Project
12D miR HR13_24
Navigation
Downloads
Results for Sox5_Sry
Z-value: 1.72
Transcription factors associated with Sox5_Sry
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
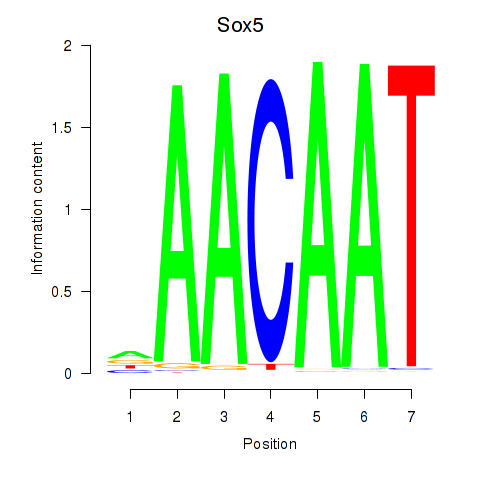
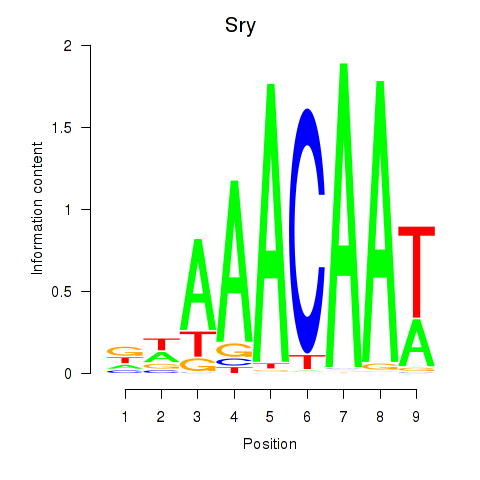
Sox5
|
ENSMUSG00000041540.10 | SRY (sex determining region Y)-box 5 |
|
Sry
|
ENSMUSG00000069036.3 | sex determining region of Chr Y |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox5 | mm10_v2_chr6_-_143947092_143947111 | 0.76 | 6.7e-03 | Click! |
Activity profile of Sox5_Sry motif
Sorted Z-values of Sox5_Sry motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_129121889 | 6.71 |
ENSMUST00000139450.1
ENSMUST00000125931.1 ENSMUST00000116444.2 |
Hpca
|
hippocalcin |
| chr10_+_88091070 | 4.42 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr4_+_122995944 | 3.89 |
ENSMUST00000106252.2
|
Mycl
|
v-myc myelocytomatosis viral oncogene homolog, lung carcinoma derived (avian) |
| chr4_+_122996035 | 3.73 |
ENSMUST00000030407.7
|
Mycl
|
v-myc myelocytomatosis viral oncogene homolog, lung carcinoma derived (avian) |
| chr10_-_45470201 | 3.61 |
ENSMUST00000079390.6
|
Lin28b
|
lin-28 homolog B (C. elegans) |
| chrX_-_104201126 | 3.49 |
ENSMUST00000056502.6
ENSMUST00000118314.1 |
C77370
|
expressed sequence C77370 |
| chr5_+_30711564 | 3.18 |
ENSMUST00000114729.1
|
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr5_+_30711849 | 3.18 |
ENSMUST00000088081.4
ENSMUST00000101442.3 |
Dpysl5
|
dihydropyrimidinase-like 5 |
| chr9_+_47530173 | 2.96 |
ENSMUST00000114548.1
ENSMUST00000152459.1 ENSMUST00000143026.1 ENSMUST00000085909.2 ENSMUST00000114547.1 ENSMUST00000034581.3 |
Cadm1
|
cell adhesion molecule 1 |
| chr9_+_26733728 | 2.87 |
ENSMUST00000160899.1
ENSMUST00000161431.1 ENSMUST00000159799.1 |
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr12_+_40446050 | 2.57 |
ENSMUST00000037488.6
|
Dock4
|
dedicator of cytokinesis 4 |
| chr9_-_35116804 | 2.52 |
ENSMUST00000034537.6
|
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr9_+_26733845 | 2.41 |
ENSMUST00000115269.2
|
B3gat1
|
beta-1,3-glucuronyltransferase 1 (glucuronosyltransferase P) |
| chr11_+_94044111 | 2.29 |
ENSMUST00000132079.1
|
Spag9
|
sperm associated antigen 9 |
| chr1_+_53061637 | 1.99 |
ENSMUST00000027269.5
|
Mstn
|
myostatin |
| chr18_-_43687695 | 1.95 |
ENSMUST00000082254.6
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr11_+_44617310 | 1.90 |
ENSMUST00000081265.5
ENSMUST00000101326.3 ENSMUST00000109268.1 |
Ebf1
|
early B cell factor 1 |
| chr3_+_68584154 | 1.88 |
ENSMUST00000182997.1
|
Schip1
|
schwannomin interacting protein 1 |
| chr11_+_94044241 | 1.87 |
ENSMUST00000103168.3
|
Spag9
|
sperm associated antigen 9 |
| chr11_+_94044194 | 1.86 |
ENSMUST00000092777.4
ENSMUST00000075695.6 |
Spag9
|
sperm associated antigen 9 |
| chr6_-_87335758 | 1.80 |
ENSMUST00000042025.9
|
Antxr1
|
anthrax toxin receptor 1 |
| chr17_-_70849644 | 1.77 |
ENSMUST00000134654.1
ENSMUST00000172229.1 ENSMUST00000127719.1 |
Tgif1
|
TGFB-induced factor homeobox 1 |
| chr19_+_53310495 | 1.76 |
ENSMUST00000003870.7
|
Mxi1
|
Max interacting protein 1 |
| chr16_-_57754707 | 1.69 |
ENSMUST00000089332.4
|
Col8a1
|
collagen, type VIII, alpha 1 |
| chr8_-_84773381 | 1.69 |
ENSMUST00000109764.1
|
Nfix
|
nuclear factor I/X |
| chr18_-_43059418 | 1.59 |
ENSMUST00000025377.7
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform |
| chrX_-_104201099 | 1.54 |
ENSMUST00000087879.4
|
C77370
|
expressed sequence C77370 |
| chr2_+_25180737 | 1.50 |
ENSMUST00000104999.2
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr1_+_34005872 | 1.48 |
ENSMUST00000182296.1
|
Dst
|
dystonin |
| chr18_+_69593361 | 1.44 |
ENSMUST00000114978.2
ENSMUST00000114977.1 |
Tcf4
|
transcription factor 4 |
| chr1_+_55406163 | 1.42 |
ENSMUST00000042986.8
|
Plcl1
|
phospholipase C-like 1 |
| chr11_+_94044331 | 1.38 |
ENSMUST00000024979.8
|
Spag9
|
sperm associated antigen 9 |
| chr7_+_82174796 | 1.36 |
ENSMUST00000032874.7
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr7_+_16309577 | 1.34 |
ENSMUST00000002152.6
|
Bbc3
|
BCL2 binding component 3 |
| chrX_+_93654863 | 1.31 |
ENSMUST00000113933.2
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr10_+_69706326 | 1.28 |
ENSMUST00000182992.1
|
Ank3
|
ankyrin 3, epithelial |
| chr1_+_158362261 | 1.27 |
ENSMUST00000046110.9
|
Astn1
|
astrotactin 1 |
| chr1_+_6730135 | 1.25 |
ENSMUST00000155921.1
|
St18
|
suppression of tumorigenicity 18 |
| chr6_+_15185456 | 1.23 |
ENSMUST00000115472.1
ENSMUST00000115474.1 ENSMUST00000031545.7 ENSMUST00000137628.1 |
Foxp2
|
forkhead box P2 |
| chr4_+_17853451 | 1.22 |
ENSMUST00000029881.3
|
Mmp16
|
matrix metallopeptidase 16 |
| chr6_+_122513583 | 1.20 |
ENSMUST00000032210.7
ENSMUST00000148517.1 |
Mfap5
|
microfibrillar associated protein 5 |
| chr3_-_27896360 | 1.20 |
ENSMUST00000058077.3
|
Tmem212
|
transmembrane protein 212 |
| chr16_+_57549232 | 1.16 |
ENSMUST00000159414.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr5_-_138171248 | 1.14 |
ENSMUST00000153867.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr18_+_11633276 | 1.13 |
ENSMUST00000115861.2
|
Rbbp8
|
retinoblastoma binding protein 8 |
| chr12_+_29528382 | 1.11 |
ENSMUST00000049784.9
|
Myt1l
|
myelin transcription factor 1-like |
| chr5_-_90640464 | 1.11 |
ENSMUST00000031317.6
|
Rassf6
|
Ras association (RalGDS/AF-6) domain family member 6 |
| chr2_+_102658640 | 1.10 |
ENSMUST00000080210.3
|
Slc1a2
|
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr5_-_138170992 | 1.09 |
ENSMUST00000139983.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr10_+_18407658 | 1.09 |
ENSMUST00000037341.7
|
Nhsl1
|
NHS-like 1 |
| chr6_+_34709610 | 1.08 |
ENSMUST00000031775.6
|
Cald1
|
caldesmon 1 |
| chr16_-_78576649 | 1.06 |
ENSMUST00000114220.1
ENSMUST00000114219.1 ENSMUST00000114218.1 |
D16Ertd472e
|
DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr17_-_35838208 | 1.05 |
ENSMUST00000134978.2
|
Tubb5
|
tubulin, beta 5 class I |
| chr7_+_16310412 | 1.04 |
ENSMUST00000136781.1
|
Bbc3
|
BCL2 binding component 3 |
| chr16_-_4559720 | 1.03 |
ENSMUST00000005862.7
|
Tfap4
|
transcription factor AP4 |
| chr6_+_34709442 | 1.02 |
ENSMUST00000115021.1
|
Cald1
|
caldesmon 1 |
| chr3_-_92083132 | 1.00 |
ENSMUST00000058150.6
|
Lor
|
loricrin |
| chr7_-_101921186 | 1.00 |
ENSMUST00000106965.1
ENSMUST00000106968.1 ENSMUST00000106967.1 |
Lrrc51
|
leucine rich repeat containing 51 |
| chrX_+_49470450 | 0.98 |
ENSMUST00000114904.3
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chrX_+_49470555 | 0.97 |
ENSMUST00000042444.6
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr6_-_12749193 | 0.95 |
ENSMUST00000046121.6
ENSMUST00000172356.1 |
Thsd7a
|
thrombospondin, type I, domain containing 7A |
| chr1_+_51289106 | 0.95 |
ENSMUST00000051572.6
|
Sdpr
|
serum deprivation response |
| chr4_+_108460000 | 0.94 |
ENSMUST00000097925.2
|
Zcchc11
|
zinc finger, CCHC domain containing 11 |
| chr2_+_3713478 | 0.94 |
ENSMUST00000115053.2
|
Fam107b
|
family with sequence similarity 107, member B |
| chr4_+_11579647 | 0.94 |
ENSMUST00000180239.1
|
Fsbp
|
fibrinogen silencer binding protein |
| chr2_-_152398046 | 0.92 |
ENSMUST00000063332.8
ENSMUST00000182625.1 |
Sox12
|
SRY-box containing gene 12 |
| chr12_-_34528844 | 0.91 |
ENSMUST00000110819.2
|
Hdac9
|
histone deacetylase 9 |
| chr2_+_3713449 | 0.91 |
ENSMUST00000027965.4
|
Fam107b
|
family with sequence similarity 107, member B |
| chr2_-_26208281 | 0.90 |
ENSMUST00000054099.9
|
Lhx3
|
LIM homeobox protein 3 |
| chr1_+_66386968 | 0.90 |
ENSMUST00000145419.1
|
Map2
|
microtubule-associated protein 2 |
| chr8_-_84937347 | 0.89 |
ENSMUST00000109741.2
ENSMUST00000119820.1 |
Mast1
|
microtubule associated serine/threonine kinase 1 |
| chr3_+_101377074 | 0.89 |
ENSMUST00000043983.5
|
Igsf3
|
immunoglobulin superfamily, member 3 |
| chrX_-_23285532 | 0.89 |
ENSMUST00000115319.2
|
Klhl13
|
kelch-like 13 |
| chr7_+_92819892 | 0.89 |
ENSMUST00000107180.1
ENSMUST00000107179.1 |
Rab30
|
RAB30, member RAS oncogene family |
| chr5_-_138171216 | 0.89 |
ENSMUST00000147920.1
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr1_-_12991109 | 0.88 |
ENSMUST00000115403.2
ENSMUST00000115402.1 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr16_-_95459245 | 0.88 |
ENSMUST00000176345.1
ENSMUST00000121809.2 ENSMUST00000118113.1 ENSMUST00000122199.1 |
Erg
|
avian erythroblastosis virus E-26 (v-ets) oncogene related |
| chr9_-_48835932 | 0.86 |
ENSMUST00000093852.3
|
Zbtb16
|
zinc finger and BTB domain containing 16 |
| chr2_+_92185438 | 0.86 |
ENSMUST00000128781.2
|
Phf21a
|
PHD finger protein 21A |
| chr3_+_129532386 | 0.85 |
ENSMUST00000071402.2
|
Elovl6
|
ELOVL family member 6, elongation of long chain fatty acids (yeast) |
| chr1_+_187997835 | 0.83 |
ENSMUST00000110938.1
|
Esrrg
|
estrogen-related receptor gamma |
| chr1_+_187997821 | 0.83 |
ENSMUST00000027906.6
|
Esrrg
|
estrogen-related receptor gamma |
| chr1_+_158362330 | 0.82 |
ENSMUST00000170718.1
|
Astn1
|
astrotactin 1 |
| chr17_-_35838259 | 0.81 |
ENSMUST00000001566.8
|
Tubb5
|
tubulin, beta 5 class I |
| chr9_+_27790947 | 0.80 |
ENSMUST00000115243.2
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr1_-_64121389 | 0.79 |
ENSMUST00000055001.3
|
Klf7
|
Kruppel-like factor 7 (ubiquitous) |
| chr3_-_33082004 | 0.79 |
ENSMUST00000108225.3
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chr6_+_134035691 | 0.77 |
ENSMUST00000081028.6
ENSMUST00000111963.1 |
Etv6
|
ets variant gene 6 (TEL oncogene) |
| chr2_+_124610573 | 0.76 |
ENSMUST00000103239.3
ENSMUST00000103240.2 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr14_+_116925379 | 0.76 |
ENSMUST00000088483.3
|
Gpc6
|
glypican 6 |
| chr19_+_53329413 | 0.76 |
ENSMUST00000025998.7
|
Mxi1
|
Max interacting protein 1 |
| chr10_-_68278713 | 0.75 |
ENSMUST00000020106.7
|
Arid5b
|
AT rich interactive domain 5B (MRF1-like) |
| chr6_-_136781718 | 0.75 |
ENSMUST00000078095.6
ENSMUST00000032338.7 |
Gucy2c
|
guanylate cyclase 2c |
| chr4_-_91376433 | 0.75 |
ENSMUST00000107109.2
ENSMUST00000107111.2 ENSMUST00000107120.1 |
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| chr6_-_52165413 | 0.75 |
ENSMUST00000014848.8
|
Hoxa2
|
homeobox A2 |
| chr4_-_91376490 | 0.74 |
ENSMUST00000107124.3
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) |
| chr7_-_101921175 | 0.73 |
ENSMUST00000098236.2
|
Lrrc51
|
leucine rich repeat containing 51 |
| chr15_+_83779999 | 0.73 |
ENSMUST00000046168.5
|
Mpped1
|
metallophosphoesterase domain containing 1 |
| chr9_-_32541589 | 0.72 |
ENSMUST00000016231.7
|
Fli1
|
Friend leukemia integration 1 |
| chr4_+_13743424 | 0.72 |
ENSMUST00000006761.3
|
Runx1t1
|
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr1_-_133921393 | 0.71 |
ENSMUST00000048432.5
|
Prelp
|
proline arginine-rich end leucine-rich repeat |
| chr6_+_15196949 | 0.71 |
ENSMUST00000151301.1
ENSMUST00000131414.1 ENSMUST00000140557.1 ENSMUST00000115469.1 |
Foxp2
|
forkhead box P2 |
| chr9_+_22099271 | 0.71 |
ENSMUST00000001384.4
|
Cnn1
|
calponin 1 |
| chr2_+_4882204 | 0.70 |
ENSMUST00000115019.1
|
Sephs1
|
selenophosphate synthetase 1 |
| chr4_-_116405986 | 0.69 |
ENSMUST00000123072.1
ENSMUST00000144281.1 |
Mast2
|
microtubule associated serine/threonine kinase 2 |
| chr1_+_87404916 | 0.69 |
ENSMUST00000173152.1
ENSMUST00000173663.1 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr1_-_119837613 | 0.65 |
ENSMUST00000064091.5
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr4_-_83486178 | 0.65 |
ENSMUST00000130626.1
|
Psip1
|
PC4 and SFRS1 interacting protein 1 |
| chr11_-_102026924 | 0.64 |
ENSMUST00000107167.1
ENSMUST00000062801.4 |
Mpp3
|
membrane protein, palmitoylated 3 (MAGUK p55 subfamily member 3) |
| chr2_+_181763315 | 0.63 |
ENSMUST00000081125.4
|
Myt1
|
myelin transcription factor 1 |
| chr14_+_45219993 | 0.62 |
ENSMUST00000146150.1
|
Gpr137c
|
G protein-coupled receptor 137C |
| chr16_-_16560046 | 0.62 |
ENSMUST00000172181.2
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr6_+_122513676 | 0.62 |
ENSMUST00000142896.1
ENSMUST00000121656.1 |
Mfap5
|
microfibrillar associated protein 5 |
| chr19_-_41848076 | 0.62 |
ENSMUST00000059231.2
|
Frat2
|
frequently rearranged in advanced T cell lymphomas 2 |
| chr2_-_65567505 | 0.60 |
ENSMUST00000100069.2
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr3_+_37639945 | 0.60 |
ENSMUST00000108109.1
ENSMUST00000038569.1 |
Spry1
|
sprouty homolog 1 (Drosophila) |
| chr3_+_37639985 | 0.59 |
ENSMUST00000108107.1
|
Spry1
|
sprouty homolog 1 (Drosophila) |
| chr18_+_69344503 | 0.59 |
ENSMUST00000114985.3
|
Tcf4
|
transcription factor 4 |
| chr12_-_71136611 | 0.59 |
ENSMUST00000021486.8
ENSMUST00000166120.1 |
Timm9
|
translocase of inner mitochondrial membrane 9 |
| chr1_+_6730051 | 0.58 |
ENSMUST00000043578.6
ENSMUST00000131467.1 ENSMUST00000150761.1 ENSMUST00000151281.1 |
St18
|
suppression of tumorigenicity 18 |
| chr4_+_109343029 | 0.58 |
ENSMUST00000030281.5
|
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr10_-_128704978 | 0.57 |
ENSMUST00000026416.7
ENSMUST00000026415.7 |
Cdk2
|
cyclin-dependent kinase 2 |
| chr5_+_31251678 | 0.56 |
ENSMUST00000054829.7
ENSMUST00000114570.1 ENSMUST00000075611.7 |
Krtcap3
|
keratinocyte associated protein 3 |
| chr12_-_101819048 | 0.56 |
ENSMUST00000021603.8
|
Fbln5
|
fibulin 5 |
| chr7_+_24907618 | 0.56 |
ENSMUST00000151121.1
|
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr9_-_72111172 | 0.55 |
ENSMUST00000183992.1
|
Tcf12
|
transcription factor 12 |
| chr19_-_46039621 | 0.55 |
ENSMUST00000056931.7
|
Ldb1
|
LIM domain binding 1 |
| chrX_+_71555918 | 0.55 |
ENSMUST00000072699.6
ENSMUST00000114582.2 ENSMUST00000015361.4 ENSMUST00000088874.3 |
Hmgb3
|
high mobility group box 3 |
| chr5_+_65348386 | 0.53 |
ENSMUST00000031096.7
|
Klb
|
klotho beta |
| chr2_-_62573813 | 0.53 |
ENSMUST00000174234.1
ENSMUST00000000402.9 ENSMUST00000174448.1 |
Fap
|
fibroblast activation protein |
| chr13_+_44840686 | 0.53 |
ENSMUST00000173906.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr1_-_158356258 | 0.53 |
ENSMUST00000004133.8
|
Brinp2
|
bone morphogenic protein/retinoic acid inducible neural-specific 2 |
| chr2_-_65567465 | 0.53 |
ENSMUST00000066432.5
|
Scn3a
|
sodium channel, voltage-gated, type III, alpha |
| chr1_+_135232045 | 0.52 |
ENSMUST00000110798.3
|
Gm4204
|
predicted gene 4204 |
| chr14_-_89898466 | 0.51 |
ENSMUST00000081204.4
|
Gm10110
|
predicted gene 10110 |
| chr7_+_126760591 | 0.51 |
ENSMUST00000091328.2
|
Mapk3
|
mitogen-activated protein kinase 3 |
| chr11_-_3863895 | 0.50 |
ENSMUST00000070552.7
|
Osbp2
|
oxysterol binding protein 2 |
| chr13_-_115101909 | 0.49 |
ENSMUST00000061673.7
|
Itga1
|
integrin alpha 1 |
| chr15_-_77153772 | 0.48 |
ENSMUST00000166610.1
ENSMUST00000111581.2 |
Rbfox2
|
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr12_+_51348265 | 0.47 |
ENSMUST00000119211.1
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr1_-_97761538 | 0.46 |
ENSMUST00000171129.1
|
Ppip5k2
|
diphosphoinositol pentakisphosphate kinase 2 |
| chr4_-_83486453 | 0.46 |
ENSMUST00000107214.2
ENSMUST00000107215.2 ENSMUST00000030207.8 |
Psip1
|
PC4 and SFRS1 interacting protein 1 |
| chr10_-_18234930 | 0.46 |
ENSMUST00000052648.8
ENSMUST00000080860.6 ENSMUST00000173243.1 |
Ccdc28a
|
coiled-coil domain containing 28A |
| chr13_+_99100698 | 0.45 |
ENSMUST00000181742.1
|
Gm807
|
predicted gene 807 |
| chr4_+_118620799 | 0.45 |
ENSMUST00000030501.8
|
Ebna1bp2
|
EBNA1 binding protein 2 |
| chr9_+_70207342 | 0.45 |
ENSMUST00000034745.7
|
Myo1e
|
myosin IE |
| chr4_+_123282778 | 0.45 |
ENSMUST00000106243.1
ENSMUST00000106241.1 ENSMUST00000080178.6 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chrM_+_7759 | 0.45 |
ENSMUST00000082407.1
ENSMUST00000082408.1 |
mt-Atp8
mt-Atp6
|
mitochondrially encoded ATP synthase 8 mitochondrially encoded ATP synthase 6 |
| chr17_+_12584183 | 0.44 |
ENSMUST00000046959.7
|
Slc22a2
|
solute carrier family 22 (organic cation transporter), member 2 |
| chrX_+_163911401 | 0.44 |
ENSMUST00000140845.1
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr13_+_44731281 | 0.44 |
ENSMUST00000174086.1
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr2_+_65620829 | 0.44 |
ENSMUST00000028377.7
|
Scn2a1
|
sodium channel, voltage-gated, type II, alpha 1 |
| chr2_+_153031852 | 0.44 |
ENSMUST00000037235.6
|
Xkr7
|
X Kell blood group precursor related family member 7 homolog |
| chr8_-_56550791 | 0.44 |
ENSMUST00000134162.1
ENSMUST00000140107.1 ENSMUST00000040330.8 ENSMUST00000135337.1 |
Cep44
|
centrosomal protein 44 |
| chr12_-_72236692 | 0.44 |
ENSMUST00000021497.9
ENSMUST00000137990.1 |
Rtn1
|
reticulon 1 |
| chr16_-_17125106 | 0.44 |
ENSMUST00000093336.6
|
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene |
| chr15_+_99702278 | 0.43 |
ENSMUST00000023759.4
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr2_+_3114220 | 0.43 |
ENSMUST00000072955.5
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr2_-_62573905 | 0.43 |
ENSMUST00000102732.3
|
Fap
|
fibroblast activation protein |
| chr13_-_40733768 | 0.41 |
ENSMUST00000110193.2
|
Tfap2a
|
transcription factor AP-2, alpha |
| chr15_-_76243401 | 0.41 |
ENSMUST00000165738.1
ENSMUST00000075689.6 |
Parp10
|
poly (ADP-ribose) polymerase family, member 10 |
| chr12_+_100779055 | 0.41 |
ENSMUST00000069782.4
|
9030617O03Rik
|
RIKEN cDNA 9030617O03 gene |
| chr5_+_42067960 | 0.40 |
ENSMUST00000087332.4
|
Gm16223
|
predicted gene 16223 |
| chr2_+_31670714 | 0.40 |
ENSMUST00000038474.7
ENSMUST00000137156.1 |
Exosc2
|
exosome component 2 |
| chr1_-_165934900 | 0.39 |
ENSMUST00000069609.5
ENSMUST00000111427.2 ENSMUST00000111426.4 |
Pou2f1
|
POU domain, class 2, transcription factor 1 |
| chr7_+_82175156 | 0.39 |
ENSMUST00000180243.1
|
Sh3gl3
|
SH3-domain GRB2-like 3 |
| chr12_+_100779074 | 0.39 |
ENSMUST00000110073.1
ENSMUST00000110070.1 |
9030617O03Rik
|
RIKEN cDNA 9030617O03 gene |
| chr11_-_5950018 | 0.39 |
ENSMUST00000102920.3
|
Gck
|
glucokinase |
| chr4_-_103215147 | 0.39 |
ENSMUST00000150285.1
|
Slc35d1
|
solute carrier family 35 (UDP-glucuronic acid/UDP-N-acetylgalactosamine dual transporter), member D1 |
| chr7_+_119900099 | 0.38 |
ENSMUST00000106516.1
|
Lyrm1
|
LYR motif containing 1 |
| chr12_+_100779088 | 0.38 |
ENSMUST00000110069.1
|
9030617O03Rik
|
RIKEN cDNA 9030617O03 gene |
| chr9_-_29411736 | 0.37 |
ENSMUST00000115236.1
|
Ntm
|
neurotrimin |
| chr10_+_116301374 | 0.37 |
ENSMUST00000092167.5
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr9_+_122923050 | 0.37 |
ENSMUST00000051667.7
ENSMUST00000148851.1 |
Zfp105
|
zinc finger protein 105 |
| chr3_+_18054258 | 0.36 |
ENSMUST00000026120.6
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr3_+_26331150 | 0.36 |
ENSMUST00000099182.2
|
A830092H15Rik
|
RIKEN cDNA A830092H15 gene |
| chr6_-_3494587 | 0.36 |
ENSMUST00000049985.8
|
Hepacam2
|
HEPACAM family member 2 |
| chr7_-_127876777 | 0.36 |
ENSMUST00000106262.1
ENSMUST00000106263.1 ENSMUST00000054415.5 |
Zfp668
|
zinc finger protein 668 |
| chr18_-_75697639 | 0.36 |
ENSMUST00000165559.1
|
Ctif
|
CBP80/20-dependent translation initiation factor |
| chr4_+_86053887 | 0.36 |
ENSMUST00000107178.2
ENSMUST00000048885.5 ENSMUST00000141889.1 ENSMUST00000120678.1 |
Adamtsl1
|
ADAMTS-like 1 |
| chr6_+_122513643 | 0.36 |
ENSMUST00000118626.1
|
Mfap5
|
microfibrillar associated protein 5 |
| chr17_+_46646225 | 0.35 |
ENSMUST00000002844.7
ENSMUST00000113429.1 ENSMUST00000113430.1 |
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr4_-_135873546 | 0.35 |
ENSMUST00000142585.1
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr12_+_51348370 | 0.35 |
ENSMUST00000121521.1
|
G2e3
|
G2/M-phase specific E3 ubiquitin ligase |
| chr15_-_10713537 | 0.35 |
ENSMUST00000090339.3
|
Rai14
|
retinoic acid induced 14 |
| chr9_+_74976096 | 0.34 |
ENSMUST00000081746.5
|
Fam214a
|
family with sequence similarity 214, member A |
| chr4_-_82505749 | 0.33 |
ENSMUST00000107245.2
ENSMUST00000107246.1 |
Nfib
|
nuclear factor I/B |
| chr6_-_120364344 | 0.33 |
ENSMUST00000146667.1
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr6_-_113934679 | 0.33 |
ENSMUST00000101044.2
|
Atp2b2
|
ATPase, Ca++ transporting, plasma membrane 2 |
| chrX_-_12128386 | 0.32 |
ENSMUST00000145872.1
|
Bcor
|
BCL6 interacting corepressor |
| chr4_+_115059507 | 0.32 |
ENSMUST00000162489.1
|
Tal1
|
T cell acute lymphocytic leukemia 1 |
| chr5_+_65131184 | 0.32 |
ENSMUST00000031089.5
ENSMUST00000101191.3 |
Klhl5
|
kelch-like 5 |
| chr13_+_108860072 | 0.31 |
ENSMUST00000177907.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr6_+_42264983 | 0.31 |
ENSMUST00000031895.6
|
Casp2
|
caspase 2 |
| chr12_-_99393010 | 0.31 |
ENSMUST00000177451.1
|
Foxn3
|
forkhead box N3 |
| chr7_+_127876796 | 0.31 |
ENSMUST00000131000.1
|
Zfp646
|
zinc finger protein 646 |
| chr14_+_54936456 | 0.30 |
ENSMUST00000037814.6
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chr9_-_60838200 | 0.30 |
ENSMUST00000063858.7
|
Gm9869
|
predicted gene 9869 |
| chr15_+_101266839 | 0.30 |
ENSMUST00000023779.6
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr10_+_69534208 | 0.29 |
ENSMUST00000182439.1
ENSMUST00000092434.5 ENSMUST00000092432.5 ENSMUST00000092431.5 ENSMUST00000054167.8 ENSMUST00000047061.6 |
Ank3
|
ankyrin 3, epithelial |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox5_Sry
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.2 | 6.7 | GO:0030824 | negative regulation of cGMP metabolic process(GO:0030824) negative regulation of cGMP biosynthetic process(GO:0030827) negative regulation of guanylate cyclase activity(GO:0031283) |
| 1.0 | 3.0 | GO:0042271 | unidimensional cell growth(GO:0009826) susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.9 | 7.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.8 | 5.0 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.7 | 7.6 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.6 | 2.6 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.6 | 4.4 | GO:0042320 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) circadian sleep/wake cycle, REM sleep(GO:0042747) |
| 0.6 | 2.4 | GO:1900739 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.5 | 4.6 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.5 | 2.0 | GO:0014732 | skeletal muscle atrophy(GO:0014732) |
| 0.5 | 2.8 | GO:1903054 | negative regulation of extracellular matrix organization(GO:1903054) |
| 0.3 | 3.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.3 | 0.9 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.2 | 2.9 | GO:2001269 | positive regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001269) |
| 0.2 | 1.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.2 | 2.5 | GO:0010650 | positive regulation of cell communication by electrical coupling(GO:0010650) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.2 | 1.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) |
| 0.2 | 1.1 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.2 | 5.2 | GO:0050650 | chondroitin sulfate proteoglycan biosynthetic process(GO:0050650) |
| 0.2 | 1.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.2 | 1.4 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.2 | 1.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.2 | 2.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.2 | 0.6 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 0.7 | GO:0021658 | rhombomere 3 morphogenesis(GO:0021658) |
| 0.1 | 0.7 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.1 | 2.0 | GO:0021684 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.1 | 0.9 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.1 | 0.4 | GO:2000616 | negative regulation of histone H3-K9 acetylation(GO:2000616) |
| 0.1 | 1.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.1 | 0.5 | GO:0043973 | histone H3-K4 acetylation(GO:0043973) |
| 0.1 | 0.4 | GO:0021623 | oculomotor nerve morphogenesis(GO:0021622) oculomotor nerve formation(GO:0021623) |
| 0.1 | 0.4 | GO:1902915 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) |
| 0.1 | 1.8 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.5 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.6 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.5 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.1 | 1.1 | GO:0097475 | motor neuron migration(GO:0097475) |
| 0.1 | 1.8 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 0.6 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.1 | 0.8 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.1 | 0.5 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.3 | GO:0060217 | hemangioblast cell differentiation(GO:0060217) |
| 0.1 | 0.4 | GO:0071051 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.1 | 0.8 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.3 | GO:1903659 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 2.0 | GO:1900746 | regulation of vascular endothelial growth factor signaling pathway(GO:1900746) |
| 0.1 | 0.9 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 0.3 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.1 | 0.9 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.1 | 0.4 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.1 | 0.7 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.1 | 0.2 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.1 | 0.5 | GO:0000189 | MAPK import into nucleus(GO:0000189) |
| 0.1 | 0.3 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.1 | 0.8 | GO:0060613 | fat pad development(GO:0060613) |
| 0.1 | 0.2 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.1 | 0.2 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.1 | 0.2 | GO:0014870 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 0.6 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 1.0 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.1 | 1.9 | GO:0097435 | fibril organization(GO:0097435) |
| 0.1 | 0.4 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 3.3 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.1 | 0.3 | GO:0048691 | regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.1 | 1.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.1 | 1.5 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.1 | 0.8 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.1 | 0.3 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.1 | 0.1 | GO:0098928 | presynaptic signal transduction(GO:0098928) presynapse to nucleus signaling pathway(GO:0099526) |
| 0.1 | 0.2 | GO:0033128 | positive regulation of reciprocal meiotic recombination(GO:0010845) negative regulation of histone phosphorylation(GO:0033128) |
| 0.1 | 0.2 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.1 | 0.6 | GO:2000121 | regulation of removal of superoxide radicals(GO:2000121) |
| 0.1 | 0.4 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.1 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.1 | 0.2 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.1 | 0.6 | GO:0019065 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.1 | 0.5 | GO:0021527 | spinal cord association neuron differentiation(GO:0021527) |
| 0.1 | 0.9 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 2.7 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.1 | 0.2 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.1 | 0.7 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.1 | 0.1 | GO:0035441 | cell migration involved in vasculogenesis(GO:0035441) |
| 0.1 | 0.2 | GO:1901536 | negative regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045869) regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.0 | 0.2 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 1.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.0 | GO:0048665 | neuron fate specification(GO:0048665) |
| 0.0 | 0.5 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.6 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.0 | 1.6 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.3 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.0 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:0001928 | regulation of exocyst assembly(GO:0001928) |
| 0.0 | 0.1 | GO:1900039 | positive regulation of cellular response to hypoxia(GO:1900039) |
| 0.0 | 0.2 | GO:0032290 | peripheral nervous system myelin formation(GO:0032290) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.4 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.2 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.1 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.3 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.2 | GO:0046552 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) |
| 0.0 | 0.7 | GO:0001967 | suckling behavior(GO:0001967) |
| 0.0 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.7 | GO:0031571 | mitotic G1 DNA damage checkpoint(GO:0031571) |
| 0.0 | 0.1 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.7 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.4 | GO:0015697 | quaternary ammonium group transport(GO:0015697) |
| 0.0 | 0.4 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.0 | 2.0 | GO:0006940 | regulation of smooth muscle contraction(GO:0006940) |
| 0.0 | 0.1 | GO:0031161 | phosphatidylinositol catabolic process(GO:0031161) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.8 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.7 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.2 | GO:0038031 | non-canonical Wnt signaling pathway via JNK cascade(GO:0038031) |
| 0.0 | 0.1 | GO:0060368 | regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060368) |
| 0.0 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.9 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.2 | GO:1903142 | blood vessel maturation(GO:0001955) positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0050904 | diapedesis(GO:0050904) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.2 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.0 | GO:1905063 | regulation of vascular smooth muscle cell differentiation(GO:1905063) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.5 | GO:0042311 | vasodilation(GO:0042311) |
| 0.0 | 0.1 | GO:1901837 | negative regulation of transcription from RNA polymerase I promoter(GO:0016479) negative regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901837) |
| 0.0 | 1.6 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.2 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.9 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 2.9 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.2 | GO:0014823 | response to activity(GO:0014823) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.2 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 1.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0070933 | histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.4 | GO:0045604 | regulation of epidermal cell differentiation(GO:0045604) |
| 0.0 | 0.2 | GO:1902043 | positive regulation of extrinsic apoptotic signaling pathway via death domain receptors(GO:1902043) |
| 0.0 | 0.3 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 1.5 | GO:1990830 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.4 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 5.0 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.5 | 6.7 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.3 | 1.5 | GO:0031673 | H zone(GO:0031673) |
| 0.2 | 2.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.2 | 3.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.2 | 3.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.2 | 0.6 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.2 | 1.9 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 2.2 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.1 | 1.8 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.1 | 1.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.6 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.1 | 2.5 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.1 | 0.8 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 0.3 | GO:0033193 | Lsd1/2 complex(GO:0033193) |
| 0.1 | 0.9 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.1 | 1.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.1 | 1.0 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.1 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.4 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.1 | 1.1 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.1 | 0.7 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0044317 | rod spherule(GO:0044317) |
| 0.0 | 2.8 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 1.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.6 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 1.2 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.8 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.5 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.5 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.4 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.6 | GO:0008074 | guanylate cyclase complex, soluble(GO:0008074) |
| 0.0 | 7.9 | GO:0000139 | Golgi membrane(GO:0000139) |
| 0.0 | 2.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 3.3 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.2 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.6 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.6 | GO:0044798 | nuclear transcription factor complex(GO:0044798) |
| 0.0 | 0.8 | GO:1990391 | DNA repair complex(GO:1990391) |
| 0.0 | 0.9 | GO:0035097 | histone methyltransferase complex(GO:0035097) |
| 0.0 | 0.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.0 | 0.1 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 2.5 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.5 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 1.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.5 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 8.0 | GO:0005815 | microtubule organizing center(GO:0005815) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 5.3 | GO:0015018 | galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase activity(GO:0015018) |
| 0.6 | 2.5 | GO:0047288 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.5 | 2.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.5 | 2.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.5 | 7.2 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.3 | 1.0 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.3 | 1.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.3 | 1.3 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 1.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.2 | 0.2 | GO:0035851 | Krueppel-associated box domain binding(GO:0035851) |
| 0.2 | 4.4 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.1 | 0.4 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.1 | 0.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.9 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.1 | 1.9 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.1 | 0.5 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 2.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 1.4 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 1.2 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.1 | 0.7 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.1 | 0.5 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.1 | 1.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.1 | 3.1 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 1.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.1 | 1.6 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.8 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.1 | 1.2 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
| 0.0 | 0.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.0 | 0.3 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.2 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.0 | 0.1 | GO:0016211 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) |
| 0.0 | 0.7 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0015165 | pyrimidine nucleotide-sugar transmembrane transporter activity(GO:0015165) |
| 0.0 | 7.4 | GO:0016810 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds(GO:0016810) |
| 0.0 | 0.2 | GO:0008761 | UDP-N-acetylglucosamine 2-epimerase activity(GO:0008761) |
| 0.0 | 0.3 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.4 | GO:0004340 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.9 | GO:0031402 | sodium ion binding(GO:0031402) |
| 0.0 | 0.7 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.3 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 2.3 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.6 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.8 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.7 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 2.1 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 1.7 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.0 | 2.6 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.8 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.2 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.4 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 2.6 | GO:0001158 | enhancer sequence-specific DNA binding(GO:0001158) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:1990247 | N6-methyladenosine-containing RNA binding(GO:1990247) |
| 0.0 | 0.3 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 1.5 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 2.4 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.4 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.5 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.9 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.2 | GO:0003678 | DNA helicase activity(GO:0003678) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 1.0 | GO:0003705 | transcription factor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0003705) |
| 0.0 | 0.1 | GO:0042166 | acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 7.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.0 | 0.0 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.2 | GO:0001671 | ATPase activator activity(GO:0001671) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.0 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.1 | 5.3 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 2.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 3.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 4.6 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 1.1 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 2.8 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 1.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.3 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 2.7 | PID PDGFRB PATHWAY | PDGFR-beta signaling pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.5 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 2.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.2 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.2 | 3.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.2 | 6.0 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.1 | 2.5 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.1 | 2.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.7 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.6 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 2.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.1 | 2.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 2.0 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.7 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 1.2 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.4 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 4.4 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.4 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.6 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 1.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.3 | REACTOME MEIOTIC RECOMBINATION | Genes involved in Meiotic Recombination |
| 0.0 | 1.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.5 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.7 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 2.4 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.3 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 1.3 | REACTOME EXTRACELLULAR MATRIX ORGANIZATION | Genes involved in Extracellular matrix organization |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 3 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 3 Promoter |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.7 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |