Project
12D miR HR13_24
Navigation
Downloads
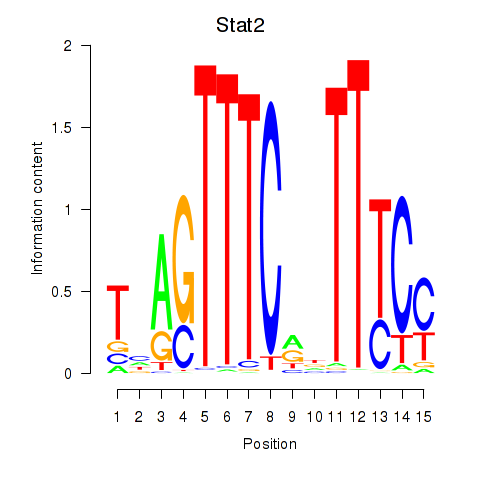
Results for Stat2
Z-value: 1.87
Transcription factors associated with Stat2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat2
|
ENSMUSG00000040033.9 | signal transducer and activator of transcription 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat2 | mm10_v2_chr10_+_128270546_128270577 | 0.57 | 6.9e-02 | Click! |
Activity profile of Stat2 motif
Sorted Z-values of Stat2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_61928081 | 4.08 |
ENSMUST00000154398.1
ENSMUST00000093485.2 ENSMUST00000156980.1 ENSMUST00000070631.7 |
Ddx60
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 |
| chr14_+_41105359 | 3.93 |
ENSMUST00000047286.6
|
Mat1a
|
methionine adenosyltransferase I, alpha |
| chr11_+_61065798 | 3.49 |
ENSMUST00000041944.2
ENSMUST00000108717.2 |
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr12_+_26469204 | 2.84 |
ENSMUST00000020969.3
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr19_+_34640871 | 2.60 |
ENSMUST00000102824.3
|
Ifit1
|
interferon-induced protein with tetratricopeptide repeats 1 |
| chr5_-_139813237 | 2.56 |
ENSMUST00000110832.1
|
Tmem184a
|
transmembrane protein 184a |
| chr7_+_104244449 | 2.36 |
ENSMUST00000106849.2
ENSMUST00000060315.5 |
Trim34a
|
tripartite motif-containing 34A |
| chr3_-_137981523 | 2.32 |
ENSMUST00000136613.1
ENSMUST00000029806.6 |
Dapp1
|
dual adaptor for phosphotyrosine and 3-phosphoinositides 1 |
| chr7_+_104244496 | 2.30 |
ENSMUST00000106854.1
ENSMUST00000143414.1 |
Trim34a
|
tripartite motif-containing 34A |
| chr12_-_78980758 | 2.29 |
ENSMUST00000174072.1
|
Tmem229b
|
transmembrane protein 229B |
| chr6_+_39381175 | 2.25 |
ENSMUST00000031986.4
|
Rab19
|
RAB19, member RAS oncogene family |
| chr8_-_111338152 | 2.24 |
ENSMUST00000056157.7
ENSMUST00000120432.1 |
Mlkl
|
mixed lineage kinase domain-like |
| chr7_+_104244465 | 2.14 |
ENSMUST00000106848.1
|
Trim34a
|
tripartite motif-containing 34A |
| chr16_-_22439719 | 2.12 |
ENSMUST00000079601.6
|
Etv5
|
ets variant gene 5 |
| chr12_-_26456423 | 2.02 |
ENSMUST00000020970.7
|
Rsad2
|
radical S-adenosyl methionine domain containing 2 |
| chr5_+_147269959 | 1.97 |
ENSMUST00000085591.5
|
Pdx1
|
pancreatic and duodenal homeobox 1 |
| chr1_-_156674290 | 1.96 |
ENSMUST00000079625.4
|
Tor3a
|
torsin family 3, member A |
| chr5_-_139814025 | 1.90 |
ENSMUST00000146780.1
|
Tmem184a
|
transmembrane protein 184a |
| chr19_+_55741810 | 1.90 |
ENSMUST00000111657.3
ENSMUST00000061496.9 ENSMUST00000041717.7 ENSMUST00000111662.4 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chr16_-_24393588 | 1.79 |
ENSMUST00000181640.1
|
1110054M08Rik
|
RIKEN cDNA 1110054M08 gene |
| chr3_-_59262825 | 1.78 |
ENSMUST00000050360.7
|
P2ry12
|
purinergic receptor P2Y, G-protein coupled 12 |
| chr4_+_42255767 | 1.70 |
ENSMUST00000178864.1
|
Ccl21b
|
chemokine (C-C motif) ligand 21B (leucine) |
| chr11_+_115834314 | 1.67 |
ENSMUST00000173289.1
ENSMUST00000137900.1 |
Llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr7_+_78913436 | 1.59 |
ENSMUST00000121645.1
|
Isg20
|
interferon-stimulated protein |
| chr3_+_127791374 | 1.57 |
ENSMUST00000171621.1
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr4_+_45972233 | 1.53 |
ENSMUST00000102929.1
|
Tdrd7
|
tudor domain containing 7 |
| chr16_-_35871544 | 1.51 |
ENSMUST00000042665.8
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr11_-_47379405 | 1.50 |
ENSMUST00000077221.5
|
Sgcd
|
sarcoglycan, delta (dystrophin-associated glycoprotein) |
| chr5_-_92348871 | 1.47 |
ENSMUST00000038816.6
ENSMUST00000118006.1 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr17_+_33919332 | 1.46 |
ENSMUST00000025161.7
|
Tapbp
|
TAP binding protein |
| chr8_+_10006656 | 1.46 |
ENSMUST00000033892.7
|
Tnfsf13b
|
tumor necrosis factor (ligand) superfamily, member 13b |
| chr15_-_98728120 | 1.45 |
ENSMUST00000003445.6
|
Fkbp11
|
FK506 binding protein 11 |
| chr11_+_93099284 | 1.41 |
ENSMUST00000092780.3
ENSMUST00000107863.2 |
Car10
|
carbonic anhydrase 10 |
| chr6_+_41521782 | 1.40 |
ENSMUST00000070380.4
|
Prss2
|
protease, serine, 2 |
| chr7_+_78913765 | 1.38 |
ENSMUST00000038142.8
|
Isg20
|
interferon-stimulated protein |
| chr1_+_130826762 | 1.33 |
ENSMUST00000133792.1
|
Pigr
|
polymeric immunoglobulin receptor |
| chr9_+_107975529 | 1.32 |
ENSMUST00000035216.4
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr1_+_130826676 | 1.31 |
ENSMUST00000027675.7
|
Pigr
|
polymeric immunoglobulin receptor |
| chr4_-_42773993 | 1.29 |
ENSMUST00000095114.4
|
Ccl21a
|
chemokine (C-C motif) ligand 21A (serine) |
| chr6_-_13839916 | 1.23 |
ENSMUST00000060442.7
|
Gpr85
|
G protein-coupled receptor 85 |
| chr6_+_90550789 | 1.20 |
ENSMUST00000130418.1
ENSMUST00000032175.8 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr17_-_34187219 | 1.16 |
ENSMUST00000173831.1
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr4_+_41903610 | 1.15 |
ENSMUST00000098128.3
|
Gm21541
|
predicted gene, 21541 |
| chr6_+_112696772 | 1.15 |
ENSMUST00000180959.1
|
Gm26799
|
predicted gene, 26799 |
| chr2_-_84743655 | 1.15 |
ENSMUST00000181711.1
|
Gm19426
|
predicted gene, 19426 |
| chr12_-_79007276 | 1.14 |
ENSMUST00000056660.6
ENSMUST00000174721.1 |
Tmem229b
|
transmembrane protein 229B |
| chr8_+_127064107 | 1.12 |
ENSMUST00000162536.1
ENSMUST00000026921.6 ENSMUST00000162665.1 ENSMUST00000160766.1 ENSMUST00000162602.1 ENSMUST00000162531.1 ENSMUST00000160581.1 ENSMUST00000161355.1 ENSMUST00000159537.1 |
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr13_-_100201961 | 1.10 |
ENSMUST00000167986.2
ENSMUST00000117913.1 |
Naip2
|
NLR family, apoptosis inhibitory protein 2 |
| chr5_-_120887582 | 1.10 |
ENSMUST00000086368.5
|
Oas1g
|
2'-5' oligoadenylate synthetase 1G |
| chr19_+_34607927 | 1.09 |
ENSMUST00000076249.5
|
I830012O16Rik
|
RIKEN cDNA I830012O16 gene |
| chr4_+_42114817 | 1.08 |
ENSMUST00000098123.3
|
Gm13304
|
predicted gene 13304 |
| chr5_-_120907510 | 1.04 |
ENSMUST00000080322.7
|
Oas1a
|
2'-5' oligoadenylate synthetase 1A |
| chr19_-_46148369 | 1.04 |
ENSMUST00000026259.9
|
Pitx3
|
paired-like homeodomain transcription factor 3 |
| chr3_+_142560351 | 1.03 |
ENSMUST00000106221.1
|
Gbp3
|
guanylate binding protein 3 |
| chr10_+_128270546 | 1.02 |
ENSMUST00000105238.3
ENSMUST00000085708.2 |
Stat2
|
signal transducer and activator of transcription 2 |
| chr3_-_58525867 | 1.02 |
ENSMUST00000029385.7
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr5_-_120749848 | 1.01 |
ENSMUST00000053909.6
ENSMUST00000081491.6 |
Oas2
|
2'-5' oligoadenylate synthetase 2 |
| chr4_+_138972885 | 1.01 |
ENSMUST00000123636.1
ENSMUST00000043042.3 ENSMUST00000050949.2 |
Tmco4
|
transmembrane and coiled-coil domains 4 |
| chr2_-_51972990 | 1.00 |
ENSMUST00000145481.1
ENSMUST00000112705.2 |
Nmi
|
N-myc (and STAT) interactor |
| chr11_-_100261021 | 0.98 |
ENSMUST00000080893.6
|
Krt17
|
keratin 17 |
| chr2_+_4559742 | 0.97 |
ENSMUST00000176828.1
|
Frmd4a
|
FERM domain containing 4A |
| chr14_-_11162008 | 0.97 |
ENSMUST00000162278.1
ENSMUST00000160340.1 ENSMUST00000160956.1 |
Fhit
|
fragile histidine triad gene |
| chr3_+_142560108 | 0.95 |
ENSMUST00000128609.1
ENSMUST00000029935.7 |
Gbp3
|
guanylate binding protein 3 |
| chr18_-_3299537 | 0.94 |
ENSMUST00000129435.1
ENSMUST00000122958.1 |
Crem
|
cAMP responsive element modulator |
| chr1_+_143640664 | 0.94 |
ENSMUST00000038252.2
|
B3galt2
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
| chr4_-_40239779 | 0.93 |
ENSMUST00000037907.6
|
Ddx58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr15_+_39745926 | 0.93 |
ENSMUST00000022913.4
|
Dcstamp
|
dentrocyte expressed seven transmembrane protein |
| chr6_-_125231772 | 0.92 |
ENSMUST00000043422.7
|
Tapbpl
|
TAP binding protein-like |
| chr4_-_46536134 | 0.92 |
ENSMUST00000046897.6
|
Trim14
|
tripartite motif-containing 14 |
| chr16_+_24393350 | 0.90 |
ENSMUST00000038053.6
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr7_+_78914216 | 0.90 |
ENSMUST00000120331.2
|
Isg20
|
interferon-stimulated protein |
| chr5_-_134229581 | 0.89 |
ENSMUST00000111275.1
ENSMUST00000016094.6 ENSMUST00000144086.1 |
Ncf1
|
neutrophil cytosolic factor 1 |
| chr6_+_41458923 | 0.89 |
ENSMUST00000031910.7
|
Prss1
|
protease, serine, 1 (trypsin 1) |
| chr18_-_3299452 | 0.88 |
ENSMUST00000126578.1
|
Crem
|
cAMP responsive element modulator |
| chr7_+_142460834 | 0.87 |
ENSMUST00000018963.4
ENSMUST00000105967.1 |
Lsp1
|
lymphocyte specific 1 |
| chr11_+_58215028 | 0.87 |
ENSMUST00000108836.1
|
Irgm2
|
immunity-related GTPase family M member 2 |
| chr2_-_62646146 | 0.86 |
ENSMUST00000112459.3
ENSMUST00000028259.5 |
Ifih1
|
interferon induced with helicase C domain 1 |
| chr10_+_39612934 | 0.86 |
ENSMUST00000019987.6
|
Traf3ip2
|
TRAF3 interacting protein 2 |
| chr5_+_114923234 | 0.86 |
ENSMUST00000031540.4
ENSMUST00000112143.3 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr6_-_39118211 | 0.85 |
ENSMUST00000038398.6
|
Parp12
|
poly (ADP-ribose) polymerase family, member 12 |
| chr9_+_118040475 | 0.84 |
ENSMUST00000044454.5
|
Azi2
|
5-azacytidine induced gene 2 |
| chr17_-_34862473 | 0.82 |
ENSMUST00000025229.4
ENSMUST00000176203.2 ENSMUST00000128767.1 |
Cfb
|
complement factor B |
| chr11_+_58199556 | 0.82 |
ENSMUST00000035266.4
ENSMUST00000094169.4 ENSMUST00000168280.1 ENSMUST00000058704.8 |
Igtp
Irgm2
|
interferon gamma induced GTPase immunity-related GTPase family M member 2 |
| chr13_+_42866247 | 0.81 |
ENSMUST00000131942.1
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr3_+_81932601 | 0.81 |
ENSMUST00000029649.2
|
Ctso
|
cathepsin O |
| chr1_+_180942452 | 0.81 |
ENSMUST00000027800.8
|
Tmem63a
|
transmembrane protein 63a |
| chrX_-_9469288 | 0.81 |
ENSMUST00000015484.3
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr9_+_118040509 | 0.81 |
ENSMUST00000133580.1
|
Azi2
|
5-azacytidine induced gene 2 |
| chr3_+_142560052 | 0.81 |
ENSMUST00000106222.2
|
Gbp3
|
guanylate binding protein 3 |
| chr7_-_102565425 | 0.80 |
ENSMUST00000106913.1
ENSMUST00000033264.4 |
Trim21
|
tripartite motif-containing 21 |
| chr1_+_52119438 | 0.79 |
ENSMUST00000070968.7
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr1_+_153749414 | 0.79 |
ENSMUST00000086209.3
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr1_+_157526127 | 0.78 |
ENSMUST00000111700.1
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr8_+_127064022 | 0.78 |
ENSMUST00000160272.1
ENSMUST00000079777.5 ENSMUST00000162907.1 |
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chr4_-_40239700 | 0.78 |
ENSMUST00000142055.1
|
Ddx58
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chrX_-_49886401 | 0.78 |
ENSMUST00000070304.2
|
Olfr1322
|
olfactory receptor 1322 |
| chr14_+_77904365 | 0.76 |
ENSMUST00000169978.1
|
Epsti1
|
epithelial stromal interaction 1 (breast) |
| chr18_+_37725706 | 0.75 |
ENSMUST00000066149.6
|
Pcdhga9
|
protocadherin gamma subfamily A, 9 |
| chr19_-_11050500 | 0.71 |
ENSMUST00000099676.4
|
AW112010
|
expressed sequence AW112010 |
| chr13_-_23710714 | 0.71 |
ENSMUST00000091707.6
ENSMUST00000006787.7 ENSMUST00000091706.6 |
Hfe
|
hemochromatosis |
| chr1_+_153751859 | 0.70 |
ENSMUST00000182538.1
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr6_+_34863130 | 0.70 |
ENSMUST00000074949.3
|
Tmem140
|
transmembrane protein 140 |
| chr2_+_155382186 | 0.69 |
ENSMUST00000134218.1
|
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr11_-_51857624 | 0.68 |
ENSMUST00000020655.7
ENSMUST00000109090.1 |
Phf15
|
PHD finger protein 15 |
| chr5_-_137116177 | 0.68 |
ENSMUST00000054384.5
ENSMUST00000152207.1 |
Trim56
|
tripartite motif-containing 56 |
| chr12_+_103434211 | 0.68 |
ENSMUST00000079294.5
ENSMUST00000076788.5 ENSMUST00000076702.5 ENSMUST00000066701.6 ENSMUST00000085065.5 ENSMUST00000140838.1 |
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr17_-_34862122 | 0.68 |
ENSMUST00000154526.1
|
Cfb
|
complement factor B |
| chr1_-_174031712 | 0.68 |
ENSMUST00000059226.6
|
Ifi205
|
interferon activated gene 205 |
| chr2_-_167062981 | 0.67 |
ENSMUST00000048988.7
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr5_-_120812506 | 0.66 |
ENSMUST00000117193.1
ENSMUST00000130045.1 |
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr9_-_78443204 | 0.66 |
ENSMUST00000070742.7
ENSMUST00000034898.7 |
Mb21d1
|
Mab-21 domain containing 1 |
| chr3_-_59130610 | 0.65 |
ENSMUST00000065220.6
ENSMUST00000091112.4 |
P2ry14
|
purinergic receptor P2Y, G-protein coupled, 14 |
| chr18_+_37400845 | 0.65 |
ENSMUST00000057228.1
|
Pcdhb9
|
protocadherin beta 9 |
| chr19_+_29367447 | 0.63 |
ENSMUST00000016640.7
|
Cd274
|
CD274 antigen |
| chr5_-_120795530 | 0.63 |
ENSMUST00000100785.3
|
Oas1e
|
2'-5' oligoadenylate synthetase 1E |
| chr7_+_142460809 | 0.63 |
ENSMUST00000105968.1
|
Lsp1
|
lymphocyte specific 1 |
| chr10_+_78069351 | 0.62 |
ENSMUST00000105393.1
|
Icosl
|
icos ligand |
| chr14_+_30716377 | 0.61 |
ENSMUST00000112177.1
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr16_+_43235856 | 0.60 |
ENSMUST00000146708.1
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr12_+_52699297 | 0.60 |
ENSMUST00000095737.3
|
Akap6
|
A kinase (PRKA) anchor protein 6 |
| chr1_+_153749496 | 0.59 |
ENSMUST00000182722.1
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr19_+_36409719 | 0.57 |
ENSMUST00000062389.5
|
Pcgf5
|
polycomb group ring finger 5 |
| chr1_+_180942500 | 0.57 |
ENSMUST00000159436.1
|
Tmem63a
|
transmembrane protein 63a |
| chr19_-_21472552 | 0.56 |
ENSMUST00000087600.3
|
Gda
|
guanine deaminase |
| chr5_-_120812484 | 0.56 |
ENSMUST00000125547.1
|
Oas1c
|
2'-5' oligoadenylate synthetase 1C |
| chr14_+_55604550 | 0.56 |
ENSMUST00000138037.1
|
Irf9
|
interferon regulatory factor 9 |
| chr13_+_14063776 | 0.55 |
ENSMUST00000129488.1
ENSMUST00000110536.1 ENSMUST00000110534.1 ENSMUST00000039538.8 ENSMUST00000110533.1 |
Arid4b
|
AT rich interactive domain 4B (RBP1-like) |
| chr6_-_140041409 | 0.54 |
ENSMUST00000032356.6
|
Plcz1
|
phospholipase C, zeta 1 |
| chr9_+_121777607 | 0.54 |
ENSMUST00000098272.2
|
Klhl40
|
kelch-like 40 |
| chr4_+_94739518 | 0.53 |
ENSMUST00000071168.5
|
Tek
|
endothelial-specific receptor tyrosine kinase |
| chr1_+_153751946 | 0.53 |
ENSMUST00000183241.1
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr19_+_6164433 | 0.53 |
ENSMUST00000045042.7
|
Batf2
|
basic leucine zipper transcription factor, ATF-like 2 |
| chr7_-_141266415 | 0.53 |
ENSMUST00000106023.1
ENSMUST00000097952.2 ENSMUST00000026571.4 |
Irf7
|
interferon regulatory factor 7 |
| chr2_-_51934644 | 0.52 |
ENSMUST00000165313.1
|
Rbm43
|
RNA binding motif protein 43 |
| chr8_-_105938384 | 0.52 |
ENSMUST00000034369.8
|
Psmb10
|
proteasome (prosome, macropain) subunit, beta type 10 |
| chr11_-_100704217 | 0.52 |
ENSMUST00000017974.6
|
Dhx58
|
DEXH (Asp-Glu-X-His) box polypeptide 58 |
| chr13_+_74639866 | 0.52 |
ENSMUST00000169114.1
|
Erap1
|
endoplasmic reticulum aminopeptidase 1 |
| chr5_+_114896936 | 0.51 |
ENSMUST00000031542.9
ENSMUST00000146072.1 ENSMUST00000150361.1 |
Oasl2
|
2'-5' oligoadenylate synthetase-like 2 |
| chr3_-_32616479 | 0.50 |
ENSMUST00000108234.1
ENSMUST00000155737.1 |
Gnb4
|
guanine nucleotide binding protein (G protein), beta 4 |
| chr13_+_37345338 | 0.50 |
ENSMUST00000021860.5
|
Ly86
|
lymphocyte antigen 86 |
| chr6_-_38354243 | 0.49 |
ENSMUST00000114900.1
|
Zc3hav1
|
zinc finger CCCH type, antiviral 1 |
| chr12_-_80260356 | 0.49 |
ENSMUST00000021554.8
|
Actn1
|
actinin, alpha 1 |
| chr12_-_78861636 | 0.48 |
ENSMUST00000021536.7
|
Atp6v1d
|
ATPase, H+ transporting, lysosomal V1 subunit D |
| chr16_+_35938470 | 0.48 |
ENSMUST00000114878.1
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr11_-_48871344 | 0.48 |
ENSMUST00000049519.3
|
Irgm1
|
immunity-related GTPase family M member 1 |
| chr9_-_103761820 | 0.48 |
ENSMUST00000049452.8
|
Tmem108
|
transmembrane protein 108 |
| chr11_+_31872100 | 0.48 |
ENSMUST00000020543.6
ENSMUST00000109412.2 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr1_-_173912904 | 0.48 |
ENSMUST00000009340.8
|
Mnda
|
myeloid cell nuclear differentiation antigen |
| chr11_+_46810792 | 0.48 |
ENSMUST00000068877.6
|
Timd4
|
T cell immunoglobulin and mucin domain containing 4 |
| chr17_-_23645264 | 0.48 |
ENSMUST00000024696.7
|
Mmp25
|
matrix metallopeptidase 25 |
| chr8_+_107150621 | 0.46 |
ENSMUST00000034400.3
|
Cyb5b
|
cytochrome b5 type B |
| chr6_-_120822680 | 0.46 |
ENSMUST00000019354.8
|
Atp6v1e1
|
ATPase, H+ transporting, lysosomal V1 subunit E1 |
| chr8_+_67494843 | 0.45 |
ENSMUST00000093470.5
ENSMUST00000163856.1 |
Nat2
|
N-acetyltransferase 2 (arylamine N-acetyltransferase) |
| chr1_+_13668739 | 0.45 |
ENSMUST00000088542.3
|
Xkr9
|
X Kell blood group precursor related family member 9 homolog |
| chr2_-_181241955 | 0.44 |
ENSMUST00000121484.1
|
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr4_-_46536096 | 0.44 |
ENSMUST00000102924.2
|
Trim14
|
tripartite motif-containing 14 |
| chr6_-_23839137 | 0.43 |
ENSMUST00000166458.2
ENSMUST00000142913.2 ENSMUST00000115357.1 ENSMUST00000069074.7 ENSMUST00000115361.2 ENSMUST00000018122.7 ENSMUST00000115355.1 ENSMUST00000115356.2 |
Cadps2
|
Ca2+-dependent activator protein for secretion 2 |
| chr1_-_86110672 | 0.43 |
ENSMUST00000155077.1
|
Htr2b
|
5-hydroxytryptamine (serotonin) receptor 2B |
| chr10_-_30655859 | 0.42 |
ENSMUST00000092610.4
|
Ncoa7
|
nuclear receptor coactivator 7 |
| chr2_-_51934943 | 0.41 |
ENSMUST00000102767.1
ENSMUST00000102768.1 |
Rbm43
|
RNA binding motif protein 43 |
| chr2_-_36104060 | 0.40 |
ENSMUST00000112961.3
ENSMUST00000112966.3 |
Lhx6
|
LIM homeobox protein 6 |
| chr17_+_34187545 | 0.40 |
ENSMUST00000170086.1
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_-_48871408 | 0.39 |
ENSMUST00000097271.2
|
Irgm1
|
immunity-related GTPase family M member 1 |
| chr2_-_167062607 | 0.39 |
ENSMUST00000128676.1
|
Znfx1
|
zinc finger, NFX1-type containing 1 |
| chr9_-_58158498 | 0.39 |
ENSMUST00000168864.2
|
Islr
|
immunoglobulin superfamily containing leucine-rich repeat |
| chr16_+_23609895 | 0.38 |
ENSMUST00000038423.5
|
Rtp4
|
receptor transporter protein 4 |
| chr6_+_71543900 | 0.38 |
ENSMUST00000065364.2
|
Chmp3
|
charged multivesicular body protein 3 |
| chr11_-_118401826 | 0.38 |
ENSMUST00000106290.3
ENSMUST00000043722.3 |
Lgals3bp
|
lectin, galactoside-binding, soluble, 3 binding protein |
| chr16_+_35938972 | 0.38 |
ENSMUST00000023622.6
ENSMUST00000114877.1 |
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr7_+_75701965 | 0.37 |
ENSMUST00000094307.3
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr2_-_181242020 | 0.36 |
ENSMUST00000094203.4
ENSMUST00000108831.1 |
Helz2
|
helicase with zinc finger 2, transcriptional coactivator |
| chr12_+_104406704 | 0.36 |
ENSMUST00000021506.5
|
Serpina3n
|
serine (or cysteine) peptidase inhibitor, clade A, member 3N |
| chr11_-_83530505 | 0.35 |
ENSMUST00000035938.2
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr10_+_51491018 | 0.35 |
ENSMUST00000078778.3
|
Lilrb4
|
leukocyte immunoglobulin-like receptor, subfamily B, member 4 |
| chr8_-_111337793 | 0.34 |
ENSMUST00000145862.1
|
Mlkl
|
mixed lineage kinase domain-like |
| chr1_+_127774164 | 0.34 |
ENSMUST00000027587.8
ENSMUST00000112570.1 |
Ccnt2
|
cyclin T2 |
| chr10_+_128225830 | 0.33 |
ENSMUST00000026455.7
|
Mip
|
major intrinsic protein of eye lens fiber |
| chr10_-_128180265 | 0.33 |
ENSMUST00000099139.1
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr9_+_118040576 | 0.31 |
ENSMUST00000134433.1
|
Azi2
|
5-azacytidine induced gene 2 |
| chr6_+_41392356 | 0.31 |
ENSMUST00000049079.7
|
Gm5771
|
predicted gene 5771 |
| chr11_-_48992226 | 0.31 |
ENSMUST00000059930.2
ENSMUST00000068063.3 |
Gm12185
Tgtp1
|
predicted gene 12185 T cell specific GTPase 1 |
| chr8_-_45333189 | 0.30 |
ENSMUST00000095328.4
|
Cyp4v3
|
cytochrome P450, family 4, subfamily v, polypeptide 3 |
| chr2_+_122147680 | 0.30 |
ENSMUST00000102476.4
|
B2m
|
beta-2 microglobulin |
| chr4_+_94739276 | 0.29 |
ENSMUST00000073939.6
ENSMUST00000102798.1 |
Tek
|
endothelial-specific receptor tyrosine kinase |
| chr15_-_77411034 | 0.29 |
ENSMUST00000089452.5
ENSMUST00000081776.3 |
Apol9a
|
apolipoprotein L 9a |
| chr17_+_36042956 | 0.29 |
ENSMUST00000097331.1
|
Gm6034
|
predicted gene 6034 |
| chr17_+_34187789 | 0.29 |
ENSMUST00000041633.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr11_+_106751255 | 0.29 |
ENSMUST00000183111.1
ENSMUST00000106794.2 |
Milr1
|
mast cell immunoglobulin like receptor 1 |
| chr6_+_71543797 | 0.28 |
ENSMUST00000059462.5
|
Chmp3
|
charged multivesicular body protein 3 |
| chr19_-_6084873 | 0.28 |
ENSMUST00000160977.1
ENSMUST00000159859.1 |
Zfpl1
|
zinc finger like protein 1 |
| chr3_+_60501252 | 0.28 |
ENSMUST00000099087.2
|
Mbnl1
|
muscleblind-like 1 (Drosophila) |
| chr13_-_100317674 | 0.28 |
ENSMUST00000118574.1
|
Naip6
|
NLR family, apoptosis inhibitory protein 6 |
| chr19_+_29410919 | 0.27 |
ENSMUST00000112576.2
|
Pdcd1lg2
|
programmed cell death 1 ligand 2 |
| chr14_+_48446128 | 0.27 |
ENSMUST00000124720.1
|
Tmem260
|
transmembrane protein 260 |
| chr11_+_48978889 | 0.26 |
ENSMUST00000179282.1
ENSMUST00000056759.4 |
Olfr56
|
olfactory receptor 56 |
| chr17_+_36837123 | 0.26 |
ENSMUST00000179968.1
ENSMUST00000130367.1 ENSMUST00000130801.1 ENSMUST00000144182.1 ENSMUST00000123715.1 ENSMUST00000053434.8 |
Trim26
|
tripartite motif-containing 26 |
| chr3_+_19957240 | 0.26 |
ENSMUST00000108325.2
|
Cp
|
ceruloplasmin |
| chr7_-_46795661 | 0.25 |
ENSMUST00000123725.1
|
Hps5
|
Hermansky-Pudlak syndrome 5 homolog (human) |
| chr14_+_28511344 | 0.24 |
ENSMUST00000112272.1
|
Wnt5a
|
wingless-related MMTV integration site 5A |
| chr17_-_78882508 | 0.24 |
ENSMUST00000024884.4
|
Eif2ak2
|
eukaryotic translation initiation factor 2-alpha kinase 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 1.0 | 3.9 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.9 | 2.8 | GO:0009138 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.8 | 3.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.7 | 4.5 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.7 | 2.0 | GO:0034165 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.7 | 2.6 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.5 | 2.1 | GO:0044565 | dendritic cell proliferation(GO:0044565) |
| 0.5 | 2.6 | GO:0035546 | interferon-beta secretion(GO:0035546) regulation of interferon-beta secretion(GO:0035547) positive regulation of interferon-beta secretion(GO:0035549) |
| 0.5 | 1.5 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.5 | 1.9 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.4 | 1.3 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.4 | 1.7 | GO:0061762 | CAMKK-AMPK signaling cascade(GO:0061762) |
| 0.4 | 1.7 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.4 | 1.2 | GO:0009258 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.4 | 2.0 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.4 | 3.0 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.4 | 1.5 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.4 | 2.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.3 | 1.7 | GO:0034239 | regulation of macrophage fusion(GO:0034239) |
| 0.3 | 1.0 | GO:1902524 | positive regulation of protein K48-linked ubiquitination(GO:1902524) |
| 0.3 | 1.6 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.3 | 1.9 | GO:0003383 | apical constriction(GO:0003383) |
| 0.3 | 0.9 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.2 | 0.7 | GO:0061763 | multivesicular body-lysosome fusion(GO:0061763) |
| 0.2 | 1.9 | GO:0048619 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) embryonic hindgut morphogenesis(GO:0048619) |
| 0.2 | 0.6 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 0.6 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.2 | 0.8 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.2 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 1.2 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.6 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 1.0 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.1 | 2.0 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.1 | 0.4 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 0.5 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.1 | 2.6 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.1 | 2.8 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.1 | 0.9 | GO:1901098 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) positive regulation of autophagosome maturation(GO:1901098) |
| 0.1 | 1.0 | GO:0015961 | diadenosine polyphosphate catabolic process(GO:0015961) |
| 0.1 | 0.6 | GO:1902261 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.1 | 0.5 | GO:0060022 | hard palate development(GO:0060022) |
| 0.1 | 1.4 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 0.3 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.1 | 0.5 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.5 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.1 | 0.9 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.1 | 0.8 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 1.5 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.1 | 1.4 | GO:0070269 | pyroptosis(GO:0070269) |
| 0.1 | 3.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.5 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 1.0 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 1.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.1 | 0.2 | GO:0060809 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.1 | 0.4 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.8 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 1.1 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.1 | 1.9 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.1 | 0.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.2 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.1 | 1.3 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.1 | 0.2 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.1 | 0.5 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 0.3 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.1 | 0.5 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.2 | GO:1902867 | negative regulation of neural retina development(GO:0061076) negative regulation of retina development in camera-type eye(GO:1902867) negative regulation of amacrine cell differentiation(GO:1902870) |
| 0.1 | 0.1 | GO:0072733 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.1 | 0.7 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.1 | 0.4 | GO:1990504 | dense core granule exocytosis(GO:1990504) |
| 0.1 | 0.6 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.3 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.4 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.5 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 1.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.3 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.6 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.0 | 1.0 | GO:0048525 | negative regulation of viral process(GO:0048525) |
| 0.0 | 6.9 | GO:0051607 | defense response to virus(GO:0051607) |
| 0.0 | 0.5 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.4 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 1.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.2 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.3 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 1.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.6 | GO:0070266 | necroptotic process(GO:0070266) |
| 0.0 | 0.1 | GO:1904075 | regulation of oligodendrocyte apoptotic process(GO:1900141) negative regulation of oligodendrocyte apoptotic process(GO:1900142) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.2 | GO:0010528 | regulation of transposition(GO:0010528) negative regulation of transposition(GO:0010529) |
| 0.0 | 0.5 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0007442 | hindgut morphogenesis(GO:0007442) |
| 0.0 | 0.5 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.0 | 1.5 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.1 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.7 | GO:0046825 | regulation of protein export from nucleus(GO:0046825) |
| 0.0 | 0.1 | GO:0051204 | protein insertion into mitochondrial membrane(GO:0051204) |
| 0.0 | 0.2 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.4 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 1.3 | GO:0007608 | sensory perception of smell(GO:0007608) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.3 | 4.5 | GO:0020003 | symbiont-containing vacuole(GO:0020003) |
| 0.3 | 2.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 1.9 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.2 | 1.5 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.2 | 1.9 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.2 | 1.4 | GO:0072557 | IPAF inflammasome complex(GO:0072557) |
| 0.2 | 0.5 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.2 | 1.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.3 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 1.7 | GO:0045495 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.1 | 1.0 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.1 | 2.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.1 | 3.9 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 4.4 | GO:0031901 | early endosome membrane(GO:0031901) |
| 0.1 | 0.7 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.1 | 0.9 | GO:0044754 | autolysosome(GO:0044754) |
| 0.1 | 6.7 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 0.2 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 4.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.3 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.5 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 4.1 | GO:0045111 | intermediate filament cytoskeleton(GO:0045111) |
| 0.0 | 1.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0033176 | proton-transporting V-type ATPase complex(GO:0033176) |
| 0.0 | 2.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.4 | GO:0033178 | proton-transporting two-sector ATPase complex, catalytic domain(GO:0033178) |
| 0.0 | 0.7 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 2.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 1.0 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 0.3 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 1.3 | GO:0010008 | endosome membrane(GO:0010008) |
| 0.0 | 1.4 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.7 | GO:0005776 | autophagosome(GO:0005776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.9 | 2.8 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.6 | 6.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.4 | 1.2 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.3 | 1.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.3 | 2.2 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.3 | 1.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.3 | 2.5 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.2 | 1.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.2 | 1.0 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.2 | 0.9 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.2 | 2.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.2 | 1.7 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.2 | 0.5 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 1.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 3.5 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.1 | 0.3 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.1 | 0.8 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 1.5 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.1 | 8.2 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.1 | 1.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 3.9 | GO:0043531 | ADP binding(GO:0043531) |
| 0.1 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.1 | 0.7 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.1 | 2.3 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 1.5 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 0.2 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.1 | 0.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 0.5 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.1 | 1.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.1 | 0.5 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.1 | 0.2 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.1 | 0.3 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.1 | 0.2 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 1.9 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.3 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 2.0 | GO:0051539 | 4 iron, 4 sulfur cluster binding(GO:0051539) |
| 0.0 | 0.7 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.0 | 0.6 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.8 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 2.1 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 2.6 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.7 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 1.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.8 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 4.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.5 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 3.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 0.9 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.2 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 5.7 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.2 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.3 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 9.5 | GO:0008270 | zinc ion binding(GO:0008270) |
| 0.0 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 1.5 | GO:0047485 | protein N-terminus binding(GO:0047485) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.4 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 1.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.1 | 4.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.1 | 1.5 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 2.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 1.9 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 1.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 3.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 1.5 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 2.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 1.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.7 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.5 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 1.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.8 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.1 | PID TELOMERASE PATHWAY | Regulation of Telomerase |
| 0.0 | 0.5 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.4 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.2 | 1.4 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.2 | 8.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.1 | 3.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.1 | 1.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 0.9 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.1 | 3.5 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.1 | 1.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.1 | 0.5 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.1 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.1 | 2.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.1 | 1.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 1.7 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.6 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.4 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.8 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.5 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.7 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 1.0 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.4 | REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | Genes involved in RIG-I/MDA5 mediated induction of IFN-alpha/beta pathways |
| 0.0 | 0.3 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |