
| chr10_-_81472859 | 2.79 |
ENSMUST00000147524.1
ENSMUST00000119060.1 |
Celf5
|
CUGBP, Elav-like family member 5 |
| chr7_+_126847908 | 2.64 |
ENSMUST00000147257.1
ENSMUST00000139174.1 |
Doc2a
|
double C2, alpha |
| chr4_+_129960760 | 2.58 |
ENSMUST00000139884.1
|
1700003M07Rik
|
RIKEN cDNA 1700003M07 gene |
| chr15_+_98167806 | 2.53 |
ENSMUST00000031914.4
|
AI836003
|
expressed sequence AI836003 |
| chr11_-_97352016 | 2.37 |
ENSMUST00000093942.4
|
Gpr179
|
G protein-coupled receptor 179 |
| chr5_+_30711849 | 2.17 |
ENSMUST00000088081.4
ENSMUST00000101442.3 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr5_+_30711564 | 2.16 |
ENSMUST00000114729.1
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr2_+_163506808 | 2.10 |
ENSMUST00000143911.1
|
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr9_+_44066993 | 2.07 |
ENSMUST00000034508.7
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr13_+_55464237 | 2.06 |
ENSMUST00000046533.7
|
Prr7
|
proline rich 7 (synaptic) |
| chr15_+_82256023 | 2.04 |
ENSMUST00000143238.1
|
1500009C09Rik
|
RIKEN cDNA 1500009C09 gene |
| chr10_+_81070035 | 2.01 |
ENSMUST00000005057.6
|
Thop1
|
thimet oligopeptidase 1 |
| chr11_-_61453992 | 1.93 |
ENSMUST00000060255.7
ENSMUST00000054927.7 ENSMUST00000102661.3 |
Rnf112
|
ring finger protein 112 |
| chr15_+_78428564 | 1.93 |
ENSMUST00000166142.2
ENSMUST00000162517.1 ENSMUST00000089414.4 |
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr15_+_78428650 | 1.77 |
ENSMUST00000159771.1
|
Kctd17
|
potassium channel tetramerisation domain containing 17 |
| chr11_-_71033462 | 1.77 |
ENSMUST00000156068.2
|
6330403K07Rik
|
RIKEN cDNA 6330403K07 gene |
| chr14_-_118923070 | 1.77 |
ENSMUST00000047208.5
|
Dzip1
|
DAZ interacting protein 1 |
| chr2_-_21205342 | 1.69 |
ENSMUST00000027992.2
|
Enkur
|
enkurin, TRPC channel interacting protein |
| chr2_+_152962485 | 1.65 |
ENSMUST00000099197.2
ENSMUST00000103155.3 |
Ttll9
|
tubulin tyrosine ligase-like family, member 9 |
| chr5_+_21424934 | 1.64 |
ENSMUST00000056045.4
|
Fam185a
|
family with sequence similarity 185, member A |
| chr4_+_135495979 | 1.61 |
ENSMUST00000063707.2
|
Stpg1
|
sperm tail PG rich repeat containing 1 |
| chr6_-_39557830 | 1.54 |
ENSMUST00000036877.3
ENSMUST00000154149.1 |
Dennd2a
|
DENN/MADD domain containing 2A |
| chr7_-_109439076 | 1.53 |
ENSMUST00000106745.2
ENSMUST00000090414.4 |
Stk33
|
serine/threonine kinase 33 |
| chr12_+_80518990 | 1.53 |
ENSMUST00000021558.6
|
Galnt16
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 16 |
| chr1_-_172329261 | 1.45 |
ENSMUST00000062387.2
|
Kcnj9
|
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr4_-_43031370 | 1.44 |
ENSMUST00000138030.1
|
Stoml2
|
stomatin (Epb7.2)-like 2 |
| chr1_+_66386968 | 1.43 |
ENSMUST00000145419.1
|
Map2
|
microtubule-associated protein 2 |
| chr1_+_191821444 | 1.41 |
ENSMUST00000027931.7
|
Nek2
|
NIMA (never in mitosis gene a)-related expressed kinase 2 |
| chr17_+_34629533 | 1.40 |
ENSMUST00000015620.6
|
Prrt1
|
proline-rich transmembrane protein 1 |
| chr4_-_128962420 | 1.39 |
ENSMUST00000119354.1
ENSMUST00000106068.1 ENSMUST00000030581.3 |
Adc
|
arginine decarboxylase |
| chr11_+_70029742 | 1.28 |
ENSMUST00000132597.2
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr7_-_109438786 | 1.28 |
ENSMUST00000121748.1
|
Stk33
|
serine/threonine kinase 33 |
| chr19_-_46327121 | 1.27 |
ENSMUST00000041391.4
ENSMUST00000096029.5 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr10_+_39133981 | 1.24 |
ENSMUST00000019991.7
|
Tube1
|
epsilon-tubulin 1 |
| chr9_+_44067072 | 1.21 |
ENSMUST00000177054.1
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr16_+_17646464 | 1.21 |
ENSMUST00000056962.4
|
Ccdc74a
|
coiled-coil domain containing 74A |
| chr9_+_47530173 | 1.20 |
ENSMUST00000114548.1
ENSMUST00000152459.1 ENSMUST00000143026.1 ENSMUST00000085909.2 ENSMUST00000114547.1 ENSMUST00000034581.3 |
Cadm1
|
cell adhesion molecule 1 |
| chr16_+_17646564 | 1.19 |
ENSMUST00000182117.1
ENSMUST00000182671.1 ENSMUST00000182344.1 |
Ccdc74a
|
coiled-coil domain containing 74A |
| chr9_-_61946768 | 1.18 |
ENSMUST00000034815.7
|
Kif23
|
kinesin family member 23 |
| chr17_+_12119274 | 1.18 |
ENSMUST00000024594.2
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) |
| chr5_-_142550965 | 1.18 |
ENSMUST00000129212.1
ENSMUST00000110785.1 ENSMUST00000063635.8 |
Radil
|
Ras association and DIL domains |
| chr17_+_56304313 | 1.17 |
ENSMUST00000113035.1
ENSMUST00000113039.2 ENSMUST00000142387.1 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr5_-_124187150 | 1.17 |
ENSMUST00000161938.1
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr15_+_79229140 | 1.14 |
ENSMUST00000163571.1
|
Pick1
|
protein interacting with C kinase 1 |
| chr1_+_172482199 | 1.14 |
ENSMUST00000135267.1
ENSMUST00000052629.6 ENSMUST00000111235.2 |
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr3_+_124321031 | 1.13 |
ENSMUST00000058994.4
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr8_-_27202542 | 1.13 |
ENSMUST00000038174.6
|
Got1l1
|
glutamic-oxaloacetic transaminase 1-like 1 |
| chr2_+_109280738 | 1.12 |
ENSMUST00000028527.7
|
Kif18a
|
kinesin family member 18A |
| chr4_-_43031429 | 1.11 |
ENSMUST00000136326.1
|
Stoml2
|
stomatin (Epb7.2)-like 2 |
| chr17_+_71183545 | 1.10 |
ENSMUST00000156570.1
|
Lpin2
|
lipin 2 |
| chr17_+_27685197 | 1.09 |
ENSMUST00000097360.2
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr15_+_79229363 | 1.08 |
ENSMUST00000018295.7
ENSMUST00000053926.5 |
Pick1
|
protein interacting with C kinase 1 |
| chr7_-_109438998 | 1.08 |
ENSMUST00000121378.1
|
Stk33
|
serine/threonine kinase 33 |
| chr8_-_92355764 | 1.07 |
ENSMUST00000180102.1
ENSMUST00000179421.1 ENSMUST00000179222.1 ENSMUST00000179029.1 |
4933436C20Rik
|
RIKEN cDNA 4933436C20 gene |
| chr7_-_45830776 | 1.06 |
ENSMUST00000107723.2
ENSMUST00000131384.1 |
Grwd1
|
glutamate-rich WD repeat containing 1 |
| chr5_+_33983534 | 1.05 |
ENSMUST00000114382.1
|
Gm1673
|
predicted gene 1673 |
| chr11_-_116335384 | 1.05 |
ENSMUST00000036215.7
|
Foxj1
|
forkhead box J1 |
| chr12_-_72070991 | 1.04 |
ENSMUST00000050649.4
|
Gpr135
|
G protein-coupled receptor 135 |
| chr4_-_117125618 | 1.02 |
ENSMUST00000183310.1
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr7_+_82648595 | 1.01 |
ENSMUST00000141726.1
ENSMUST00000179489.1 ENSMUST00000039881.3 |
Eftud1
|
elongation factor Tu GTP binding domain containing 1 |
| chr8_+_119575235 | 1.00 |
ENSMUST00000093100.2
|
Dnaaf1
|
dynein, axonemal assembly factor 1 |
| chr8_-_4259257 | 0.96 |
ENSMUST00000053252.7
|
Ctxn1
|
cortexin 1 |
| chr5_-_136170634 | 0.96 |
ENSMUST00000041048.1
|
Orai2
|
ORAI calcium release-activated calcium modulator 2 |
| chr13_-_100775844 | 0.94 |
ENSMUST00000075550.3
|
Cenph
|
centromere protein H |
| chr8_-_40308331 | 0.94 |
ENSMUST00000118639.1
|
Fgf20
|
fibroblast growth factor 20 |
| chr7_+_45785331 | 0.93 |
ENSMUST00000120005.1
ENSMUST00000123585.1 |
Lmtk3
|
lemur tyrosine kinase 3 |
| chr8_+_69832633 | 0.93 |
ENSMUST00000131637.2
ENSMUST00000081503.6 |
Pbx4
|
pre B cell leukemia homeobox 4 |
| chr2_-_29055051 | 0.93 |
ENSMUST00000113843.1
ENSMUST00000157048.2 |
1700101E01Rik
|
RIKEN cDNA 1700101E01 gene |
| chr12_-_103738158 | 0.92 |
ENSMUST00000095450.4
|
Serpina1b
|
serine (or cysteine) preptidase inhibitor, clade A, member 1B |
| chr7_+_99268338 | 0.92 |
ENSMUST00000107100.2
|
Map6
|
microtubule-associated protein 6 |
| chr10_+_33905015 | 0.91 |
ENSMUST00000169670.1
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr15_+_54745702 | 0.91 |
ENSMUST00000050027.8
|
Nov
|
nephroblastoma overexpressed gene |
| chr2_+_91922178 | 0.91 |
ENSMUST00000170432.1
|
Chrm4
|
cholinergic receptor, muscarinic 4 |
| chrX_+_7919816 | 0.91 |
ENSMUST00000041096.3
|
Pcsk1n
|
proprotein convertase subtilisin/kexin type 1 inhibitor |
| chr5_+_123252087 | 0.90 |
ENSMUST00000121964.1
|
Wdr66
|
WD repeat domain 66 |
| chr11_+_102248842 | 0.90 |
ENSMUST00000100392.4
|
BC030867
|
cDNA sequence BC030867 |
| chr7_-_118855984 | 0.90 |
ENSMUST00000116280.2
ENSMUST00000106550.3 ENSMUST00000063607.5 |
Knop1
|
lysine rich nucleolar protein 1 |
| chr5_-_48889531 | 0.89 |
ENSMUST00000176978.1
|
Kcnip4
|
Kv channel interacting protein 4 |
| chr11_-_101551837 | 0.89 |
ENSMUST00000017290.4
|
Brca1
|
breast cancer 1 |
| chrX_-_136993027 | 0.88 |
ENSMUST00000171738.1
ENSMUST00000056674.5 ENSMUST00000129807.1 |
Slc25a53
|
solute carrier family 25, member 53 |
| chr17_+_84626458 | 0.87 |
ENSMUST00000025101.8
|
Dync2li1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr9_+_82829806 | 0.87 |
ENSMUST00000113245.2
ENSMUST00000034783.4 |
Irak1bp1
|
interleukin-1 receptor-associated kinase 1 binding protein 1 |
| chr12_-_103773592 | 0.86 |
ENSMUST00000078869.5
|
Serpina1d
|
serine (or cysteine) peptidase inhibitor, clade A, member 1D |
| chr1_-_75180349 | 0.86 |
ENSMUST00000027396.8
|
Abcb6
|
ATP-binding cassette, sub-family B (MDR/TAP), member 6 |
| chr10_+_42860648 | 0.85 |
ENSMUST00000105495.1
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr7_-_13054665 | 0.85 |
ENSMUST00000182515.1
ENSMUST00000069289.8 |
Mzf1
|
myeloid zinc finger 1 |
| chr12_-_103956891 | 0.85 |
ENSMUST00000085054.4
|
Serpina1e
|
serine (or cysteine) peptidase inhibitor, clade A, member 1E |
| chr2_-_5714490 | 0.85 |
ENSMUST00000044009.7
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr14_-_8309770 | 0.85 |
ENSMUST00000121887.1
ENSMUST00000036070.8 ENSMUST00000137133.1 |
Fam107a
|
family with sequence similarity 107, member A |
| chr11_-_69413675 | 0.85 |
ENSMUST00000094077.4
|
Kdm6b
|
KDM1 lysine (K)-specific demethylase 6B |
| chr7_-_13054514 | 0.84 |
ENSMUST00000182087.1
|
Mzf1
|
myeloid zinc finger 1 |
| chr5_-_139460501 | 0.84 |
ENSMUST00000066052.7
|
3110082I17Rik
|
RIKEN cDNA 3110082I17 gene |
| chr16_-_4523056 | 0.83 |
ENSMUST00000090500.3
ENSMUST00000023161.7 |
Srl
|
sarcalumenin |
| chr6_-_122340200 | 0.83 |
ENSMUST00000159384.1
|
Phc1
|
polyhomeotic-like 1 (Drosophila) |
| chr5_-_138172383 | 0.83 |
ENSMUST00000000505.9
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr15_+_89532816 | 0.81 |
ENSMUST00000167173.1
|
Shank3
|
SH3/ankyrin domain gene 3 |
| chr10_+_80295930 | 0.81 |
ENSMUST00000105359.1
|
Apc2
|
adenomatosis polyposis coli 2 |
| chr12_-_103863551 | 0.81 |
ENSMUST00000085056.6
ENSMUST00000072876.5 ENSMUST00000124717.1 |
Serpina1a
|
serine (or cysteine) peptidase inhibitor, clade A, member 1A |
| chr12_+_84361968 | 0.81 |
ENSMUST00000021661.6
|
Coq6
|
coenzyme Q6 homolog (yeast) |
| chr12_+_84362029 | 0.80 |
ENSMUST00000110278.1
ENSMUST00000145522.1 |
Coq6
|
coenzyme Q6 homolog (yeast) |
| chr4_-_141933080 | 0.80 |
ENSMUST00000036701.7
|
Fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr11_+_20647149 | 0.80 |
ENSMUST00000109585.1
|
Sertad2
|
SERTA domain containing 2 |
| chr12_-_110889119 | 0.80 |
ENSMUST00000043716.8
|
Cinp
|
cyclin-dependent kinase 2 interacting protein |
| chr19_-_41896132 | 0.80 |
ENSMUST00000038677.3
|
Rrp12
|
ribosomal RNA processing 12 homolog (S. cerevisiae) |
| chr12_+_84361636 | 0.79 |
ENSMUST00000110276.1
|
Coq6
|
coenzyme Q6 homolog (yeast) |
| chr7_-_118855602 | 0.78 |
ENSMUST00000106549.1
ENSMUST00000126792.1 |
Knop1
|
lysine rich nucleolar protein 1 |
| chr18_-_43687695 | 0.77 |
ENSMUST00000082254.6
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr19_+_47178820 | 0.75 |
ENSMUST00000111808.3
|
Neurl1a
|
neuralized homolog 1A (Drosophila) |
| chr7_-_80901220 | 0.75 |
ENSMUST00000146402.1
ENSMUST00000026816.8 |
Wdr73
|
WD repeat domain 73 |
| chr9_+_122951051 | 0.75 |
ENSMUST00000040717.5
|
Kif15
|
kinesin family member 15 |
| chr11_-_66168505 | 0.74 |
ENSMUST00000080665.3
|
Dnah9
|
dynein, axonemal, heavy chain 9 |
| chr4_+_123016590 | 0.74 |
ENSMUST00000102649.3
|
Trit1
|
tRNA isopentenyltransferase 1 |
| chr5_+_33983437 | 0.74 |
ENSMUST00000114384.1
ENSMUST00000094869.5 ENSMUST00000114383.1 |
Gm1673
|
predicted gene 1673 |
| chr15_+_79141324 | 0.73 |
ENSMUST00000040077.6
|
Polr2f
|
polymerase (RNA) II (DNA directed) polypeptide F |
| chr8_-_95142477 | 0.73 |
ENSMUST00000034240.7
ENSMUST00000169748.1 |
Kifc3
|
kinesin family member C3 |
| chr9_+_74953053 | 0.73 |
ENSMUST00000170846.1
|
Fam214a
|
family with sequence similarity 214, member A |
| chr17_+_49615104 | 0.73 |
ENSMUST00000162854.1
|
Kif6
|
kinesin family member 6 |
| chr14_+_77156733 | 0.72 |
ENSMUST00000022589.7
|
Enox1
|
ecto-NOX disulfide-thiol exchanger 1 |
| chr14_-_99099701 | 0.72 |
ENSMUST00000042471.9
|
Dis3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr2_-_167492826 | 0.72 |
ENSMUST00000109211.2
ENSMUST00000057627.9 |
Spata2
|
spermatogenesis associated 2 |
| chr7_+_19176416 | 0.72 |
ENSMUST00000117338.1
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr15_+_100761741 | 0.71 |
ENSMUST00000023776.6
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr11_-_45955183 | 0.71 |
ENSMUST00000109254.1
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr7_-_48881596 | 0.71 |
ENSMUST00000119223.1
|
E2f8
|
E2F transcription factor 8 |
| chr6_-_102464667 | 0.71 |
ENSMUST00000032159.6
|
Cntn3
|
contactin 3 |
| chrX_-_152368680 | 0.69 |
ENSMUST00000070316.5
|
Gpr173
|
G-protein coupled receptor 173 |
| chr10_-_117063764 | 0.69 |
ENSMUST00000047672.7
|
Cct2
|
chaperonin containing Tcp1, subunit 2 (beta) |
| chr2_-_118256929 | 0.69 |
ENSMUST00000028820.6
ENSMUST00000028821.3 |
Fsip1
|
fibrous sheath-interacting protein 1 |
| chr12_+_29528382 | 0.68 |
ENSMUST00000049784.9
|
Myt1l
|
myelin transcription factor 1-like |
| chr1_-_133921393 | 0.67 |
ENSMUST00000048432.5
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr15_-_58324161 | 0.67 |
ENSMUST00000022985.1
|
Klhl38
|
kelch-like 38 |
| chr13_-_106936907 | 0.66 |
ENSMUST00000080856.7
|
Ipo11
|
importin 11 |
| chr15_-_103215285 | 0.66 |
ENSMUST00000122182.1
ENSMUST00000108813.3 ENSMUST00000127191.1 |
Cbx5
|
chromobox 5 |
| chr8_-_33747724 | 0.66 |
ENSMUST00000179364.1
|
Smim18
|
small integral membrane protein 18 |
| chr11_-_72207413 | 0.65 |
ENSMUST00000108505.1
|
4933427D14Rik
|
RIKEN cDNA 4933427D14 gene |
| chr2_-_131160006 | 0.65 |
ENSMUST00000103188.3
ENSMUST00000133602.1 ENSMUST00000028800.5 |
1700037H04Rik
|
RIKEN cDNA 1700037H04 gene |
| chr17_-_45592485 | 0.65 |
ENSMUST00000166119.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr9_-_27155418 | 0.64 |
ENSMUST00000167074.1
ENSMUST00000034472.8 |
Jam3
|
junction adhesion molecule 3 |
| chr10_-_79645732 | 0.64 |
ENSMUST00000095464.2
|
Odf3l2
|
outer dense fiber of sperm tails 3-like 2 |
| chr3_-_108536466 | 0.63 |
ENSMUST00000048012.6
ENSMUST00000106626.2 ENSMUST00000106625.3 |
5330417C22Rik
|
RIKEN cDNA 5330417C22 gene |
| chr2_+_145934776 | 0.62 |
ENSMUST00000126415.1
ENSMUST00000116398.1 |
4930529M08Rik
|
RIKEN cDNA 4930529M08 gene |
| chr12_-_103904887 | 0.62 |
ENSMUST00000074051.5
|
Serpina1c
|
serine (or cysteine) peptidase inhibitor, clade A, member 1C |
| chr7_-_118856254 | 0.62 |
ENSMUST00000033277.7
|
Knop1
|
lysine rich nucleolar protein 1 |
| chr10_-_79874233 | 0.61 |
ENSMUST00000166023.1
ENSMUST00000167707.1 ENSMUST00000165601.1 |
BC005764
|
cDNA sequence BC005764 |
| chr7_-_48881032 | 0.61 |
ENSMUST00000058745.8
|
E2f8
|
E2F transcription factor 8 |
| chr11_-_5950018 | 0.61 |
ENSMUST00000102920.3
|
Gck
|
glucokinase |
| chr2_-_5895319 | 0.60 |
ENSMUST00000026926.4
ENSMUST00000102981.3 |
Sec61a2
|
Sec61, alpha subunit 2 (S. cerevisiae) |
| chr6_+_48593883 | 0.60 |
ENSMUST00000154010.1
ENSMUST00000163452.1 ENSMUST00000118229.1 ENSMUST00000009420.8 |
Repin1
|
replication initiator 1 |
| chr6_+_48593927 | 0.60 |
ENSMUST00000135151.1
|
Repin1
|
replication initiator 1 |
| chr10_-_39133848 | 0.60 |
ENSMUST00000134279.1
ENSMUST00000139743.1 ENSMUST00000149949.1 ENSMUST00000124941.1 ENSMUST00000125042.1 ENSMUST00000063204.2 |
Fam229b
|
family with sequence similarity 229, member B |
| chr17_-_45592262 | 0.59 |
ENSMUST00000164769.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr17_-_45592569 | 0.58 |
ENSMUST00000163492.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr19_-_4839286 | 0.58 |
ENSMUST00000037246.5
|
Ccs
|
copper chaperone for superoxide dismutase |
| chr10_-_39133914 | 0.58 |
ENSMUST00000135785.1
|
Fam229b
|
family with sequence similarity 229, member B |
| chr1_+_172481788 | 0.57 |
ENSMUST00000127052.1
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr3_-_88459047 | 0.57 |
ENSMUST00000165898.1
ENSMUST00000127436.1 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr4_+_117550326 | 0.57 |
ENSMUST00000037127.8
|
Eri3
|
exoribonuclease 3 |
| chr10_+_42860348 | 0.57 |
ENSMUST00000063063.7
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr9_-_50617228 | 0.56 |
ENSMUST00000147671.1
ENSMUST00000145139.1 ENSMUST00000155435.1 |
AU019823
|
expressed sequence AU019823 |
| chr4_+_118620799 | 0.55 |
ENSMUST00000030501.8
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr3_-_88458876 | 0.55 |
ENSMUST00000147200.1
ENSMUST00000169222.1 |
Sema4a
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr15_-_75678732 | 0.55 |
ENSMUST00000000958.8
|
Top1mt
|
DNA topoisomerase 1, mitochondrial |
| chr10_+_87058043 | 0.55 |
ENSMUST00000169849.1
|
1700113H08Rik
|
RIKEN cDNA 1700113H08 gene |
| chr16_+_44394771 | 0.54 |
ENSMUST00000099742.2
|
Wdr52
|
WD repeat domain 52 |
| chr16_+_3909032 | 0.54 |
ENSMUST00000124849.1
|
Cluap1
|
clusterin associated protein 1 |
| chr3_+_82358056 | 0.54 |
ENSMUST00000091014.4
|
Map9
|
microtubule-associated protein 9 |
| chr7_-_44375006 | 0.53 |
ENSMUST00000107933.1
|
1700008O03Rik
|
RIKEN cDNA 1700008O03 gene |
| chr14_+_50955992 | 0.53 |
ENSMUST00000095925.4
|
Pnp2
|
purine-nucleoside phosphorylase 2 |
| chr5_+_124598749 | 0.53 |
ENSMUST00000130912.1
ENSMUST00000100706.3 |
Tctn2
|
tectonic family member 2 |
| chr2_-_25580099 | 0.52 |
ENSMUST00000114217.1
|
Gm996
|
predicted gene 996 |
| chr2_-_168601620 | 0.52 |
ENSMUST00000171689.1
ENSMUST00000137451.1 |
Nfatc2
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 |
| chr19_-_10304867 | 0.52 |
ENSMUST00000039327.4
|
Dagla
|
diacylglycerol lipase, alpha |
| chr17_-_34627365 | 0.51 |
ENSMUST00000064953.8
ENSMUST00000170345.1 ENSMUST00000171121.2 ENSMUST00000168391.2 ENSMUST00000169067.2 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr17_-_34627148 | 0.51 |
ENSMUST00000171376.1
ENSMUST00000169287.1 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr11_+_70562898 | 0.51 |
ENSMUST00000102559.4
|
Mink1
|
misshapen-like kinase 1 (zebrafish) |
| chr2_+_32629467 | 0.50 |
ENSMUST00000068271.4
|
Ak1
|
adenylate kinase 1 |
| chr17_-_80290476 | 0.50 |
ENSMUST00000086555.3
ENSMUST00000038166.7 |
Dhx57
|
DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 |
| chr5_+_66259890 | 0.50 |
ENSMUST00000065530.6
|
Nsun7
|
NOL1/NOP2/Sun domain family, member 7 |
| chr9_-_109059711 | 0.49 |
ENSMUST00000061973.4
|
Trex1
|
three prime repair exonuclease 1 |
| chr16_-_93603803 | 0.49 |
ENSMUST00000023669.5
ENSMUST00000113951.2 |
Setd4
|
SET domain containing 4 |
| chr12_-_83439910 | 0.49 |
ENSMUST00000177801.1
|
Dpf3
|
D4, zinc and double PHD fingers, family 3 |
| chr7_-_25250720 | 0.48 |
ENSMUST00000116343.2
ENSMUST00000045847.8 |
Erf
|
Ets2 repressor factor |
| chr10_+_42860543 | 0.48 |
ENSMUST00000157071.1
|
Scml4
|
sex comb on midleg-like 4 (Drosophila) |
| chr7_+_118855735 | 0.48 |
ENSMUST00000098087.2
ENSMUST00000106547.1 |
Iqck
|
IQ motif containing K |
| chr3_+_156562141 | 0.48 |
ENSMUST00000175773.1
|
Negr1
|
neuronal growth regulator 1 |
| chr5_-_112228934 | 0.48 |
ENSMUST00000181535.2
|
Miat
|
myocardial infarction associated transcript (non-protein coding) |
| chr7_+_19149722 | 0.48 |
ENSMUST00000049294.2
|
Snrpd2
|
small nuclear ribonucleoprotein D2 |
| chr2_+_121289589 | 0.47 |
ENSMUST00000094639.3
|
Map1a
|
microtubule-associated protein 1 A |
| chr11_+_70030023 | 0.47 |
ENSMUST00000143920.2
|
Dlg4
|
discs, large homolog 4 (Drosophila) |
| chr17_-_34024639 | 0.47 |
ENSMUST00000025183.8
|
Ring1
|
ring finger protein 1 |
| chr7_+_25681158 | 0.46 |
ENSMUST00000108403.3
|
B9d2
|
B9 protein domain 2 |
| chr11_-_45955465 | 0.46 |
ENSMUST00000011398.6
|
Thg1l
|
tRNA-histidine guanylyltransferase 1-like (S. cerevisiae) |
| chr2_+_36049453 | 0.46 |
ENSMUST00000028256.4
|
Morn5
|
MORN repeat containing 5 |
| chr13_+_91461050 | 0.46 |
ENSMUST00000004094.8
ENSMUST00000042122.8 |
Ssbp2
|
single-stranded DNA binding protein 2 |
| chr17_-_25837082 | 0.45 |
ENSMUST00000183929.1
ENSMUST00000184865.1 ENSMUST00000026831.7 |
Rhbdl1
|
rhomboid, veinlet-like 1 (Drosophila) |
| chr7_+_133776857 | 0.45 |
ENSMUST00000065359.5
ENSMUST00000151031.1 ENSMUST00000121560.1 |
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chr10_-_103236280 | 0.45 |
ENSMUST00000123364.1
ENSMUST00000166240.1 ENSMUST00000020043.5 |
Lrriq1
|
leucine-rich repeats and IQ motif containing 1 |
| chr19_-_3282958 | 0.45 |
ENSMUST00000119292.1
ENSMUST00000025751.3 |
Ighmbp2
|
immunoglobulin mu binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx3_Rfx1_Rfx4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:1990046 | positive regulation of mitochondrial DNA replication(GO:0090297) regulation of cardiolipin metabolic process(GO:1900208) positive regulation of cardiolipin metabolic process(GO:1900210) stress-induced mitochondrial fusion(GO:1990046) |
| 0.8 | 9.3 | GO:0098970 | receptor diffusion trapping(GO:0098953) postsynaptic neurotransmitter receptor diffusion trapping(GO:0098970) neurotransmitter receptor diffusion trapping(GO:0099628) |
| 0.7 | 2.1 | GO:0010534 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.5 | 1.6 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.4 | 1.2 | GO:0042271 | unidimensional cell growth(GO:0009826) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.4 | 1.2 | GO:0000912 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.4 | 1.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.4 | 1.4 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.3 | 0.9 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) endothelial cell-cell adhesion(GO:0071603) negative regulation of sensory perception of pain(GO:1904057) bone regeneration(GO:1990523) |
| 0.3 | 0.9 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.3 | 2.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.3 | 1.8 | GO:0015862 | uridine transport(GO:0015862) |
| 0.3 | 0.5 | GO:0098917 | retrograde trans-synaptic signaling(GO:0098917) |
| 0.2 | 0.9 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.9 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.2 | 1.1 | GO:0061141 | lung ciliated cell differentiation(GO:0061141) |
| 0.2 | 0.8 | GO:0021508 | floor plate formation(GO:0021508) |
| 0.2 | 0.6 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.2 | 0.9 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.2 | 0.7 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.2 | 0.5 | GO:0010751 | regulation of arginine metabolic process(GO:0000821) negative regulation of nitric oxide mediated signal transduction(GO:0010751) negative regulation of cellular amino acid metabolic process(GO:0045763) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.2 | 3.4 | GO:0048642 | negative regulation of skeletal muscle tissue development(GO:0048642) |
| 0.2 | 0.8 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.2 | 1.1 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.2 | 0.9 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.2 | 2.7 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.1 | 0.4 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.1 | 0.7 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.1 | 0.4 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.1 | 0.4 | GO:0018317 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 1.6 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.1 | 0.9 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.7 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.6 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.4 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.1 | 0.8 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.1 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.1 | 0.5 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.1 | 0.4 | GO:0090346 | nitrate catabolic process(GO:0043602) nitric oxide catabolic process(GO:0046210) cellular organohalogen metabolic process(GO:0090345) cellular organofluorine metabolic process(GO:0090346) |
| 0.1 | 0.9 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.1 | 0.8 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.1 | 0.4 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.9 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.1 | 0.6 | GO:0051365 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 3.7 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
| 0.1 | 0.7 | GO:0071494 | cellular response to UV-C(GO:0071494) |
| 0.1 | 0.3 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.1 | 0.4 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.1 | 0.8 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.1 | 0.5 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.1 | 0.3 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 1.2 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 0.5 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.8 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.1 | 0.6 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 1.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.3 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.1 | 0.4 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.1 | 0.6 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.1 | 1.1 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 0.8 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.2 | GO:0030862 | positive regulation of polarized epithelial cell differentiation(GO:0030862) |
| 0.1 | 0.6 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.1 | 0.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.1 | 0.5 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.1 | 0.4 | GO:0090154 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.1 | 0.3 | GO:0032423 | regulation of mismatch repair(GO:0032423) |
| 0.1 | 0.9 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.1 | 0.3 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 2.8 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.1 | 0.4 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.1 | 1.9 | GO:0071158 | positive regulation of cell cycle arrest(GO:0071158) |
| 0.1 | 0.2 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 0.1 | 0.2 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.2 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.1 | 1.0 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.7 | GO:2001223 | negative regulation of neuron migration(GO:2001223) |
| 0.0 | 0.2 | GO:0032417 | positive regulation of sodium:proton antiporter activity(GO:0032417) |
| 0.0 | 0.5 | GO:0000290 | deadenylation-dependent decapping of nuclear-transcribed mRNA(GO:0000290) |
| 0.0 | 0.4 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 1.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.4 | GO:0032011 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.3 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.7 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 1.1 | GO:0072520 | seminiferous tubule development(GO:0072520) |
| 0.0 | 0.1 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 | 0.2 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.5 | GO:0035435 | phosphate ion transmembrane transport(GO:0035435) |
| 0.0 | 0.7 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.2 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.5 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.4 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.2 | GO:0060830 | signal transduction downstream of smoothened(GO:0007227) ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 0.0 | 1.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.7 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 1.1 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.0 | 0.4 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.3 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 1.8 | GO:0051220 | cytoplasmic sequestering of protein(GO:0051220) |
| 0.0 | 1.2 | GO:2000191 | regulation of fatty acid transport(GO:2000191) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.9 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.9 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.0 | 1.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.4 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.0 | 0.1 | GO:0090283 | regulation of protein glycosylation in Golgi(GO:0090283) negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.2 | GO:0034141 | positive regulation of toll-like receptor 3 signaling pathway(GO:0034141) |
| 0.0 | 0.2 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 0.4 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 3.9 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.3 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.2 | GO:0036371 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 2.0 | GO:0042273 | ribosomal large subunit biogenesis(GO:0042273) |
| 0.0 | 0.3 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.0 | 0.1 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 2.0 | GO:0006400 | tRNA modification(GO:0006400) |
| 0.0 | 0.1 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 1.2 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.3 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 1.0 | GO:0019432 | triglyceride biosynthetic process(GO:0019432) |
| 0.0 | 0.3 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.2 | GO:2000484 | positive regulation of interleukin-8 secretion(GO:2000484) |
| 0.0 | 0.1 | GO:0051105 | regulation of DNA ligation(GO:0051105) positive regulation of DNA ligation(GO:0051106) |
| 0.0 | 0.1 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.7 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.9 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.9 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0071963 | establishment or maintenance of cell polarity regulating cell shape(GO:0071963) |
| 0.0 | 0.5 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.3 | GO:0006271 | DNA strand elongation involved in DNA replication(GO:0006271) |
| 0.0 | 0.8 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.5 | GO:0035518 | histone H2A monoubiquitination(GO:0035518) |
| 0.0 | 0.2 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.0 | GO:1902956 | regulation of mitochondrial electron transport, NADH to ubiquinone(GO:1902956) |
| 0.0 | 0.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.3 | GO:1901978 | positive regulation of cell cycle checkpoint(GO:1901978) |
| 0.0 | 0.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.1 | GO:1903898 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.4 | GO:1990845 | adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 1.5 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0035247 | peptidyl-arginine N-methylation(GO:0035246) peptidyl-arginine omega-N-methylation(GO:0035247) |
| 0.0 | 0.2 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.4 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.2 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.4 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.4 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:1902564 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) negative regulation of neutrophil activation(GO:1902564) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) |
| 0.0 | 0.1 | GO:1990403 | common bile duct development(GO:0061009) embryonic brain development(GO:1990403) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.2 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 1.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.6 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.1 | GO:0044376 | RNA polymerase II complex import to nucleus(GO:0044376) RNA polymerase III complex localization to nucleus(GO:1990022) |
| 0.0 | 0.2 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.8 | GO:0007569 | cell aging(GO:0007569) |
| 0.0 | 0.1 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.5 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.0 | 0.1 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.4 | GO:2000134 | negative regulation of G1/S transition of mitotic cell cycle(GO:2000134) |
| 0.0 | 0.2 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.4 | GO:0097484 | dendrite extension(GO:0097484) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.1 | GO:0006390 | transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 2.4 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.0 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.0 | 0.4 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.2 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.7 | 2.2 | GO:0098842 | postsynaptic early endosome(GO:0098842) |
| 0.2 | 1.2 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.2 | 1.4 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.2 | 1.4 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.2 | 2.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.1 | 0.9 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.1 | 1.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.1 | 0.6 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.1 | 0.3 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.1 | 0.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.6 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.7 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.5 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.1 | 0.4 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.4 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.1 | 2.6 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.1 | 1.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 1.5 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.9 | GO:0097542 | ciliary tip(GO:0097542) |
| 0.1 | 0.7 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.1 | 1.0 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.1 | 1.8 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.1 | 2.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.1 | 0.7 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.1 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.1 | 0.3 | GO:0034709 | methylosome(GO:0034709) |
| 0.1 | 0.8 | GO:0042555 | MCM complex(GO:0042555) |
| 0.1 | 0.8 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.1 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.1 | 0.2 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.9 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 3.7 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.6 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 1.3 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.3 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.0 | 0.3 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.9 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.2 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.3 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.7 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.4 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.2 | GO:0033588 | Elongator holoenzyme complex(GO:0033588) |
| 0.0 | 0.3 | GO:0032797 | SMN complex(GO:0032797) |
| 0.0 | 0.9 | GO:1990752 | microtubule plus-end(GO:0035371) microtubule end(GO:1990752) |
| 0.0 | 0.1 | GO:0035061 | interchromatin granule(GO:0035061) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.9 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 2.0 | GO:0005657 | replication fork(GO:0005657) |
| 0.0 | 0.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 1.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.3 | GO:0005688 | U6 snRNP(GO:0005688) |
| 0.0 | 1.3 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.3 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.7 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 1.7 | GO:0005758 | mitochondrial intermembrane space(GO:0005758) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.2 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.3 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.5 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 7.9 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.1 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.0 | 0.3 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.4 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.0 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.0 | 2.1 | GO:0022626 | cytosolic ribosome(GO:0022626) |
| 0.0 | 0.7 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 1.1 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 6.7 | GO:0030425 | dendrite(GO:0030425) |
| 0.0 | 0.2 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.7 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.0 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 3.7 | GO:0045211 | postsynaptic membrane(GO:0045211) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 1.5 | GO:0005741 | mitochondrial outer membrane(GO:0005741) |
| 0.0 | 1.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 7.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.7 | 2.1 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.4 | 1.2 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.4 | 1.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.3 | 2.6 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 1.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.2 | 1.8 | GO:0031812 | G-protein coupled nucleotide receptor binding(GO:0031811) P2Y1 nucleotide receptor binding(GO:0031812) |
| 0.2 | 0.9 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 1.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.2 | 0.8 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.2 | 1.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 0.2 | 1.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.2 | 0.9 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.2 | 1.4 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.2 | 0.9 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.2 | 0.8 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 2.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.2 | 0.5 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.1 | 3.7 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.1 | 0.4 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.1 | 0.9 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.1 | 0.4 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.7 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.7 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.1 | 0.6 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.1 | 0.4 | GO:0016708 | nitric oxide dioxygenase activity(GO:0008941) oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of two atoms of oxygen into one donor(GO:0016708) iron-cytochrome-c reductase activity(GO:0047726) |
| 0.1 | 0.4 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 1.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.1 | 0.6 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.1 | 1.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.1 | 2.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.1 | 0.4 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 1.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.1 | 1.2 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.1 | 0.2 | GO:0070736 | protein-glycine ligase activity, initiating(GO:0070736) |
| 0.1 | 0.6 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.1 | 0.4 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) |
| 0.1 | 0.3 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 2.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.7 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.1 | 0.2 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 1.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.1 | 3.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.1 | 0.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 1.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.1 | 0.9 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.2 | GO:0061513 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.6 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.1 | 0.4 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.1 | 0.8 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.1 | 0.4 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 1.1 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.1 | 0.4 | GO:0016413 | O-acetyltransferase activity(GO:0016413) |
| 0.1 | 0.5 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.1 | 0.2 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.1 | 0.7 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 0.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.1 | 0.2 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.1 | 0.2 | GO:0000700 | mismatch base pair DNA N-glycosylase activity(GO:0000700) |
| 0.0 | 0.2 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 1.4 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 1.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 1.1 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 4.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 1.1 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 9.6 | GO:0008017 | microtubule binding(GO:0008017) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0042328 | heparan sulfate N-acetylglucosaminyltransferase activity(GO:0042328) |
| 0.0 | 0.1 | GO:0001034 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) RNA polymerase III transcription factor activity, sequence-specific DNA binding(GO:0001034) |
| 0.0 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.2 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0032356 | oxidized DNA binding(GO:0032356) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.0 | 0.5 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:1990189 | peptide-serine-N-acetyltransferase activity(GO:1990189) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.7 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 1.8 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.9 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.2 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.4 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.9 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.8 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.1 | GO:0016434 | rRNA (cytosine) methyltransferase activity(GO:0016434) |
| 0.0 | 2.2 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.1 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.0 | GO:0004844 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.8 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 3.8 | GO:0001078 | transcriptional repressor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001078) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.7 | GO:0045182 | translation regulator activity(GO:0045182) |
| 0.0 | 0.0 | GO:0042171 | lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.8 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.0 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.6 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.2 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.8 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.2 | GO:0015605 | organophosphate ester transmembrane transporter activity(GO:0015605) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.4 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.1 | 3.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 3.1 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 2.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.6 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 1.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 1.0 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.5 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.3 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.4 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 7.6 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.2 | 4.3 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 2.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.1 | 2.3 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 2.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 0.7 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 1.0 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 1.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 1.4 | REACTOME APC CDC20 MEDIATED DEGRADATION OF NEK2A | Genes involved in APC-Cdc20 mediated degradation of Nek2A |
| 0.1 | 0.8 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.1 | 1.1 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.9 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 1.1 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 1.9 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.3 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.8 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 1.8 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.6 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 1.6 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 1.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.4 | REACTOME RESOLUTION OF AP SITES VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the single-nucleotide replacement pathway |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF AMPA RECEPTORS | Genes involved in Trafficking of AMPA receptors |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.7 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.4 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 1.6 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |
| 0.0 | 0.6 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.5 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.2 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.6 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |