Project
12D miR HR13_24
Navigation
Downloads
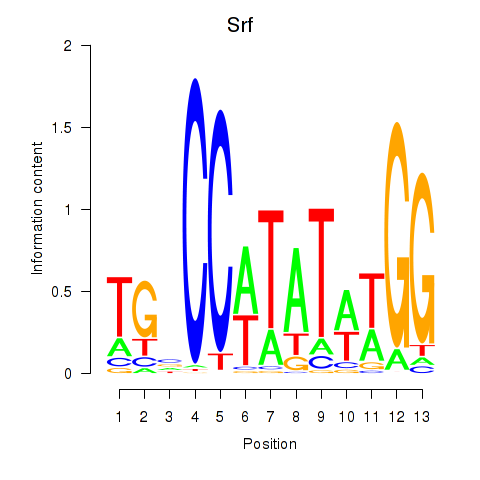
Results for Srf
Z-value: 1.39
Transcription factors associated with Srf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Srf
|
ENSMUSG00000015605.5 | serum response factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srf | mm10_v2_chr17_-_46556158_46556188 | 0.82 | 1.9e-03 | Click! |
Activity profile of Srf motif
Sorted Z-values of Srf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_14863495 | 4.03 |
ENSMUST00000029076.4
|
Car3
|
carbonic anhydrase 3 |
| chr6_-_85513586 | 3.79 |
ENSMUST00000095759.3
|
Egr4
|
early growth response 4 |
| chr18_+_34861200 | 3.43 |
ENSMUST00000165033.1
|
Egr1
|
early growth response 1 |
| chr6_-_83536215 | 3.03 |
ENSMUST00000075161.5
|
Actg2
|
actin, gamma 2, smooth muscle, enteric |
| chr1_-_43163891 | 2.86 |
ENSMUST00000008280.7
|
Fhl2
|
four and a half LIM domains 2 |
| chr2_+_127336152 | 2.74 |
ENSMUST00000028846.6
|
Dusp2
|
dual specificity phosphatase 2 |
| chr14_+_3652030 | 2.47 |
ENSMUST00000167430.1
|
Gm3020
|
predicted gene 3020 |
| chr14_+_4334763 | 2.42 |
ENSMUST00000165466.1
|
2610042L04Rik
|
RIKEN cDNA 2610042L04 gene |
| chr14_+_3412614 | 2.38 |
ENSMUST00000170123.1
|
Gm10409
|
predicted gene 10409 |
| chr2_+_156775409 | 2.32 |
ENSMUST00000088552.6
|
Myl9
|
myosin, light polypeptide 9, regulatory |
| chr14_+_4855576 | 2.30 |
ENSMUST00000166776.1
|
Gm3264
|
predicted gene 3264 |
| chr14_+_3049285 | 2.22 |
ENSMUST00000166494.1
|
Gm2897
|
predicted gene 2897 |
| chr14_-_6287250 | 2.15 |
ENSMUST00000170104.2
|
Gm3411
|
predicted gene 3411 |
| chr14_+_4415448 | 2.04 |
ENSMUST00000168866.1
|
Gm3164
|
predicted gene 3164 |
| chr14_-_7568566 | 1.93 |
ENSMUST00000163790.1
|
Gm3558
|
predicted gene 3558 |
| chr14_-_7027449 | 1.82 |
ENSMUST00000170738.2
|
Gm10406
|
predicted gene 10406 |
| chr14_+_3810074 | 1.56 |
ENSMUST00000170480.1
|
Gm3002
|
predicted gene 3002 |
| chr14_-_5389049 | 1.44 |
ENSMUST00000177986.1
|
Gm3500
|
predicted gene 3500 |
| chr11_-_73324616 | 1.40 |
ENSMUST00000021119.2
|
Aspa
|
aspartoacylase |
| chr2_+_31759993 | 1.36 |
ENSMUST00000124089.1
|
Abl1
|
c-abl oncogene 1, non-receptor tyrosine kinase |
| chr14_+_5501674 | 1.35 |
ENSMUST00000181562.1
|
Gm3488
|
predicted gene, 3488 |
| chr10_-_120899067 | 1.27 |
ENSMUST00000092143.5
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr3_+_68468162 | 1.19 |
ENSMUST00000182532.1
|
Schip1
|
schwannomin interacting protein 1 |
| chr11_-_12412136 | 1.18 |
ENSMUST00000174874.1
|
Cobl
|
cordon-bleu WH2 repeat |
| chr12_+_86678685 | 1.12 |
ENSMUST00000021681.3
|
Vash1
|
vasohibin 1 |
| chr2_-_127521358 | 1.10 |
ENSMUST00000028850.8
ENSMUST00000103215.4 |
Kcnip3
|
Kv channel interacting protein 3, calsenilin |
| chr10_-_111997204 | 1.05 |
ENSMUST00000074805.5
|
Glipr1
|
GLI pathogenesis-related 1 (glioma) |
| chr7_+_101896817 | 1.04 |
ENSMUST00000143835.1
|
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr17_+_7925990 | 1.04 |
ENSMUST00000036370.7
|
Tagap
|
T cell activation Rho GTPase activating protein |
| chr14_+_20929416 | 1.03 |
ENSMUST00000022369.7
|
Vcl
|
vinculin |
| chr3_-_152266320 | 1.01 |
ENSMUST00000046045.8
|
Nexn
|
nexilin |
| chr14_-_5741577 | 0.96 |
ENSMUST00000177556.1
|
Gm3373
|
predicted gene 3373 |
| chr12_-_103355967 | 0.95 |
ENSMUST00000021617.7
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr4_-_132351636 | 0.89 |
ENSMUST00000105951.1
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr11_+_101330605 | 0.89 |
ENSMUST00000103105.3
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr16_+_57549232 | 0.88 |
ENSMUST00000159414.1
|
Filip1l
|
filamin A interacting protein 1-like |
| chr1_+_67123015 | 0.86 |
ENSMUST00000027144.7
|
Cps1
|
carbamoyl-phosphate synthetase 1 |
| chr12_+_85473883 | 0.85 |
ENSMUST00000021674.6
|
Fos
|
FBJ osteosarcoma oncogene |
| chr7_-_137314394 | 0.79 |
ENSMUST00000168203.1
ENSMUST00000106118.2 ENSMUST00000169486.2 ENSMUST00000033378.5 |
Ebf3
|
early B cell factor 3 |
| chr11_-_94973447 | 0.71 |
ENSMUST00000100551.4
ENSMUST00000152042.1 |
Sgca
|
sarcoglycan, alpha (dystrophin-associated glycoprotein) |
| chr11_+_101331069 | 0.68 |
ENSMUST00000017316.6
|
Aoc3
|
amine oxidase, copper containing 3 |
| chr11_-_120731944 | 0.68 |
ENSMUST00000154565.1
ENSMUST00000026148.2 |
Cbr2
|
carbonyl reductase 2 |
| chr9_+_75051977 | 0.67 |
ENSMUST00000170310.1
ENSMUST00000166549.1 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr2_+_125068118 | 0.65 |
ENSMUST00000070353.3
|
Slc24a5
|
solute carrier family 24, member 5 |
| chr2_+_154548888 | 0.64 |
ENSMUST00000045116.4
ENSMUST00000109709.3 |
1700003F12Rik
|
RIKEN cDNA 1700003F12 gene |
| chr7_-_19310035 | 0.60 |
ENSMUST00000003640.2
|
Fosb
|
FBJ osteosarcoma oncogene B |
| chr7_+_128246812 | 0.56 |
ENSMUST00000164710.1
ENSMUST00000070656.5 |
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr4_+_43957401 | 0.55 |
ENSMUST00000030202.7
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr15_-_76229492 | 0.53 |
ENSMUST00000074834.5
|
Plec
|
plectin |
| chr10_+_79988584 | 0.51 |
ENSMUST00000004784.4
ENSMUST00000105374.1 |
Cnn2
|
calponin 2 |
| chr14_+_55575617 | 0.48 |
ENSMUST00000022826.5
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr11_+_33963013 | 0.48 |
ENSMUST00000020362.2
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr7_+_128246953 | 0.47 |
ENSMUST00000167965.1
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr7_+_101896340 | 0.47 |
ENSMUST00000035395.7
ENSMUST00000106973.1 ENSMUST00000144207.1 |
Anapc15
|
anaphase prompoting complex C subunit 15 |
| chr6_+_113378113 | 0.46 |
ENSMUST00000171058.1
ENSMUST00000156898.1 |
Arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr18_+_45268876 | 0.45 |
ENSMUST00000183850.1
ENSMUST00000066890.7 |
Kcnn2
|
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 |
| chr1_+_63176818 | 0.44 |
ENSMUST00000129339.1
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr15_-_77842133 | 0.44 |
ENSMUST00000016771.6
|
Myh9
|
myosin, heavy polypeptide 9, non-muscle |
| chr3_-_57575907 | 0.42 |
ENSMUST00000120977.1
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr12_-_110696289 | 0.42 |
ENSMUST00000021698.6
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr12_-_110695860 | 0.42 |
ENSMUST00000149189.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr12_-_80968075 | 0.42 |
ENSMUST00000095572.4
|
Slc10a1
|
solute carrier family 10 (sodium/bile acid cotransporter family), member 1 |
| chr10_+_67537861 | 0.42 |
ENSMUST00000048289.7
ENSMUST00000105438.2 ENSMUST00000130933.1 ENSMUST00000146986.1 |
Egr2
|
early growth response 2 |
| chr8_-_71381907 | 0.41 |
ENSMUST00000002466.8
|
Nr2f6
|
nuclear receptor subfamily 2, group F, member 6 |
| chr5_-_145191511 | 0.41 |
ENSMUST00000161845.1
|
Atp5j2
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit F2 |
| chr12_-_110696248 | 0.39 |
ENSMUST00000124156.1
|
Hsp90aa1
|
heat shock protein 90, alpha (cytosolic), class A member 1 |
| chr3_-_95891938 | 0.38 |
ENSMUST00000036360.6
ENSMUST00000090476.3 |
BC028528
|
cDNA sequence BC028528 |
| chr4_+_43957678 | 0.38 |
ENSMUST00000107855.1
|
Glipr2
|
GLI pathogenesis-related 2 |
| chr7_+_119896292 | 0.37 |
ENSMUST00000106517.1
|
Lyrm1
|
LYR motif containing 1 |
| chr3_+_90072641 | 0.37 |
ENSMUST00000121503.1
ENSMUST00000119570.1 ENSMUST00000062193.9 |
Tpm3
|
tropomyosin 3, gamma |
| chr13_+_109260481 | 0.34 |
ENSMUST00000153234.1
|
Pde4d
|
phosphodiesterase 4D, cAMP specific |
| chr10_+_4266323 | 0.33 |
ENSMUST00000045730.5
|
Akap12
|
A kinase (PRKA) anchor protein (gravin) 12 |
| chr15_-_10714612 | 0.31 |
ENSMUST00000169385.1
|
Rai14
|
retinoic acid induced 14 |
| chr9_-_66514567 | 0.30 |
ENSMUST00000056890.8
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr2_+_69380431 | 0.27 |
ENSMUST00000063690.3
|
Dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr14_-_45529964 | 0.27 |
ENSMUST00000150660.1
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr4_+_132351768 | 0.27 |
ENSMUST00000172202.1
|
Gm17300
|
predicted gene, 17300 |
| chr7_+_19411086 | 0.27 |
ENSMUST00000003643.1
|
Ckm
|
creatine kinase, muscle |
| chr3_-_95891929 | 0.26 |
ENSMUST00000171519.1
|
BC028528
|
cDNA sequence BC028528 |
| chr14_+_74732384 | 0.25 |
ENSMUST00000176957.1
|
Esd
|
esterase D/formylglutathione hydrolase |
| chr1_+_167618246 | 0.25 |
ENSMUST00000111380.1
|
Rxrg
|
retinoid X receptor gamma |
| chr1_+_78816909 | 0.24 |
ENSMUST00000057262.6
|
Kcne4
|
potassium voltage-gated channel, Isk-related subfamily, gene 4 |
| chr7_-_105600103 | 0.23 |
ENSMUST00000033185.8
|
Hpx
|
hemopexin |
| chr14_-_76010863 | 0.22 |
ENSMUST00000088922.4
|
Gtf2f2
|
general transcription factor IIF, polypeptide 2 |
| chr16_-_4880284 | 0.21 |
ENSMUST00000037843.6
|
Ubald1
|
UBA-like domain containing 1 |
| chr17_-_46153517 | 0.21 |
ENSMUST00000171172.1
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr11_-_120348475 | 0.20 |
ENSMUST00000062147.7
ENSMUST00000128055.1 |
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr11_-_120348091 | 0.20 |
ENSMUST00000106215.4
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr7_-_44997535 | 0.20 |
ENSMUST00000124232.1
ENSMUST00000003290.4 |
Bcl2l12
|
BCL2-like 12 (proline rich) |
| chr7_+_119895836 | 0.20 |
ENSMUST00000106518.1
ENSMUST00000054440.3 |
Lyrm1
|
LYR motif containing 1 |
| chrX_-_166518567 | 0.19 |
ENSMUST00000112187.1
|
Tceanc
|
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr2_+_127208358 | 0.17 |
ENSMUST00000103220.3
|
Snrnp200
|
small nuclear ribonucleoprotein 200 (U5) |
| chr19_+_5447692 | 0.17 |
ENSMUST00000025850.5
|
Fosl1
|
fos-like antigen 1 |
| chr4_-_63403330 | 0.16 |
ENSMUST00000035724.4
|
Akna
|
AT-hook transcription factor |
| chr2_+_164940742 | 0.16 |
ENSMUST00000137626.1
|
Mmp9
|
matrix metallopeptidase 9 |
| chr7_+_120677579 | 0.15 |
ENSMUST00000060175.6
|
BC030336
|
cDNA sequence BC030336 |
| chr1_-_33757711 | 0.15 |
ENSMUST00000044691.7
|
Bag2
|
BCL2-associated athanogene 2 |
| chr11_-_120348513 | 0.14 |
ENSMUST00000071555.6
|
Actg1
|
actin, gamma, cytoplasmic 1 |
| chr17_-_46556158 | 0.14 |
ENSMUST00000015749.5
|
Srf
|
serum response factor |
| chr2_+_164833841 | 0.14 |
ENSMUST00000152721.1
|
Ctsa
|
cathepsin A |
| chr7_+_19131686 | 0.13 |
ENSMUST00000165913.1
|
Fbxo46
|
F-box protein 46 |
| chr9_+_53537021 | 0.12 |
ENSMUST00000035850.7
|
Npat
|
nuclear protein in the AT region |
| chr7_-_119720742 | 0.11 |
ENSMUST00000033236.7
|
Thumpd1
|
THUMP domain containing 1 |
| chr1_+_180942325 | 0.10 |
ENSMUST00000161847.1
|
Tmem63a
|
transmembrane protein 63a |
| chr15_-_36794498 | 0.09 |
ENSMUST00000110361.1
ENSMUST00000022894.7 ENSMUST00000110359.1 |
Ywhaz
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide |
| chr2_-_164833438 | 0.09 |
ENSMUST00000042775.4
|
Neurl2
|
neuralized-like 2 (Drosophila) |
| chr9_-_58313189 | 0.09 |
ENSMUST00000061799.8
|
Loxl1
|
lysyl oxidase-like 1 |
| chr1_-_131097535 | 0.09 |
ENSMUST00000016672.4
|
Mapkapk2
|
MAP kinase-activated protein kinase 2 |
| chr11_-_99493112 | 0.07 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chr1_+_38987806 | 0.07 |
ENSMUST00000027247.5
|
Pdcl3
|
phosducin-like 3 |
| chr19_+_25406661 | 0.07 |
ENSMUST00000146647.1
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr10_+_90576872 | 0.07 |
ENSMUST00000182550.1
ENSMUST00000099364.5 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr2_-_13271268 | 0.05 |
ENSMUST00000137670.1
ENSMUST00000114791.2 |
Rsu1
|
Ras suppressor protein 1 |
| chr5_-_112896350 | 0.04 |
ENSMUST00000086617.4
|
Myo18b
|
myosin XVIIIb |
| chr5_-_142905928 | 0.03 |
ENSMUST00000106216.2
|
Actb
|
actin, beta |
| chr14_-_45530118 | 0.02 |
ENSMUST00000045905.6
|
Fermt2
|
fermitin family homolog 2 (Drosophila) |
| chr2_+_20737306 | 0.00 |
ENSMUST00000114606.1
ENSMUST00000114608.1 |
Etl4
|
enhancer trap locus 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Srf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 3.4 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) positive regulation of metanephric glomerulus development(GO:0072300) |
| 0.6 | 2.4 | GO:0060025 | regulation of synaptic activity(GO:0060025) |
| 0.4 | 1.3 | GO:1900275 | negative regulation of phospholipase C activity(GO:1900275) |
| 0.3 | 1.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.3 | 1.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.2 | 1.4 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.2 | 1.2 | GO:0045585 | regulation of cytotoxic T cell differentiation(GO:0045583) positive regulation of cytotoxic T cell differentiation(GO:0045585) |
| 0.2 | 2.9 | GO:0055015 | ventricular cardiac muscle cell development(GO:0055015) |
| 0.2 | 1.6 | GO:0035902 | response to immobilization stress(GO:0035902) |
| 0.2 | 1.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.2 | 0.6 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.1 | 1.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.4 | GO:1903919 | regulation of actin filament severing(GO:1903918) negative regulation of actin filament severing(GO:1903919) |
| 0.1 | 0.9 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.7 | GO:0006116 | NADH oxidation(GO:0006116) |
| 0.1 | 0.4 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.1 | 1.5 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.1 | 4.0 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.1 | 0.9 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 0.5 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.3 | GO:0046294 | formaldehyde catabolic process(GO:0046294) |
| 0.1 | 0.4 | GO:0072257 | metanephric nephron tubule epithelial cell differentiation(GO:0072257) regulation of metanephric nephron tubule epithelial cell differentiation(GO:0072307) |
| 0.1 | 0.5 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.1 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.1 | 1.0 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 1.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.5 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.9 | GO:0035994 | response to muscle stretch(GO:0035994) |
| 0.0 | 0.5 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.3 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 2.9 | GO:0070527 | platelet aggregation(GO:0070527) |
| 0.0 | 1.4 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 1.1 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.3 | GO:1901844 | regulation of cell communication by electrical coupling involved in cardiac conduction(GO:1901844) negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.5 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.7 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.4 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.9 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.4 | GO:0043153 | entrainment of circadian clock by photoperiod(GO:0043153) |
| 0.0 | 0.2 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0044351 | macropinocytosis(GO:0044351) |
| 0.0 | 0.2 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.5 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.5 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.6 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.1 | 1.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.1 | 0.9 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.9 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.1 | 2.9 | GO:0031430 | M band(GO:0031430) |
| 0.1 | 0.7 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 0.4 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.4 | GO:0005826 | actomyosin contractile ring(GO:0005826) myosin II filament(GO:0097513) |
| 0.1 | 1.9 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.1 | 1.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.1 | 1.0 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.5 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.4 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.4 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
| 0.0 | 0.9 | GO:0042645 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.7 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 3.1 | GO:0031965 | nuclear membrane(GO:0031965) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0052596 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.5 | 3.4 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.5 | 1.4 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.3 | 4.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.3 | 0.9 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.2 | 0.6 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.2 | 1.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.2 | 1.2 | GO:0002135 | CTP binding(GO:0002135) |
| 0.2 | 0.7 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.1 | 0.9 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.1 | 1.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 2.3 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 1.3 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.1 | 1.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.1 | 0.3 | GO:0018738 | S-formylglutathione hydrolase activity(GO:0018738) |
| 0.1 | 1.0 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.4 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.1 | 0.3 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.1 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.1 | 0.3 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 1.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.6 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.3 | GO:0015459 | potassium channel regulator activity(GO:0015459) |
| 0.0 | 0.4 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0000990 | transcription factor activity, core RNA polymerase binding(GO:0000990) |
| 0.0 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.9 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.4 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.3 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.7 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 2.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.1 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.6 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 2.3 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 1.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 6.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.1 | 1.2 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.1 | 0.4 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 2.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.9 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 4.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.5 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.5 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.4 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |