Project
12D miR HR13_24
Navigation
Downloads
Results for Runx2_Bcl11a
Z-value: 1.41
Transcription factors associated with Runx2_Bcl11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
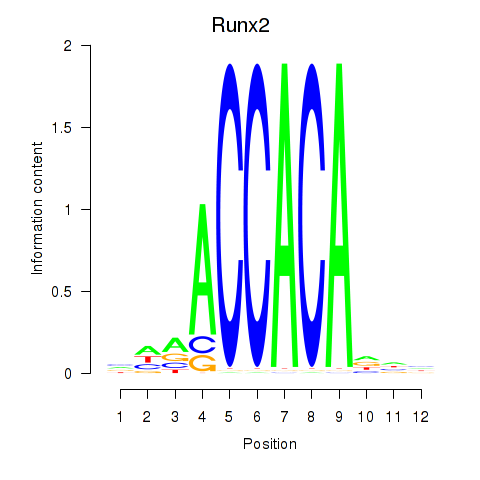
Runx2
|
ENSMUSG00000039153.10 | runt related transcription factor 2 |
|
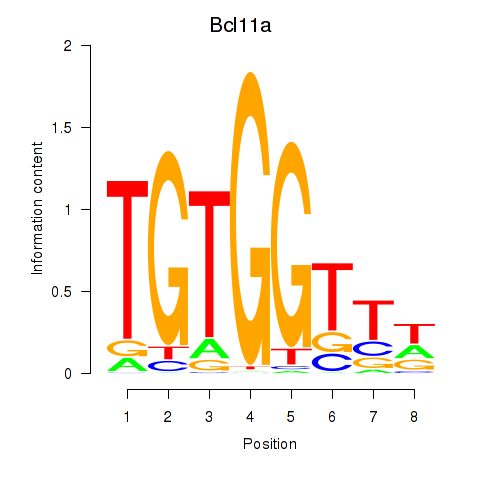
Bcl11a
|
ENSMUSG00000000861.9 | B cell CLL/lymphoma 11A (zinc finger protein) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl11a | mm10_v2_chr11_+_24078173_24078219 | -0.86 | 7.3e-04 | Click! |
| Runx2 | mm10_v2_chr17_-_44814581_44814595 | -0.65 | 3.0e-02 | Click! |
Activity profile of Runx2_Bcl11a motif
Sorted Z-values of Runx2_Bcl11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_17838173 | 2.92 |
ENSMUST00000118960.1
|
Car15
|
carbonic anhydrase 15 |
| chr8_-_109579056 | 2.76 |
ENSMUST00000074898.6
|
Hp
|
haptoglobin |
| chr13_+_89540636 | 2.66 |
ENSMUST00000022108.7
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr2_+_21367532 | 2.56 |
ENSMUST00000055946.7
|
Gpr158
|
G protein-coupled receptor 158 |
| chr4_-_57916283 | 2.36 |
ENSMUST00000063816.5
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr7_+_121865070 | 2.28 |
ENSMUST00000033161.5
|
Scnn1b
|
sodium channel, nonvoltage-gated 1 beta |
| chr10_-_81291227 | 2.20 |
ENSMUST00000045744.6
|
Tjp3
|
tight junction protein 3 |
| chr3_-_90514250 | 2.16 |
ENSMUST00000107340.1
ENSMUST00000060738.8 |
S100a1
|
S100 calcium binding protein A1 |
| chr3_+_28697901 | 2.13 |
ENSMUST00000029240.7
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr15_-_75567176 | 2.10 |
ENSMUST00000156032.1
ENSMUST00000127095.1 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr9_+_53301571 | 2.00 |
ENSMUST00000051014.1
|
Exph5
|
exophilin 5 |
| chr15_+_9436028 | 1.99 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chrX_+_159697308 | 1.84 |
ENSMUST00000123433.1
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr11_+_96849863 | 1.78 |
ENSMUST00000018816.7
|
Copz2
|
coatomer protein complex, subunit zeta 2 |
| chr14_+_55853997 | 1.60 |
ENSMUST00000100529.3
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr4_-_137118135 | 1.52 |
ENSMUST00000154285.1
|
Gm13001
|
predicted gene 13001 |
| chr7_+_49974864 | 1.41 |
ENSMUST00000081872.5
ENSMUST00000151721.1 |
Nell1
|
NEL-like 1 |
| chr11_-_101967005 | 1.36 |
ENSMUST00000001534.6
|
Sost
|
sclerostin |
| chr15_-_54278420 | 1.33 |
ENSMUST00000079772.3
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr3_-_89093358 | 1.30 |
ENSMUST00000090929.5
ENSMUST00000052539.6 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr12_-_110682606 | 1.26 |
ENSMUST00000070659.5
|
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr2_-_144332146 | 1.25 |
ENSMUST00000037423.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr10_+_127866457 | 1.23 |
ENSMUST00000092058.3
|
BC089597
|
cDNA sequence BC089597 |
| chr4_+_128058962 | 1.22 |
ENSMUST00000184063.1
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr2_+_70562007 | 1.21 |
ENSMUST00000094934.4
|
Gad1
|
glutamate decarboxylase 1 |
| chr2_+_70562147 | 1.21 |
ENSMUST00000148210.1
|
Gad1
|
glutamate decarboxylase 1 |
| chr9_+_53405280 | 1.20 |
ENSMUST00000005262.1
|
4930550C14Rik
|
RIKEN cDNA 4930550C14 gene |
| chr11_-_31824518 | 1.19 |
ENSMUST00000134944.1
|
D630024D03Rik
|
RIKEN cDNA D630024D03 gene |
| chr12_+_116077720 | 1.19 |
ENSMUST00000011315.3
|
Vipr2
|
vasoactive intestinal peptide receptor 2 |
| chr13_+_38151324 | 1.18 |
ENSMUST00000127906.1
|
Dsp
|
desmoplakin |
| chr13_+_38151343 | 1.16 |
ENSMUST00000124830.1
|
Dsp
|
desmoplakin |
| chr7_+_80246375 | 1.15 |
ENSMUST00000058266.6
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chrX_-_8090442 | 1.10 |
ENSMUST00000033505.6
|
Was
|
Wiskott-Aldrich syndrome homolog (human) |
| chr16_-_17132377 | 1.08 |
ENSMUST00000023453.7
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr6_+_49036518 | 1.08 |
ENSMUST00000031840.7
|
Gpnmb
|
glycoprotein (transmembrane) nmb |
| chr3_+_96181151 | 1.08 |
ENSMUST00000035371.8
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr2_-_32387760 | 1.07 |
ENSMUST00000050785.8
|
Lcn2
|
lipocalin 2 |
| chr5_-_117319242 | 1.04 |
ENSMUST00000100834.1
|
Gm10399
|
predicted gene 10399 |
| chr7_-_100863373 | 1.01 |
ENSMUST00000142885.1
ENSMUST00000008462.3 |
Relt
|
RELT tumor necrosis factor receptor |
| chr9_-_62510498 | 1.01 |
ENSMUST00000164246.2
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr3_+_90514435 | 0.99 |
ENSMUST00000048138.6
ENSMUST00000181271.1 |
S100a13
|
S100 calcium binding protein A13 |
| chr11_-_58502554 | 0.99 |
ENSMUST00000170501.2
ENSMUST00000081743.2 |
Olfr331
|
olfactory receptor 331 |
| chr8_+_60655540 | 0.97 |
ENSMUST00000034066.3
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr1_+_125676969 | 0.96 |
ENSMUST00000027581.6
|
Gpr39
|
G protein-coupled receptor 39 |
| chr16_+_32756336 | 0.96 |
ENSMUST00000135753.1
|
Muc4
|
mucin 4 |
| chr4_+_40920047 | 0.94 |
ENSMUST00000030122.4
|
Spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr2_-_94264713 | 0.93 |
ENSMUST00000129661.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chr11_+_58580837 | 0.93 |
ENSMUST00000169428.2
|
Olfr325
|
olfactory receptor 325 |
| chr3_+_3634145 | 0.90 |
ENSMUST00000108394.1
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr9_-_53706211 | 0.90 |
ENSMUST00000068449.3
|
Rab39
|
RAB39, member RAS oncogene family |
| chr8_+_124576105 | 0.87 |
ENSMUST00000093033.5
ENSMUST00000133086.1 |
Capn9
|
calpain 9 |
| chr7_-_90129339 | 0.82 |
ENSMUST00000181189.1
|
2310010J17Rik
|
RIKEN cDNA 2310010J17 gene |
| chr14_+_31641051 | 0.82 |
ENSMUST00000090147.6
|
Btd
|
biotinidase |
| chr3_-_54915867 | 0.82 |
ENSMUST00000070342.3
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr8_+_21776567 | 0.81 |
ENSMUST00000051017.8
|
Defb1
|
defensin beta 1 |
| chr2_-_94264745 | 0.81 |
ENSMUST00000134563.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chr6_-_83121385 | 0.80 |
ENSMUST00000146328.1
ENSMUST00000113936.3 ENSMUST00000032111.4 |
Wbp1
|
WW domain binding protein 1 |
| chr10_-_89506631 | 0.80 |
ENSMUST00000058126.8
ENSMUST00000105296.2 |
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr12_-_112860886 | 0.79 |
ENSMUST00000021729.7
|
Gpr132
|
G protein-coupled receptor 132 |
| chr18_-_11051479 | 0.79 |
ENSMUST00000180789.1
|
1010001N08Rik
|
RIKEN cDNA 1010001N08 gene |
| chr15_-_95791642 | 0.78 |
ENSMUST00000180649.1
|
A130051J06Rik
|
RIKEN cDNA A130051J0 gene |
| chr18_-_3299537 | 0.78 |
ENSMUST00000129435.1
ENSMUST00000122958.1 |
Crem
|
cAMP responsive element modulator |
| chr11_+_78322965 | 0.78 |
ENSMUST00000017534.8
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr11_-_83649349 | 0.78 |
ENSMUST00000001008.5
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr7_-_100656953 | 0.78 |
ENSMUST00000107046.1
ENSMUST00000107045.1 ENSMUST00000139708.1 |
Plekhb1
|
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr10_+_43579161 | 0.78 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chr18_-_35649349 | 0.77 |
ENSMUST00000025211.4
|
Mzb1
|
marginal zone B and B1 cell-specific protein 1 |
| chr3_+_27317028 | 0.77 |
ENSMUST00000046383.5
ENSMUST00000174840.1 |
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr18_-_3299452 | 0.77 |
ENSMUST00000126578.1
|
Crem
|
cAMP responsive element modulator |
| chr15_-_34356421 | 0.75 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr5_+_37047464 | 0.75 |
ENSMUST00000137019.1
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr9_-_95845215 | 0.75 |
ENSMUST00000093800.2
|
Pls1
|
plastin 1 (I-isoform) |
| chr19_-_36736653 | 0.73 |
ENSMUST00000087321.2
|
Ppp1r3c
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C |
| chrX_+_106027300 | 0.72 |
ENSMUST00000055941.6
|
Atp7a
|
ATPase, Cu++ transporting, alpha polypeptide |
| chr14_-_31640878 | 0.71 |
ENSMUST00000167066.1
ENSMUST00000127204.2 ENSMUST00000022437.8 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr1_+_165302625 | 0.71 |
ENSMUST00000111450.1
|
Gpr161
|
G protein-coupled receptor 161 |
| chr13_+_76579681 | 0.71 |
ENSMUST00000109589.2
|
Mctp1
|
multiple C2 domains, transmembrane 1 |
| chr1_-_10009098 | 0.70 |
ENSMUST00000176398.1
ENSMUST00000027049.3 |
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr2_-_152831112 | 0.70 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr7_+_141078188 | 0.70 |
ENSMUST00000106039.2
|
Pkp3
|
plakophilin 3 |
| chr1_-_156674290 | 0.70 |
ENSMUST00000079625.4
|
Tor3a
|
torsin family 3, member A |
| chr4_-_115133977 | 0.70 |
ENSMUST00000051400.7
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr11_+_87663087 | 0.69 |
ENSMUST00000165679.1
|
Rnf43
|
ring finger protein 43 |
| chr18_+_65800543 | 0.69 |
ENSMUST00000025394.6
ENSMUST00000153193.1 |
Sec11c
|
SEC11 homolog C (S. cerevisiae) |
| chr11_+_114851814 | 0.69 |
ENSMUST00000053361.5
ENSMUST00000021071.7 ENSMUST00000136785.1 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr1_-_169747634 | 0.69 |
ENSMUST00000027991.5
ENSMUST00000111357.1 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr8_-_11008458 | 0.68 |
ENSMUST00000040514.6
|
Irs2
|
insulin receptor substrate 2 |
| chr4_-_106799779 | 0.67 |
ENSMUST00000145061.1
ENSMUST00000102762.3 |
Acot11
|
acyl-CoA thioesterase 11 |
| chr1_+_16688405 | 0.67 |
ENSMUST00000026881.4
|
Ly96
|
lymphocyte antigen 96 |
| chr5_-_71548190 | 0.67 |
ENSMUST00000050129.5
|
Cox7b2
|
cytochrome c oxidase subunit VIIb2 |
| chr2_+_126552407 | 0.66 |
ENSMUST00000061491.7
|
Slc27a2
|
solute carrier family 27 (fatty acid transporter), member 2 |
| chr5_-_107726017 | 0.66 |
ENSMUST00000159263.2
|
Gfi1
|
growth factor independent 1 |
| chr13_-_37050237 | 0.65 |
ENSMUST00000164727.1
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr4_-_131672133 | 0.65 |
ENSMUST00000144212.1
|
Gm12962
|
predicted gene 12962 |
| chr7_+_143005677 | 0.64 |
ENSMUST00000082008.5
ENSMUST00000105925.1 ENSMUST00000105924.1 |
Tspan32
|
tetraspanin 32 |
| chr1_+_88087802 | 0.64 |
ENSMUST00000113139.1
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr13_+_23870284 | 0.64 |
ENSMUST00000006785.7
|
Slc17a1
|
solute carrier family 17 (sodium phosphate), member 1 |
| chr1_+_180935022 | 0.63 |
ENSMUST00000037361.8
|
Lefty1
|
left right determination factor 1 |
| chr5_-_24351604 | 0.63 |
ENSMUST00000036092.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr16_-_92826004 | 0.62 |
ENSMUST00000023673.7
|
Runx1
|
runt related transcription factor 1 |
| chr7_-_141214080 | 0.62 |
ENSMUST00000026573.5
ENSMUST00000170841.1 |
1600016N20Rik
|
RIKEN cDNA 1600016N20 gene |
| chr19_+_53677286 | 0.61 |
ENSMUST00000095969.3
ENSMUST00000164202.1 |
Rbm20
|
RNA binding motif protein 20 |
| chr8_-_107065632 | 0.61 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr5_-_149184063 | 0.61 |
ENSMUST00000180733.1
|
5730422E09Rik
|
RIKEN cDNA 5730422E09 gene |
| chr11_+_82045705 | 0.61 |
ENSMUST00000021011.2
|
Ccl7
|
chemokine (C-C motif) ligand 7 |
| chr1_+_74409376 | 0.60 |
ENSMUST00000027366.6
|
Vil1
|
villin 1 |
| chr7_+_44572370 | 0.60 |
ENSMUST00000002274.8
|
Napsa
|
napsin A aspartic peptidase |
| chr7_+_30565410 | 0.60 |
ENSMUST00000043850.7
|
Igflr1
|
IGF-like family receptor 1 |
| chr4_-_133263042 | 0.60 |
ENSMUST00000105908.3
ENSMUST00000030674.7 |
Sytl1
|
synaptotagmin-like 1 |
| chr1_+_135132693 | 0.60 |
ENSMUST00000049449.4
|
Ptpn7
|
protein tyrosine phosphatase, non-receptor type 7 |
| chr11_-_109473220 | 0.60 |
ENSMUST00000070872.6
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr6_+_56017489 | 0.60 |
ENSMUST00000052827.4
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr10_+_76562270 | 0.58 |
ENSMUST00000009259.4
ENSMUST00000105414.1 |
Spatc1l
|
spermatogenesis and centriole associated 1 like |
| chr10_-_24101951 | 0.58 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr5_+_24413406 | 0.58 |
ENSMUST00000049346.5
|
Asic3
|
acid-sensing (proton-gated) ion channel 3 |
| chr5_+_37050854 | 0.58 |
ENSMUST00000043794.4
|
Jakmip1
|
janus kinase and microtubule interacting protein 1 |
| chr1_+_128244122 | 0.57 |
ENSMUST00000027592.3
|
Ubxn4
|
UBX domain protein 4 |
| chr4_-_143299463 | 0.57 |
ENSMUST00000119654.1
|
Pdpn
|
podoplanin |
| chr10_-_40025253 | 0.57 |
ENSMUST00000163705.2
|
AI317395
|
expressed sequence AI317395 |
| chr2_+_86041317 | 0.57 |
ENSMUST00000111589.1
|
Olfr1033
|
olfactory receptor 1033 |
| chr8_+_70724064 | 0.57 |
ENSMUST00000034307.7
ENSMUST00000110095.2 |
Pde4c
|
phosphodiesterase 4C, cAMP specific |
| chr6_-_124840192 | 0.56 |
ENSMUST00000024206.5
|
Gnb3
|
guanine nucleotide binding protein (G protein), beta 3 |
| chr9_-_95815389 | 0.56 |
ENSMUST00000119760.1
|
Pls1
|
plastin 1 (I-isoform) |
| chr5_+_117319292 | 0.56 |
ENSMUST00000086464.4
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr10_-_61147659 | 0.56 |
ENSMUST00000092498.5
ENSMUST00000137833.1 ENSMUST00000155919.1 |
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr4_+_118527229 | 0.56 |
ENSMUST00000030261.5
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chr15_-_9529868 | 0.56 |
ENSMUST00000003981.4
|
Il7r
|
interleukin 7 receptor |
| chr4_+_145696161 | 0.56 |
ENSMUST00000180014.1
|
Gm13242
|
predicted gene 13242 |
| chr4_-_114908892 | 0.56 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr10_+_116986314 | 0.56 |
ENSMUST00000020378.4
|
Best3
|
bestrophin 3 |
| chr11_-_100414829 | 0.55 |
ENSMUST00000066489.6
|
Leprel4
|
leprecan-like 4 |
| chr15_+_99393610 | 0.55 |
ENSMUST00000159531.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr2_-_28699661 | 0.55 |
ENSMUST00000124840.1
|
1700026L06Rik
|
RIKEN cDNA 1700026L06 gene |
| chr6_-_122602345 | 0.55 |
ENSMUST00000147760.1
ENSMUST00000112585.1 |
Apobec1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chrX_-_102908672 | 0.55 |
ENSMUST00000119624.1
ENSMUST00000033686.1 |
Dmrtc1a
|
DMRT-like family C1a |
| chr16_-_10395476 | 0.55 |
ENSMUST00000043415.6
|
Tekt5
|
tektin 5 |
| chr19_+_37436707 | 0.54 |
ENSMUST00000128184.1
|
Hhex
|
hematopoietically expressed homeobox |
| chr15_+_99393574 | 0.54 |
ENSMUST00000162624.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr7_+_143005638 | 0.54 |
ENSMUST00000075172.5
ENSMUST00000105923.1 |
Tspan32
|
tetraspanin 32 |
| chr6_-_122602404 | 0.54 |
ENSMUST00000112586.1
|
Apobec1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr19_+_5568002 | 0.53 |
ENSMUST00000096318.3
|
Ap5b1
|
adaptor-related protein complex 5, beta 1 subunit |
| chr4_+_102589687 | 0.53 |
ENSMUST00000097949.4
ENSMUST00000106901.1 |
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr6_-_113719880 | 0.53 |
ENSMUST00000064993.5
|
Ghrl
|
ghrelin |
| chr6_+_87778084 | 0.53 |
ENSMUST00000032133.3
|
Gp9
|
glycoprotein 9 (platelet) |
| chrX_-_162643629 | 0.53 |
ENSMUST00000112334.1
|
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr16_-_94370994 | 0.52 |
ENSMUST00000113914.1
ENSMUST00000113905.1 |
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr4_-_143299498 | 0.52 |
ENSMUST00000030317.7
|
Pdpn
|
podoplanin |
| chr18_+_37496997 | 0.52 |
ENSMUST00000059571.5
|
Pcdhb19
|
protocadherin beta 19 |
| chr8_-_122678072 | 0.52 |
ENSMUST00000006525.7
ENSMUST00000064674.6 |
Cbfa2t3
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 3 (human) |
| chr1_+_161395409 | 0.52 |
ENSMUST00000028024.4
|
Tnfsf4
|
tumor necrosis factor (ligand) superfamily, member 4 |
| chr3_-_131303144 | 0.51 |
ENSMUST00000106337.2
|
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr11_-_79523760 | 0.51 |
ENSMUST00000179322.1
|
Evi2b
|
ecotropic viral integration site 2b |
| chr8_+_105269788 | 0.51 |
ENSMUST00000036127.2
ENSMUST00000163734.2 |
Hsf4
|
heat shock transcription factor 4 |
| chr19_-_46672883 | 0.51 |
ENSMUST00000026012.7
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr1_-_162813926 | 0.51 |
ENSMUST00000144916.1
ENSMUST00000140274.1 |
Fmo4
|
flavin containing monooxygenase 4 |
| chr7_-_142176001 | 0.50 |
ENSMUST00000067978.5
|
Krtap5-2
|
keratin associated protein 5-2 |
| chr11_+_114851507 | 0.50 |
ENSMUST00000177952.1
|
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr11_-_46389454 | 0.49 |
ENSMUST00000101306.3
|
Itk
|
IL2 inducible T cell kinase |
| chr10_-_62507737 | 0.49 |
ENSMUST00000020271.6
|
Srgn
|
serglycin |
| chr7_-_143074561 | 0.49 |
ENSMUST00000148715.1
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr6_-_142507805 | 0.49 |
ENSMUST00000134191.1
ENSMUST00000032373.5 |
Ldhb
|
lactate dehydrogenase B |
| chr5_+_114146525 | 0.49 |
ENSMUST00000102582.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr15_-_76660108 | 0.48 |
ENSMUST00000066677.8
ENSMUST00000177359.1 |
Cyhr1
|
cysteine and histidine rich 1 |
| chr5_+_117319258 | 0.48 |
ENSMUST00000111967.1
|
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr2_-_28699636 | 0.48 |
ENSMUST00000102877.1
|
1700026L06Rik
|
RIKEN cDNA 1700026L06 gene |
| chr2_-_3422608 | 0.47 |
ENSMUST00000064685.7
|
Meig1
|
meiosis expressed gene 1 |
| chr2_-_168741752 | 0.47 |
ENSMUST00000029060.4
|
Atp9a
|
ATPase, class II, type 9A |
| chr4_+_115057410 | 0.47 |
ENSMUST00000136946.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr15_-_79834323 | 0.47 |
ENSMUST00000177316.2
ENSMUST00000175858.2 |
Nptxr
|
neuronal pentraxin receptor |
| chr4_-_43040279 | 0.46 |
ENSMUST00000107958.1
ENSMUST00000107959.1 ENSMUST00000152846.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr11_-_116238077 | 0.46 |
ENSMUST00000037007.3
|
Evpl
|
envoplakin |
| chr11_-_100207507 | 0.46 |
ENSMUST00000007272.7
|
Krt14
|
keratin 14 |
| chr3_+_28263205 | 0.46 |
ENSMUST00000159236.2
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr6_-_38637220 | 0.46 |
ENSMUST00000096030.3
|
Klrg2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr14_-_64949838 | 0.45 |
ENSMUST00000067843.3
ENSMUST00000176489.1 ENSMUST00000175905.1 ENSMUST00000022544.7 ENSMUST00000175744.1 ENSMUST00000176128.1 |
Hmbox1
|
homeobox containing 1 |
| chr6_-_72361396 | 0.45 |
ENSMUST00000130064.1
|
Rnf181
|
ring finger protein 181 |
| chr11_-_72550255 | 0.45 |
ENSMUST00000021154.6
|
Spns3
|
spinster homolog 3 |
| chr15_+_25758755 | 0.45 |
ENSMUST00000131834.1
ENSMUST00000124966.1 |
Myo10
|
myosin X |
| chr1_+_75549581 | 0.45 |
ENSMUST00000154101.1
|
Slc4a3
|
solute carrier family 4 (anion exchanger), member 3 |
| chr9_-_70657121 | 0.45 |
ENSMUST00000049031.5
|
Fam63b
|
family with sequence similarity 63, member B |
| chr13_-_9878998 | 0.44 |
ENSMUST00000063093.9
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr7_+_30169861 | 0.44 |
ENSMUST00000085668.4
|
Gm5113
|
predicted gene 5113 |
| chr7_-_104353328 | 0.44 |
ENSMUST00000130139.1
ENSMUST00000059037.8 |
Trim12c
|
tripartite motif-containing 12C |
| chr17_+_56717759 | 0.44 |
ENSMUST00000002452.6
|
Ndufa11
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 11 |
| chr9_+_120149733 | 0.44 |
ENSMUST00000068698.7
ENSMUST00000093773.1 ENSMUST00000111627.1 |
Mobp
|
myelin-associated oligodendrocytic basic protein |
| chr3_-_122619442 | 0.44 |
ENSMUST00000162947.1
|
Fnbp1l
|
formin binding protein 1-like |
| chr4_+_3940747 | 0.44 |
ENSMUST00000119403.1
|
Chchd7
|
coiled-coil-helix-coiled-coil-helix domain containing 7 |
| chr18_+_76059458 | 0.43 |
ENSMUST00000167921.1
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr5_-_139819906 | 0.43 |
ENSMUST00000147328.1
|
Tmem184a
|
transmembrane protein 184a |
| chr6_+_42245907 | 0.43 |
ENSMUST00000031897.5
|
Gstk1
|
glutathione S-transferase kappa 1 |
| chr14_+_55854115 | 0.43 |
ENSMUST00000168479.1
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr15_-_100584075 | 0.43 |
ENSMUST00000184908.1
|
POU6F1
|
POU domain, class 6, transcription factor 1 (Pou6f1), mRNA |
| chr7_+_80246529 | 0.43 |
ENSMUST00000107381.1
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr11_-_3931960 | 0.43 |
ENSMUST00000109990.1
ENSMUST00000020710.4 ENSMUST00000109989.3 ENSMUST00000109991.1 ENSMUST00000109993.2 |
Tcn2
|
transcobalamin 2 |
| chr1_-_162859684 | 0.43 |
ENSMUST00000131058.1
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr7_-_45238794 | 0.43 |
ENSMUST00000098461.1
ENSMUST00000107797.1 |
Cd37
|
CD37 antigen |
| chr11_-_103208542 | 0.42 |
ENSMUST00000021323.4
ENSMUST00000107026.2 |
1700023F06Rik
|
RIKEN cDNA 1700023F06 gene |
| chr2_-_35100677 | 0.42 |
ENSMUST00000045776.4
ENSMUST00000113050.3 |
AI182371
|
expressed sequence AI182371 |
| chr2_+_126034967 | 0.42 |
ENSMUST00000110442.1
|
Fgf7
|
fibroblast growth factor 7 |
| chr11_-_97115327 | 0.42 |
ENSMUST00000001484.2
|
Tbx21
|
T-box 21 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx2_Bcl11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 0.5 | 1.9 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.4 | 2.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.4 | 1.5 | GO:0035482 | gastric motility(GO:0035482) |
| 0.4 | 1.1 | GO:1990764 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.3 | 2.4 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 1.0 | GO:1904732 | elastin biosynthetic process(GO:0051542) regulation of electron carrier activity(GO:1904732) regulation of cytochrome-c oxidase activity(GO:1904959) |
| 0.3 | 1.0 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.3 | 2.3 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.3 | 2.0 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.3 | 0.6 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.3 | 0.8 | GO:0043382 | positive regulation of memory T cell differentiation(GO:0043382) |
| 0.3 | 0.8 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.3 | 1.4 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.3 | 0.5 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.3 | 1.1 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.3 | 0.8 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.3 | 2.3 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.3 | 2.0 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 1.0 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.2 | 1.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.2 | 2.3 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 0.7 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.2 | 1.1 | GO:0031438 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.2 | 0.6 | GO:1903465 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.2 | 0.6 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.2 | 1.0 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 | 1.2 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.2 | 1.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 1.1 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.2 | 0.7 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.2 | 1.7 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.2 | 0.7 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.3 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.2 | 0.5 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.2 | 1.0 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 0.5 | GO:0033373 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.2 | 0.5 | GO:2001293 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 0.6 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.2 | 1.0 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.2 | 1.2 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.2 | 0.5 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 1.5 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.1 | 1.5 | GO:0033689 | negative regulation of osteoblast proliferation(GO:0033689) |
| 0.1 | 0.4 | GO:0035938 | androgen catabolic process(GO:0006710) estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.1 | 0.7 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.8 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 0.4 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 1.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.4 | GO:0046271 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.2 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.1 | 0.4 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.1 | 0.7 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.3 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.1 | 0.6 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.1 | 0.5 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.5 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.3 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.1 | 0.3 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.3 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 0.4 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.1 | 0.3 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.1 | 0.4 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.3 | GO:1903660 | negative regulation of NAD(P)H oxidase activity(GO:0033861) pancreatic stellate cell proliferation(GO:0072343) complement-dependent cytotoxicity(GO:0097278) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.1 | 0.3 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.1 | 0.7 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.3 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.1 | 0.6 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.1 | 0.6 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.2 | GO:0070602 | regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.1 | 0.4 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.7 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.1 | 0.2 | GO:1903969 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) |
| 0.1 | 0.4 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.1 | 0.3 | GO:0072221 | distal convoluted tubule development(GO:0072025) metanephric distal convoluted tubule development(GO:0072221) metanephric distal tubule development(GO:0072235) |
| 0.1 | 1.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.3 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.1 | 0.3 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.1 | 1.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 1.3 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.1 | 0.5 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.5 | GO:0006363 | termination of RNA polymerase I transcription(GO:0006363) |
| 0.1 | 1.8 | GO:1905144 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.2 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.5 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.2 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.1 | 0.5 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.1 | 1.1 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 0.4 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.1 | 0.2 | GO:0045872 | positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.1 | 2.1 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.1 | 0.2 | GO:0071895 | negative regulation of interleukin-13 production(GO:0032696) odontoblast differentiation(GO:0071895) |
| 0.1 | 0.3 | GO:0046722 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.4 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.3 | GO:0051611 | regulation of serotonin uptake(GO:0051611) |
| 0.1 | 0.2 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.1 | 0.7 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.3 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.1 | 0.7 | GO:1901621 | negative regulation of smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:1901621) |
| 0.1 | 0.2 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.1 | GO:0032752 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0042253) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045423) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.1 | 0.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.1 | 0.1 | GO:0015866 | ADP transport(GO:0015866) |
| 0.1 | 0.7 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 0.6 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 0.3 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 0.4 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.1 | 1.0 | GO:0017144 | drug metabolic process(GO:0017144) |
| 0.1 | 0.2 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.1 | GO:0072054 | renal outer medulla development(GO:0072054) |
| 0.1 | 1.1 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.1 | 0.2 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 1.0 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.1 | 0.5 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.1 | 0.4 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.2 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 0.3 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.2 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.1 | 0.3 | GO:0006680 | glucosylceramide catabolic process(GO:0006680) |
| 0.1 | 0.2 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.1 | 0.1 | GO:0070858 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.1 | 0.8 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.6 | GO:0060837 | blood vessel endothelial cell differentiation(GO:0060837) |
| 0.0 | 0.2 | GO:0036506 | maintenance of unfolded protein(GO:0036506) tail-anchored membrane protein insertion into ER membrane(GO:0071816) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.2 | GO:0045896 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.6 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:2001280 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.5 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.3 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.2 | GO:0061188 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.0 | 0.1 | GO:2000170 | positive regulation of peptidyl-cysteine S-nitrosylation(GO:2000170) |
| 0.0 | 0.3 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.7 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.6 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.3 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.0 | 0.8 | GO:0030149 | sphingolipid catabolic process(GO:0030149) |
| 0.0 | 0.3 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.2 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.1 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.0 | 0.2 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 1.1 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.6 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.0 | 0.4 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.1 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.2 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.6 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.1 | GO:0061428 | negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.0 | 0.3 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.6 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.3 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.2 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.2 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.6 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.2 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.4 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.7 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 0.1 | GO:0019323 | pentose catabolic process(GO:0019323) |
| 0.0 | 0.2 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0048298 | positive regulation of germinal center formation(GO:0002636) positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.3 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.1 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) fatty-acyl-CoA catabolic process(GO:0036115) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.4 | GO:0034242 | negative regulation of syncytium formation by plasma membrane fusion(GO:0034242) |
| 0.0 | 0.4 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:1900245 | negative regulation of MDA-5 signaling pathway(GO:0039534) positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.3 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.3 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.0 | 0.1 | GO:0045415 | negative regulation of interleukin-8 biosynthetic process(GO:0045415) |
| 0.0 | 0.3 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.3 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0071503 | corticospinal neuron axon guidance through spinal cord(GO:0021972) positive regulation of negative chemotaxis(GO:0050924) induction of negative chemotaxis(GO:0050929) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) negative regulation of mononuclear cell migration(GO:0071676) negative regulation of retinal ganglion cell axon guidance(GO:0090260) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.4 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.2 | GO:0043313 | regulation of neutrophil degranulation(GO:0043313) |
| 0.0 | 0.5 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.4 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:2000318 | positive regulation of T-helper 17 type immune response(GO:2000318) |
| 0.0 | 0.8 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 2.4 | GO:0007608 | sensory perception of smell(GO:0007608) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 1.0 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 1.0 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.4 | GO:0046130 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 1.2 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.3 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0060213 | regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.2 | GO:0036371 | T-tubule organization(GO:0033292) protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.3 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.0 | 0.2 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.3 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.1 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.0 | 0.2 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:1990440 | positive regulation of transcription from RNA polymerase II promoter in response to endoplasmic reticulum stress(GO:1990440) |
| 0.0 | 0.2 | GO:1902170 | cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.1 | GO:0042148 | strand invasion(GO:0042148) |
| 0.0 | 0.3 | GO:0071545 | inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.2 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.2 | GO:0048484 | enteric nervous system development(GO:0048484) |
| 0.0 | 0.1 | GO:1900194 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:1903546 | protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.2 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 0.9 | GO:0030433 | ER-associated ubiquitin-dependent protein catabolic process(GO:0030433) |
| 0.0 | 0.1 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.1 | GO:2000360 | negative regulation of binding of sperm to zona pellucida(GO:2000360) |
| 0.0 | 0.3 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.0 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.5 | GO:0001569 | patterning of blood vessels(GO:0001569) |
| 0.0 | 0.2 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.0 | 0.5 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.0 | GO:1904016 | response to Thyroglobulin triiodothyronine(GO:1904016) cellular response to Thyroglobulin triiodothyronine(GO:1904017) |
| 0.0 | 0.4 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.5 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.1 | GO:0014004 | microglia differentiation(GO:0014004) microglia development(GO:0014005) |
| 0.0 | 0.1 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.0 | 0.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0003418 | growth plate cartilage chondrocyte differentiation(GO:0003418) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.2 | 2.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 1.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.2 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 0.5 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.2 | 2.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.1 | 0.7 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.1 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.9 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.4 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.4 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 1.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.3 | GO:0097637 | platelet dense granule membrane(GO:0031088) intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 0.7 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 0.5 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.6 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 2.3 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.7 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 4.2 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.2 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.1 | 3.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.4 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.1 | 0.7 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.7 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.3 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.1 | 0.7 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.6 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.7 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 0.3 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.1 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.1 | 0.3 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 2.4 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 1.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.4 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.5 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.2 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.2 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.4 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.4 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.4 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 1.0 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.7 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.4 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.1 | GO:0070522 | ERCC4-ERCC1 complex(GO:0070522) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.4 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.6 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0044753 | amphisome(GO:0044753) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.3 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 1.0 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.3 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.5 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.9 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.0 | 0.4 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 1.4 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.3 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0030119 | AP-type membrane coat adaptor complex(GO:0030119) |
| 0.0 | 0.6 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.5 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.1 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 1.1 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.6 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.2 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.1 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.4 | 1.2 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.4 | 1.5 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.3 | 2.4 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.3 | 2.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.3 | 2.9 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.3 | 1.0 | GO:0043682 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 1.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.2 | 0.7 | GO:0005152 | interleukin-1, Type II receptor binding(GO:0005151) interleukin-1 receptor antagonist activity(GO:0005152) interleukin-1 Type I receptor antagonist activity(GO:0045352) interleukin-1 Type II receptor antagonist activity(GO:0045353) |
| 0.2 | 0.6 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 0.8 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.2 | 0.6 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.2 | 1.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 1.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 1.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.2 | 0.8 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.2 | 1.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.6 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 1.0 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 2.3 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 1.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 1.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.5 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.1 | 0.4 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.4 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 2.9 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.6 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.8 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.8 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 0.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.6 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.3 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.1 | 1.6 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 0.8 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.1 | 0.9 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 1.1 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.1 | 0.5 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.1 | 1.7 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.1 | 0.3 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.4 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.4 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.3 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.5 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.6 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.7 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.3 | GO:0004921 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 0.3 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.1 | 1.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 0.3 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 1.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.3 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.1 | 0.7 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 0.6 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.2 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.5 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.6 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.1 | 1.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.7 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 2.4 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.6 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.3 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 0.3 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.8 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.1 | 0.3 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.3 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) IkappaB kinase activity(GO:0008384) |
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.3 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.1 | 0.8 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.1 | 0.2 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.1 | 0.9 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.3 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.5 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.2 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.5 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.4 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0031711 | bradykinin receptor binding(GO:0031711) |
| 0.0 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.3 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 2.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.6 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.2 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.4 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.0 | 0.1 | GO:0008988 | rRNA (adenine-N6-)-methyltransferase activity(GO:0008988) U6 snRNA 3'-end binding(GO:0030629) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.4 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.1 | GO:0005008 | hepatocyte growth factor-activated receptor activity(GO:0005008) |
| 0.0 | 0.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.4 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.4 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.9 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.6 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.4 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0046403 | polynucleotide 3'-phosphatase activity(GO:0046403) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.3 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.7 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0031531 | thyrotropin-releasing hormone receptor binding(GO:0031531) |
| 0.0 | 0.1 | GO:1902379 | chemoattractant activity involved in axon guidance(GO:1902379) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.9 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.7 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.2 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 1.1 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.2 | GO:0016401 | palmitoyl-CoA oxidase activity(GO:0016401) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.8 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0070001 | aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 2.2 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.0 | 0.5 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.3 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 1.5 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.7 | GO:0030546 | receptor activator activity(GO:0030546) |
| 0.0 | 0.2 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 1.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.3 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0019767 | IgE receptor activity(GO:0019767) |
| 0.0 | 0.4 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.1 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.7 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 0.6 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004373 | glycogen (starch) synthase activity(GO:0004373) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.2 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.7 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.4 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.2 | GO:0022842 | leak channel activity(GO:0022840) narrow pore channel activity(GO:0022842) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.3 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.5 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.6 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.1 | GO:0016755 | transferase activity, transferring amino-acyl groups(GO:0016755) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.5 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.9 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 0.5 | GO:0008028 | monocarboxylic acid transmembrane transporter activity(GO:0008028) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 1.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.4 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.2 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.9 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.8 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 2.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.5 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.7 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.8 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 1.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 1.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.9 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 1.1 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 1.2 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 1.1 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.3 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.2 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 0.7 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.2 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | PID ERBB2 ERBB3 PATHWAY | ErbB2/ErbB3 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.5 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 0.8 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 2.0 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 2.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 0.9 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.1 | 0.8 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.1 | 0.4 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.1 | 1.0 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.5 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.8 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.8 | REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | Genes involved in Nucleotide-binding domain, leucine rich repeat containing receptor (NLR) signaling pathways |
| 0.0 | 0.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 1.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.7 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 1.0 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.6 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 1.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.9 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.1 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.3 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.3 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.1 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.8 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.3 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.8 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.5 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.7 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.6 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.4 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 1.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 0.2 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.4 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 1.9 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |