Project
12D miR HR13_24
Navigation
Downloads
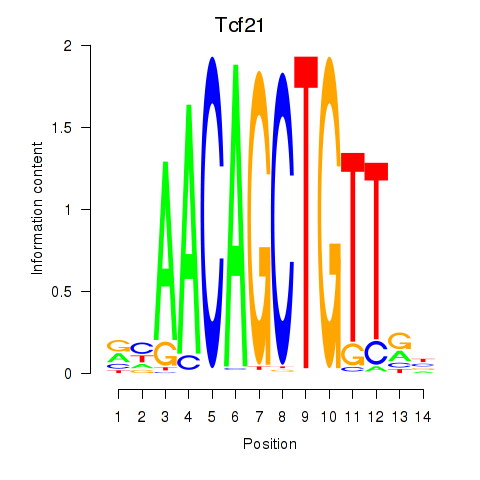
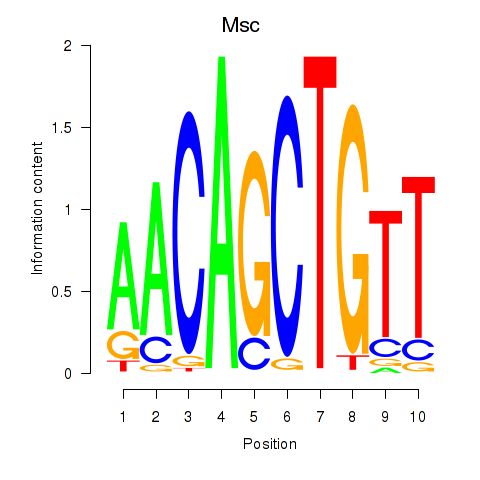
Results for Tcf21_Msc
Z-value: 1.80
Transcription factors associated with Tcf21_Msc
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tcf21
|
ENSMUSG00000045680.7 | transcription factor 21 |
|
Msc
|
ENSMUSG00000025930.5 | musculin |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tcf21 | mm10_v2_chr10_-_22820126_22820150 | -0.70 | 1.5e-02 | Click! |
| Msc | mm10_v2_chr1_-_14755966_14755998 | -0.70 | 1.7e-02 | Click! |
Activity profile of Tcf21_Msc motif
Sorted Z-values of Tcf21_Msc motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_107529717 | 5.60 |
ENSMUST00000049285.8
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chr13_-_71963713 | 4.16 |
ENSMUST00000077337.8
|
Irx1
|
Iroquois related homeobox 1 (Drosophila) |
| chr5_-_131538687 | 4.09 |
ENSMUST00000161374.1
|
Auts2
|
autism susceptibility candidate 2 |
| chr15_-_101370125 | 3.85 |
ENSMUST00000077196.4
|
Krt80
|
keratin 80 |
| chr10_+_69208546 | 3.76 |
ENSMUST00000164034.1
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr4_-_106799779 | 3.45 |
ENSMUST00000145061.1
ENSMUST00000102762.3 |
Acot11
|
acyl-CoA thioesterase 11 |
| chr16_+_36693972 | 3.45 |
ENSMUST00000023617.6
ENSMUST00000089618.3 |
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr4_-_133263042 | 3.27 |
ENSMUST00000105908.3
ENSMUST00000030674.7 |
Sytl1
|
synaptotagmin-like 1 |
| chr11_-_119086221 | 3.27 |
ENSMUST00000026665.7
|
Cbx4
|
chromobox 4 |
| chr16_+_36694024 | 2.77 |
ENSMUST00000119464.1
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chrX_-_74645635 | 2.62 |
ENSMUST00000114119.1
|
Gm5640
|
predicted gene 5640 |
| chrX_+_169036610 | 2.44 |
ENSMUST00000087016.4
ENSMUST00000112129.1 ENSMUST00000112131.2 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr10_+_127866457 | 2.25 |
ENSMUST00000092058.3
|
BC089597
|
cDNA sequence BC089597 |
| chr18_+_50051702 | 2.22 |
ENSMUST00000134348.1
ENSMUST00000153873.2 |
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr8_+_94172618 | 2.08 |
ENSMUST00000034214.6
|
Mt2
|
metallothionein 2 |
| chr2_-_104712122 | 1.99 |
ENSMUST00000111118.1
ENSMUST00000028597.3 |
Tcp11l1
|
t-complex 11 like 1 |
| chr18_+_20558038 | 1.92 |
ENSMUST00000059787.8
|
Dsg2
|
desmoglein 2 |
| chr15_-_71727815 | 1.80 |
ENSMUST00000022953.8
|
Fam135b
|
family with sequence similarity 135, member B |
| chr18_+_20558221 | 1.78 |
ENSMUST00000121837.1
|
Dsg2
|
desmoglein 2 |
| chr10_+_82985473 | 1.77 |
ENSMUST00000040110.7
|
Chst11
|
carbohydrate sulfotransferase 11 |
| chr19_+_52264323 | 1.70 |
ENSMUST00000039652.4
|
Ins1
|
insulin I |
| chr15_-_34356421 | 1.69 |
ENSMUST00000179647.1
|
9430069I07Rik
|
RIKEN cDNA 9430069I07 gene |
| chr1_+_151755339 | 1.67 |
ENSMUST00000059498.5
|
Edem3
|
ER degradation enhancer, mannosidase alpha-like 3 |
| chr3_+_106486009 | 1.66 |
ENSMUST00000183271.1
ENSMUST00000061206.3 |
Dennd2d
|
DENN/MADD domain containing 2D |
| chr19_+_58759700 | 1.65 |
ENSMUST00000026081.3
|
Pnliprp2
|
pancreatic lipase-related protein 2 |
| chr1_+_90915064 | 1.63 |
ENSMUST00000027528.6
|
Mlph
|
melanophilin |
| chr1_-_162866502 | 1.59 |
ENSMUST00000046049.7
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr11_-_31824518 | 1.59 |
ENSMUST00000134944.1
|
D630024D03Rik
|
RIKEN cDNA D630024D03 gene |
| chr14_-_56571830 | 1.57 |
ENSMUST00000065302.7
|
Cenpj
|
centromere protein J |
| chr7_-_127993831 | 1.57 |
ENSMUST00000033056.3
|
Pycard
|
PYD and CARD domain containing |
| chr1_+_188263034 | 1.55 |
ENSMUST00000060479.7
|
Ush2a
|
Usher syndrome 2A (autosomal recessive, mild) |
| chr11_-_109611417 | 1.53 |
ENSMUST00000103060.3
ENSMUST00000047186.3 ENSMUST00000106689.1 |
Wipi1
|
WD repeat domain, phosphoinositide interacting 1 |
| chr11_+_115877497 | 1.48 |
ENSMUST00000144032.1
|
Myo15b
|
myosin XVB |
| chr9_-_104063049 | 1.45 |
ENSMUST00000035166.5
|
Uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr14_+_65968483 | 1.43 |
ENSMUST00000022616.6
|
Clu
|
clusterin |
| chr4_-_106800249 | 1.43 |
ENSMUST00000148688.1
|
Acot11
|
acyl-CoA thioesterase 11 |
| chr1_-_172632931 | 1.42 |
ENSMUST00000027826.5
|
Dusp23
|
dual specificity phosphatase 23 |
| chr4_-_137409777 | 1.41 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr19_+_32389202 | 1.41 |
ENSMUST00000181612.1
|
2700046G09Rik
|
RIKEN cDNA 2700046G09 gene |
| chr12_+_11456052 | 1.40 |
ENSMUST00000124065.1
|
Rad51ap2
|
RAD51 associated protein 2 |
| chr7_+_43950614 | 1.40 |
ENSMUST00000072204.4
|
Klk1b8
|
kallikrein 1-related peptidase b8 |
| chr16_-_32797413 | 1.37 |
ENSMUST00000115116.1
ENSMUST00000041123.8 |
Muc20
|
mucin 20 |
| chr2_+_174760619 | 1.36 |
ENSMUST00000029030.2
|
Edn3
|
endothelin 3 |
| chr4_+_128883549 | 1.35 |
ENSMUST00000035667.8
|
Trim62
|
tripartite motif-containing 62 |
| chr13_-_47014814 | 1.35 |
ENSMUST00000052747.2
|
Nhlrc1
|
NHL repeat containing 1 |
| chr6_+_47244359 | 1.35 |
ENSMUST00000060839.6
|
Cntnap2
|
contactin associated protein-like 2 |
| chr2_-_162661075 | 1.33 |
ENSMUST00000109442.1
ENSMUST00000109445.2 ENSMUST00000109443.1 ENSMUST00000109441.1 |
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr8_+_36489191 | 1.33 |
ENSMUST00000171777.1
|
6430573F11Rik
|
RIKEN cDNA 6430573F11 gene |
| chr4_+_148160613 | 1.32 |
ENSMUST00000047951.8
|
Fbxo2
|
F-box protein 2 |
| chr4_-_149454971 | 1.31 |
ENSMUST00000030848.2
|
Rbp7
|
retinol binding protein 7, cellular |
| chr14_+_58072686 | 1.31 |
ENSMUST00000022545.7
|
Fgf9
|
fibroblast growth factor 9 |
| chr11_-_99979053 | 1.31 |
ENSMUST00000105051.1
|
Krtap29-1
|
keratin associated protein 29-1 |
| chr4_-_141846359 | 1.30 |
ENSMUST00000037059.10
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr2_-_170131156 | 1.28 |
ENSMUST00000063710.6
|
Zfp217
|
zinc finger protein 217 |
| chr10_+_79854658 | 1.28 |
ENSMUST00000171599.1
ENSMUST00000095457.4 |
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr12_+_52516077 | 1.25 |
ENSMUST00000110725.1
|
Arhgap5
|
Rho GTPase activating protein 5 |
| chr12_-_119238794 | 1.24 |
ENSMUST00000026360.8
|
Itgb8
|
integrin beta 8 |
| chr8_+_83165348 | 1.24 |
ENSMUST00000034145.4
|
Tbc1d9
|
TBC1 domain family, member 9 |
| chr14_-_31577318 | 1.24 |
ENSMUST00000112027.2
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr4_-_141846277 | 1.24 |
ENSMUST00000105781.1
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr18_-_20059478 | 1.24 |
ENSMUST00000075214.2
ENSMUST00000039247.4 |
Dsc2
|
desmocollin 2 |
| chr3_+_95588990 | 1.22 |
ENSMUST00000177399.1
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr10_+_79854618 | 1.22 |
ENSMUST00000165704.1
|
Ptbp1
|
polypyrimidine tract binding protein 1 |
| chr10_+_69212634 | 1.21 |
ENSMUST00000020101.5
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chrX_+_103321398 | 1.20 |
ENSMUST00000033689.2
|
Cdx4
|
caudal type homeobox 4 |
| chr4_-_137430517 | 1.18 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr10_-_83534130 | 1.17 |
ENSMUST00000020497.7
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr10_-_83533383 | 1.15 |
ENSMUST00000146640.1
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chrX_-_102906469 | 1.14 |
ENSMUST00000120808.1
ENSMUST00000121197.1 |
Dmrtc1a
|
DMRT-like family C1a |
| chr7_+_121707189 | 1.14 |
ENSMUST00000065310.2
|
1700069B07Rik
|
RIKEN cDNA 1700069B07 gene |
| chr18_-_61536522 | 1.14 |
ENSMUST00000171629.1
|
Arhgef37
|
Rho guanine nucleotide exchange factor (GEF) 37 |
| chr2_+_160888101 | 1.12 |
ENSMUST00000109455.2
ENSMUST00000040872.6 |
Lpin3
|
lipin 3 |
| chr11_-_95076657 | 1.12 |
ENSMUST00000001548.7
|
Itga3
|
integrin alpha 3 |
| chr6_+_97807014 | 1.12 |
ENSMUST00000043637.7
|
Mitf
|
microphthalmia-associated transcription factor |
| chr19_+_4510472 | 1.12 |
ENSMUST00000068004.6
|
Pcx
|
pyruvate carboxylase |
| chr17_-_87797994 | 1.12 |
ENSMUST00000055221.7
|
Kcnk12
|
potassium channel, subfamily K, member 12 |
| chr6_+_8259379 | 1.11 |
ENSMUST00000162034.1
ENSMUST00000160705.1 ENSMUST00000159433.1 |
Gm16039
|
predicted gene 16039 |
| chr3_-_53863764 | 1.11 |
ENSMUST00000122330.1
ENSMUST00000146598.1 |
Ufm1
|
ubiquitin-fold modifier 1 |
| chr3_+_95588960 | 1.10 |
ENSMUST00000176674.1
ENSMUST00000177389.1 ENSMUST00000176755.1 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr3_-_75270073 | 1.07 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr11_+_87760533 | 1.05 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr11_+_113619318 | 1.05 |
ENSMUST00000146390.2
ENSMUST00000106630.1 |
Sstr2
|
somatostatin receptor 2 |
| chr1_-_121327672 | 1.05 |
ENSMUST00000159085.1
ENSMUST00000159125.1 ENSMUST00000161818.1 |
Insig2
|
insulin induced gene 2 |
| chr11_+_96931387 | 1.05 |
ENSMUST00000107633.1
|
Prr15l
|
proline rich 15-like |
| chr15_+_98634743 | 1.05 |
ENSMUST00000003442.7
|
Cacnb3
|
calcium channel, voltage-dependent, beta 3 subunit |
| chr11_-_95076797 | 1.04 |
ENSMUST00000145671.1
ENSMUST00000120375.1 |
Itga3
|
integrin alpha 3 |
| chr8_+_127064107 | 1.04 |
ENSMUST00000162536.1
ENSMUST00000026921.6 ENSMUST00000162665.1 ENSMUST00000160766.1 ENSMUST00000162602.1 ENSMUST00000162531.1 ENSMUST00000160581.1 ENSMUST00000161355.1 ENSMUST00000159537.1 |
Pard3
|
par-3 (partitioning defective 3) homolog (C. elegans) |
| chrX_-_17319316 | 1.02 |
ENSMUST00000026014.7
|
Efhc2
|
EF-hand domain (C-terminal) containing 2 |
| chr5_+_12383156 | 1.02 |
ENSMUST00000030868.6
|
Sema3d
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
| chr14_-_72709534 | 1.01 |
ENSMUST00000162478.1
|
Fndc3a
|
fibronectin type III domain containing 3A |
| chr4_-_42168603 | 1.01 |
ENSMUST00000098121.3
|
Gm13305
|
predicted gene 13305 |
| chr13_-_103096818 | 1.01 |
ENSMUST00000166336.1
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr5_+_117357274 | 1.00 |
ENSMUST00000031309.9
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr11_+_82045705 | 1.00 |
ENSMUST00000021011.2
|
Ccl7
|
chemokine (C-C motif) ligand 7 |
| chr14_+_55853997 | 0.99 |
ENSMUST00000100529.3
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr16_+_75592844 | 0.99 |
ENSMUST00000114249.1
ENSMUST00000046378.7 ENSMUST00000114253.1 |
Rbm11
|
RNA binding motif protein 11 |
| chr10_+_116986314 | 0.99 |
ENSMUST00000020378.4
|
Best3
|
bestrophin 3 |
| chr9_-_22307638 | 0.98 |
ENSMUST00000086278.6
|
Zfp810
|
zinc finger protein 810 |
| chr12_-_72664759 | 0.98 |
ENSMUST00000021512.9
|
Dhrs7
|
dehydrogenase/reductase (SDR family) member 7 |
| chr2_+_160888156 | 0.97 |
ENSMUST00000109457.2
|
Lpin3
|
lipin 3 |
| chr8_-_25101985 | 0.96 |
ENSMUST00000128715.1
ENSMUST00000064883.6 |
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr15_+_25622525 | 0.96 |
ENSMUST00000110457.1
ENSMUST00000137601.1 |
Myo10
|
myosin X |
| chr2_-_52742169 | 0.96 |
ENSMUST00000102759.1
ENSMUST00000127316.1 |
Stam2
|
signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 |
| chrX_-_104857228 | 0.96 |
ENSMUST00000033575.5
|
Magee2
|
melanoma antigen, family E, 2 |
| chr5_-_36830647 | 0.95 |
ENSMUST00000031002.3
|
Man2b2
|
mannosidase 2, alpha B2 |
| chr1_-_121327734 | 0.95 |
ENSMUST00000160968.1
ENSMUST00000162582.1 |
Insig2
|
insulin induced gene 2 |
| chr5_+_38039224 | 0.95 |
ENSMUST00000031008.6
ENSMUST00000042146.8 ENSMUST00000154929.1 |
Stx18
|
syntaxin 18 |
| chr7_+_19094594 | 0.95 |
ENSMUST00000049454.5
|
Six5
|
sine oculis-related homeobox 5 |
| chr13_+_51846673 | 0.95 |
ENSMUST00000021903.2
|
Gadd45g
|
growth arrest and DNA-damage-inducible 45 gamma |
| chr4_+_136423524 | 0.94 |
ENSMUST00000088677.4
ENSMUST00000121571.1 ENSMUST00000117699.1 |
Htr1d
|
5-hydroxytryptamine (serotonin) receptor 1D |
| chr14_-_57104693 | 0.94 |
ENSMUST00000055698.7
|
Gjb2
|
gap junction protein, beta 2 |
| chr6_+_56017489 | 0.94 |
ENSMUST00000052827.4
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr8_+_121590361 | 0.94 |
ENSMUST00000034270.10
ENSMUST00000181948.1 |
Map1lc3b
|
microtubule-associated protein 1 light chain 3 beta |
| chr5_+_102724971 | 0.93 |
ENSMUST00000112853.1
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr3_+_95588928 | 0.92 |
ENSMUST00000177390.1
ENSMUST00000098861.4 ENSMUST00000060323.5 |
Golph3l
|
golgi phosphoprotein 3-like |
| chr11_-_69122589 | 0.92 |
ENSMUST00000180487.1
|
9130213A22Rik
|
RIKEN cDNA 9130213A22 gene |
| chr5_-_116591811 | 0.92 |
ENSMUST00000076124.5
|
Srrm4
|
serine/arginine repetitive matrix 4 |
| chrX_-_71492592 | 0.90 |
ENSMUST00000080035.4
|
Cd99l2
|
CD99 antigen-like 2 |
| chr11_+_115824029 | 0.89 |
ENSMUST00000103032.4
ENSMUST00000133250.1 ENSMUST00000177736.1 |
Llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr6_+_80018877 | 0.89 |
ENSMUST00000147663.1
ENSMUST00000128718.1 ENSMUST00000126005.1 ENSMUST00000133918.1 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr9_+_104063678 | 0.88 |
ENSMUST00000047799.5
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr7_+_44207307 | 0.88 |
ENSMUST00000077354.4
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr3_-_152166230 | 0.86 |
ENSMUST00000046614.9
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr10_+_82699007 | 0.86 |
ENSMUST00000020478.7
|
Hcfc2
|
host cell factor C2 |
| chr14_+_55854115 | 0.85 |
ENSMUST00000168479.1
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr7_-_24208093 | 0.84 |
ENSMUST00000086006.5
|
Zfp111
|
zinc finger protein 111 |
| chr7_+_50599180 | 0.84 |
ENSMUST00000119710.2
|
4933405O20Rik
|
RIKEN cDNA 4933405O20 gene |
| chr19_-_57197556 | 0.84 |
ENSMUST00000099294.2
|
Ablim1
|
actin-binding LIM protein 1 |
| chr2_-_104716379 | 0.83 |
ENSMUST00000099659.2
|
Pin1rt1
|
protein (peptidyl-prolyl cis/trans isomerase) NIMA-interacting 1, retrogene 1 |
| chr1_+_180935022 | 0.82 |
ENSMUST00000037361.8
|
Lefty1
|
left right determination factor 1 |
| chr7_+_122289297 | 0.82 |
ENSMUST00000064989.5
ENSMUST00000064921.4 |
Prkcb
|
protein kinase C, beta |
| chr3_+_92288566 | 0.82 |
ENSMUST00000090872.4
|
Sprr2a3
|
small proline-rich protein 2A3 |
| chr15_-_84855093 | 0.81 |
ENSMUST00000016768.5
|
Phf21b
|
PHD finger protein 21B |
| chr8_+_54550324 | 0.81 |
ENSMUST00000033918.2
|
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr5_-_52471534 | 0.81 |
ENSMUST00000059428.5
|
Ccdc149
|
coiled-coil domain containing 149 |
| chr19_+_58670358 | 0.81 |
ENSMUST00000057270.7
|
Pnlip
|
pancreatic lipase |
| chr17_+_55952623 | 0.80 |
ENSMUST00000003274.6
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr3_+_89436699 | 0.79 |
ENSMUST00000038942.3
ENSMUST00000130858.1 |
Pbxip1
|
pre B cell leukemia transcription factor interacting protein 1 |
| chr9_+_104063376 | 0.79 |
ENSMUST00000120854.1
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr17_-_26199008 | 0.78 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr4_+_116596672 | 0.78 |
ENSMUST00000051869.7
|
Ccdc17
|
coiled-coil domain containing 17 |
| chr6_+_29398920 | 0.78 |
ENSMUST00000181464.1
ENSMUST00000180829.1 |
Ccdc136
|
coiled-coil domain containing 136 |
| chr14_+_58070547 | 0.78 |
ENSMUST00000165526.1
|
Fgf9
|
fibroblast growth factor 9 |
| chr7_+_30977043 | 0.78 |
ENSMUST00000058093.4
|
Fam187b
|
family with sequence similarity 187, member B |
| chr17_+_29549783 | 0.78 |
ENSMUST00000048677.7
|
Tbc1d22b
|
TBC1 domain family, member 22B |
| chr6_+_80019008 | 0.77 |
ENSMUST00000126399.1
ENSMUST00000136421.1 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr7_+_30458280 | 0.77 |
ENSMUST00000126297.1
|
Nphs1
|
nephrosis 1, nephrin |
| chrX_+_143518576 | 0.76 |
ENSMUST00000033640.7
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr6_+_48448100 | 0.76 |
ENSMUST00000169350.2
ENSMUST00000043676.5 |
Sspo
|
SCO-spondin |
| chr11_-_95514570 | 0.76 |
ENSMUST00000058866.7
|
Nxph3
|
neurexophilin 3 |
| chr2_+_174760781 | 0.76 |
ENSMUST00000140908.1
|
Edn3
|
endothelin 3 |
| chr10_+_93897156 | 0.75 |
ENSMUST00000180815.1
|
4930471D02Rik
|
RIKEN cDNA 4930471D02 gene |
| chr12_+_58211772 | 0.75 |
ENSMUST00000110671.2
ENSMUST00000044299.2 |
Sstr1
|
somatostatin receptor 1 |
| chr1_+_109983737 | 0.75 |
ENSMUST00000172005.1
|
Cdh7
|
cadherin 7, type 2 |
| chr4_+_11123950 | 0.75 |
ENSMUST00000142297.1
|
Gm11827
|
predicted gene 11827 |
| chr1_-_157108632 | 0.74 |
ENSMUST00000118207.1
ENSMUST00000027884.6 ENSMUST00000121911.1 |
Tex35
|
testis expressed 35 |
| chr6_-_87809757 | 0.73 |
ENSMUST00000032134.7
|
Rab43
|
RAB43, member RAS oncogene family |
| chr15_+_59315030 | 0.72 |
ENSMUST00000022977.7
|
Sqle
|
squalene epoxidase |
| chr6_+_29853746 | 0.72 |
ENSMUST00000064872.6
ENSMUST00000152581.1 ENSMUST00000176265.1 ENSMUST00000154079.1 |
Ahcyl2
|
S-adenosylhomocysteine hydrolase-like 2 |
| chr9_-_114781986 | 0.72 |
ENSMUST00000035009.8
ENSMUST00000084867.7 |
Cmtm7
|
CKLF-like MARVEL transmembrane domain containing 7 |
| chr5_+_150952607 | 0.71 |
ENSMUST00000078856.6
|
Kl
|
klotho |
| chr2_-_27246814 | 0.70 |
ENSMUST00000149733.1
|
Sardh
|
sarcosine dehydrogenase |
| chr3_+_96181151 | 0.70 |
ENSMUST00000035371.8
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr2_-_75938407 | 0.70 |
ENSMUST00000099996.3
|
Ttc30b
|
tetratricopeptide repeat domain 30B |
| chr15_-_100599983 | 0.69 |
ENSMUST00000073837.6
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr9_+_120539801 | 0.69 |
ENSMUST00000047687.7
|
Entpd3
|
ectonucleoside triphosphate diphosphohydrolase 3 |
| chr1_+_36511867 | 0.68 |
ENSMUST00000001166.7
ENSMUST00000097776.3 |
Cnnm3
|
cyclin M3 |
| chr4_-_141933080 | 0.68 |
ENSMUST00000036701.7
|
Fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr11_-_69685537 | 0.68 |
ENSMUST00000018896.7
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chrX_+_36328353 | 0.68 |
ENSMUST00000016383.3
|
Lonrf3
|
LON peptidase N-terminal domain and ring finger 3 |
| chr6_-_54566484 | 0.68 |
ENSMUST00000019268.4
|
Scrn1
|
secernin 1 |
| chr18_-_21652362 | 0.67 |
ENSMUST00000049105.4
|
Klhl14
|
kelch-like 14 |
| chr11_-_99986593 | 0.67 |
ENSMUST00000105050.2
|
Krtap16-1
|
keratin associated protein 16-1 |
| chr15_+_80671829 | 0.67 |
ENSMUST00000023044.5
|
Fam83f
|
family with sequence similarity 83, member F |
| chr5_-_100159261 | 0.66 |
ENSMUST00000139520.1
|
Tmem150c
|
transmembrane protein 150C |
| chr12_-_110682606 | 0.66 |
ENSMUST00000070659.5
|
1700001K19Rik
|
RIKEN cDNA 1700001K19 gene |
| chr12_+_108334341 | 0.65 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr15_+_89429560 | 0.65 |
ENSMUST00000168646.1
|
C730034F03Rik
|
RIKEN cDNA C730034F03 gene |
| chr5_-_31202215 | 0.64 |
ENSMUST00000176245.1
ENSMUST00000177310.1 ENSMUST00000114590.1 |
Zfp513
|
zinc finger protein 513 |
| chr8_+_121544378 | 0.64 |
ENSMUST00000181133.1
ENSMUST00000181679.1 |
1700030M09Rik
|
RIKEN cDNA 1700030M09 gene |
| chr17_-_87265866 | 0.64 |
ENSMUST00000145895.1
ENSMUST00000129616.1 ENSMUST00000155904.1 ENSMUST00000151155.1 ENSMUST00000144236.1 ENSMUST00000024963.3 |
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr11_+_5520652 | 0.64 |
ENSMUST00000063084.9
|
Xbp1
|
X-box binding protein 1 |
| chr9_+_57130690 | 0.63 |
ENSMUST00000160147.1
ENSMUST00000161663.1 ENSMUST00000034836.9 ENSMUST00000161182.1 |
Man2c1
|
mannosidase, alpha, class 2C, member 1 |
| chrX_+_143518671 | 0.63 |
ENSMUST00000134402.1
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr9_-_78587968 | 0.63 |
ENSMUST00000117645.1
ENSMUST00000119213.1 ENSMUST00000052441.5 |
Slc17a5
|
solute carrier family 17 (anion/sugar transporter), member 5 |
| chr10_+_118860826 | 0.63 |
ENSMUST00000059966.4
|
4932442E05Rik
|
RIKEN cDNA 4932442E05 gene |
| chr5_+_115466234 | 0.63 |
ENSMUST00000145785.1
ENSMUST00000031495.4 ENSMUST00000112071.1 ENSMUST00000125568.1 |
Pla2g1b
|
phospholipase A2, group IB, pancreas |
| chr15_+_59315088 | 0.63 |
ENSMUST00000100640.4
|
Sqle
|
squalene epoxidase |
| chr12_+_32954179 | 0.63 |
ENSMUST00000020885.6
|
Sypl
|
synaptophysin-like protein |
| chr2_-_27072175 | 0.62 |
ENSMUST00000009358.2
|
Tmem8c
|
transmembrane protein 8C |
| chr11_+_71749914 | 0.62 |
ENSMUST00000150531.1
|
Wscd1
|
WSC domain containing 1 |
| chr11_-_30025915 | 0.62 |
ENSMUST00000058902.5
|
Eml6
|
echinoderm microtubule associated protein like 6 |
| chr6_+_29694204 | 0.62 |
ENSMUST00000046750.7
ENSMUST00000115250.3 |
Tspan33
|
tetraspanin 33 |
| chr10_-_53647080 | 0.62 |
ENSMUST00000169866.1
|
Fam184a
|
family with sequence similarity 184, member A |
| chr3_-_84305385 | 0.62 |
ENSMUST00000122849.1
ENSMUST00000132283.1 |
Trim2
|
tripartite motif-containing 2 |
| chr17_-_33951438 | 0.62 |
ENSMUST00000087543.2
|
B3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
| chr8_+_113635787 | 0.61 |
ENSMUST00000035777.8
|
Mon1b
|
MON1 homolog b (yeast) |
| chr2_+_69135799 | 0.61 |
ENSMUST00000041865.7
|
Nostrin
|
nitric oxide synthase trafficker |
| chr10_-_127121125 | 0.61 |
ENSMUST00000164259.1
ENSMUST00000080975.4 |
Os9
|
amplified in osteosarcoma |
| chr12_-_112942102 | 0.61 |
ENSMUST00000002881.3
|
Nudt14
|
nudix (nucleoside diphosphate linked moiety X)-type motif 14 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tcf21_Msc
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 4.1 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 1.1 | 4.2 | GO:0003165 | Purkinje myocyte development(GO:0003165) |
| 1.0 | 4.2 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.8 | 2.3 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.5 | 1.6 | GO:0002582 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.4 | 2.6 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.4 | 1.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.4 | 1.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.4 | 2.1 | GO:0014826 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) vein smooth muscle contraction(GO:0014826) |
| 0.3 | 1.7 | GO:1904382 | protein deglycosylation involved in glycoprotein catabolic process(GO:0035977) protein demannosylation(GO:0036507) protein alpha-1,2-demannosylation(GO:0036508) glycoprotein ERAD pathway(GO:0097466) mannose trimming involved in glycoprotein ERAD pathway(GO:1904382) |
| 0.3 | 1.6 | GO:0061511 | centriole elongation(GO:0061511) |
| 0.3 | 1.5 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.3 | 2.1 | GO:0010273 | detoxification of copper ion(GO:0010273) stress response to copper ion(GO:1990169) |
| 0.3 | 1.3 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.3 | 0.5 | GO:0060168 | positive regulation of adenosine receptor signaling pathway(GO:0060168) |
| 0.2 | 1.0 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.2 | 1.2 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.2 | 0.7 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 1.4 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.2 | 0.9 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.2 | 0.7 | GO:1901052 | sarcosine metabolic process(GO:1901052) sarcosine catabolic process(GO:1901053) |
| 0.2 | 1.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.7 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.2 | 0.9 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.2 | 0.6 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 0.6 | GO:0071332 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) cellular response to fructose stimulus(GO:0071332) |
| 0.2 | 1.9 | GO:0006517 | protein deglycosylation(GO:0006517) |
| 0.2 | 0.6 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.2 | 1.2 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.2 | 2.2 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.2 | 2.9 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.2 | 0.6 | GO:0010446 | response to alkaline pH(GO:0010446) |
| 0.2 | 0.7 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.2 | 0.5 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.2 | 1.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.2 | 1.0 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.2 | 2.6 | GO:0060363 | cranial suture morphogenesis(GO:0060363) |
| 0.2 | 1.0 | GO:0003383 | apical constriction(GO:0003383) |
| 0.2 | 1.2 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.2 | 0.8 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.2 | 1.6 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.2 | 2.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.2 | 0.5 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.2 | 0.3 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.2 | 1.7 | GO:0044336 | canonical Wnt signaling pathway involved in negative regulation of apoptotic process(GO:0044336) |
| 0.1 | 2.5 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.1 | 0.7 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 0.6 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.6 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.7 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.7 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.1 | 0.4 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.1 | 0.7 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 1.9 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 0.4 | GO:2000016 | mammary gland bud morphogenesis(GO:0060648) negative regulation of determination of dorsal identity(GO:2000016) |
| 0.1 | 0.3 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.1 | 1.6 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 1.3 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.1 | 1.9 | GO:0019377 | glycolipid catabolic process(GO:0019377) |
| 0.1 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.1 | 1.6 | GO:0048199 | vesicle targeting, to, from or within Golgi(GO:0048199) |
| 0.1 | 1.1 | GO:0030432 | peristalsis(GO:0030432) |
| 0.1 | 0.3 | GO:0021650 | vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.3 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.3 | GO:1900060 | negative regulation of ceramide biosynthetic process(GO:1900060) |
| 0.1 | 0.3 | GO:1903070 | positive regulation of protein lipidation(GO:1903061) negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.5 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.3 | GO:0070676 | intralumenal vesicle formation(GO:0070676) negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.1 | 0.9 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 1.3 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 0.4 | GO:1903944 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.1 | 4.9 | GO:0009409 | response to cold(GO:0009409) |
| 0.1 | 0.4 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.4 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.1 | 0.6 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.1 | 1.3 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.1 | 1.7 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.5 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.1 | 0.5 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.1 | 1.3 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.1 | 0.7 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.1 | 0.4 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.4 | GO:0009816 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.1 | 1.7 | GO:0097113 | AMPA glutamate receptor clustering(GO:0097113) |
| 0.1 | 0.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.1 | 1.0 | GO:0014857 | regulation of skeletal muscle cell proliferation(GO:0014857) |
| 0.1 | 0.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.9 | GO:0014821 | phasic smooth muscle contraction(GO:0014821) |
| 0.1 | 0.9 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.1 | 0.3 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.1 | 0.5 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.1 | 2.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.1 | 0.5 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 0.6 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 1.0 | GO:0098903 | regulation of membrane repolarization during action potential(GO:0098903) |
| 0.1 | 0.9 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.1 | 0.3 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.6 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.1 | 0.2 | GO:0048549 | positive regulation of pinocytosis(GO:0048549) |
| 0.1 | 0.2 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) L-cystine transport(GO:0015811) |
| 0.1 | 0.2 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) sensory system development(GO:0048880) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.3 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.1 | 0.7 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.1 | 1.5 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.1 | 0.3 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.2 | GO:0070340 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.8 | GO:0061365 | intestinal cholesterol absorption(GO:0030299) positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 1.4 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.1 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.1 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.1 | 0.2 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.1 | 0.7 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.1 | 0.3 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.1 | 0.1 | GO:2000832 | negative regulation of steroid hormone secretion(GO:2000832) |
| 0.1 | 0.8 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.1 | 0.6 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.1 | 5.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.1 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.1 | 0.3 | GO:2000564 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) CD8-positive, alpha-beta T cell proliferation(GO:0035740) regulation of CD8-positive, alpha-beta T cell proliferation(GO:2000564) |
| 0.1 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.1 | 0.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.4 | GO:0010909 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) |
| 0.1 | 0.2 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) negative regulation of vascular endothelial growth factor production(GO:1904046) |
| 0.0 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.2 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.7 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.6 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.6 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.4 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.3 | GO:0006083 | acetate metabolic process(GO:0006083) |
| 0.0 | 0.4 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.6 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 1.3 | GO:0046596 | regulation of viral entry into host cell(GO:0046596) |
| 0.0 | 0.3 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.5 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 8.5 | GO:1990823 | response to leukemia inhibitory factor(GO:1990823) cellular response to leukemia inhibitory factor(GO:1990830) |
| 0.0 | 0.5 | GO:0032119 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.2 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:0071718 | sodium-independent icosanoid transport(GO:0071718) |
| 0.0 | 0.1 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.0 | 1.5 | GO:0046463 | triglyceride biosynthetic process(GO:0019432) neutral lipid biosynthetic process(GO:0046460) acylglycerol biosynthetic process(GO:0046463) |
| 0.0 | 0.2 | GO:0042125 | protein glycosylation at cell surface(GO:0033575) protein galactosylation at cell surface(GO:0033580) protein galactosylation(GO:0042125) |
| 0.0 | 0.2 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.0 | 0.4 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.4 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of female gonad development(GO:2000196) positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 2.3 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 1.0 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 1.9 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.6 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.0 | 0.3 | GO:0046865 | retinol transport(GO:0034633) isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) alveolar primary septum development(GO:0061143) |
| 0.0 | 0.5 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0043385 | mycotoxin metabolic process(GO:0043385) mycotoxin catabolic process(GO:0043387) aflatoxin metabolic process(GO:0046222) aflatoxin catabolic process(GO:0046223) organic heteropentacyclic compound metabolic process(GO:1901376) organic heteropentacyclic compound catabolic process(GO:1901377) |
| 0.0 | 0.3 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.5 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 1.1 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.8 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.9 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.0 | 0.5 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.3 | GO:0015791 | polyol transport(GO:0015791) |
| 0.0 | 0.3 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 1.2 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.5 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 1.9 | GO:0032611 | interleukin-1 beta production(GO:0032611) |
| 0.0 | 0.4 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of autophagosome maturation(GO:1901097) negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:0042450 | arginine biosynthetic process via ornithine(GO:0042450) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.2 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.2 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.0 | 1.2 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.5 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.0 | 0.3 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 1.6 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.2 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 1.0 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.2 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.4 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.3 | GO:1900112 | regulation of histone H3-K9 trimethylation(GO:1900112) negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.4 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.5 | GO:0090502 | RNA phosphodiester bond hydrolysis, endonucleolytic(GO:0090502) |
| 0.0 | 0.2 | GO:1901386 | negative regulation of voltage-gated calcium channel activity(GO:1901386) |
| 0.0 | 0.2 | GO:2000463 | positive regulation of excitatory postsynaptic potential(GO:2000463) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.5 | GO:1903432 | regulation of TORC1 signaling(GO:1903432) |
| 0.0 | 0.9 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 2.0 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.1 | GO:0002036 | regulation of L-glutamate transport(GO:0002036) |
| 0.0 | 0.3 | GO:0019370 | leukotriene biosynthetic process(GO:0019370) |
| 0.0 | 0.8 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.4 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.0 | 0.1 | GO:1902237 | positive regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902237) |
| 0.0 | 1.6 | GO:0008203 | cholesterol metabolic process(GO:0008203) |
| 0.0 | 0.0 | GO:0071799 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.2 | GO:0090003 | regulation of Golgi to plasma membrane protein transport(GO:0042996) regulation of establishment of protein localization to plasma membrane(GO:0090003) |
| 0.0 | 0.2 | GO:0007635 | chemosensory behavior(GO:0007635) |
| 0.0 | 0.2 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.2 | GO:0005980 | glycogen catabolic process(GO:0005980) glucan catabolic process(GO:0009251) cellular polysaccharide catabolic process(GO:0044247) |
| 0.0 | 0.3 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.7 | GO:0042073 | intraciliary transport(GO:0042073) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.6 | GO:1901264 | carbohydrate derivative transport(GO:1901264) |
| 0.0 | 0.3 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.9 | GO:0009060 | aerobic respiration(GO:0009060) |
| 0.0 | 1.0 | GO:0030216 | keratinocyte differentiation(GO:0030216) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.3 | GO:0048268 | clathrin coat assembly(GO:0048268) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 6.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.7 | 2.2 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.4 | 1.6 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.4 | 1.5 | GO:1990696 | USH2 complex(GO:1990696) |
| 0.3 | 2.6 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.2 | 1.0 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.2 | 5.5 | GO:0030057 | desmosome(GO:0030057) |
| 0.2 | 3.3 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.2 | 4.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 1.0 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.1 | 0.4 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.1 | 1.6 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.1 | 1.4 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.5 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 2.5 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.1 | 1.6 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.0 | GO:1990454 | L-type voltage-gated calcium channel complex(GO:1990454) |
| 0.1 | 1.4 | GO:0035748 | myelin sheath abaxonal region(GO:0035748) |
| 0.1 | 2.5 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.1 | 0.3 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.1 | 0.3 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.1 | 0.5 | GO:0098651 | collagen type IV trimer(GO:0005587) network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) basement membrane collagen trimer(GO:0098651) |
| 0.1 | 0.5 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.1 | 0.6 | GO:0000835 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.1 | 0.8 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.1 | 1.3 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.1 | 0.6 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 4.1 | GO:0031985 | Golgi cisterna(GO:0031985) |
| 0.1 | 0.6 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.2 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.1 | 0.4 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.1 | 1.2 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.1 | 0.5 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.8 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.1 | 0.4 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0036019 | endolysosome(GO:0036019) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.4 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.7 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 2.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.4 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.5 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.3 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 1.5 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.3 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.4 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 1.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.3 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.3 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.5 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.3 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.8 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.5 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.3 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.7 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.5 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.3 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 1.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0098560 | cytoplasmic side of endosome membrane(GO:0010009) cytoplasmic side of late endosome membrane(GO:0098560) cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 1.3 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.5 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 2.8 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 1.3 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 1.5 | GO:0036064 | ciliary basal body(GO:0036064) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 2.4 | GO:0044306 | neuron projection terminus(GO:0044306) |
| 0.0 | 0.5 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.8 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.6 | 6.2 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.8 | 2.3 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.6 | 1.8 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.6 | 1.7 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.5 | 2.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.4 | 2.5 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 1.8 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.3 | 2.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.3 | 0.8 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 1.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.2 | 1.9 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.2 | 3.6 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.2 | 0.7 | GO:0046997 | sarcosine dehydrogenase activity(GO:0008480) oxidoreductase activity, acting on the CH-NH group of donors, flavin as acceptor(GO:0046997) |
| 0.2 | 1.6 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 1.3 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.2 | 1.6 | GO:0009374 | biotin binding(GO:0009374) |
| 0.2 | 0.6 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.2 | 0.5 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.2 | 0.7 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.2 | 1.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.5 | GO:0071633 | dihydroceramidase activity(GO:0071633) |
| 0.2 | 2.4 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.2 | 1.6 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.2 | 0.5 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.2 | 0.5 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.2 | 0.6 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 1.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 3.3 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.1 | 0.8 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 3.7 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.1 | 2.1 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.4 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.1 | 3.3 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.3 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 3.6 | GO:0043236 | laminin binding(GO:0043236) |
| 0.1 | 0.7 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 3.6 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.6 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.1 | 1.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.1 | 0.4 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.1 | 0.3 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 1.6 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.1 | 0.3 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 1.3 | GO:0016918 | retinal binding(GO:0016918) retinol binding(GO:0019841) |
| 0.1 | 1.6 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.7 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.5 | GO:0033265 | choline binding(GO:0033265) |
| 0.1 | 1.3 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.8 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.1 | 0.6 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.1 | 0.5 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.1 | 0.6 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 2.6 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.1 | 0.9 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) serotonin binding(GO:0051378) |
| 0.1 | 0.3 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.2 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.1 | 0.6 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 0.7 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.4 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 1.4 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.7 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.1 | 0.5 | GO:0016661 | oxidoreductase activity, acting on other nitrogenous compounds as donors(GO:0016661) |
| 0.1 | 0.3 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.1 | 0.2 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 1.0 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 1.4 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.1 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 2.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.5 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.1 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.1 | 0.3 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.3 | GO:0070728 | leucine binding(GO:0070728) |
| 0.1 | 0.5 | GO:0046790 | virion binding(GO:0046790) |
| 0.1 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.1 | 0.2 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.1 | 0.5 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.1 | 1.8 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.4 | GO:0036122 | BMP binding(GO:0036122) |
| 0.0 | 0.3 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.4 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.6 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.6 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.5 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 2.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 7.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.6 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.7 | GO:0016805 | dipeptidase activity(GO:0016805) |
| 0.0 | 1.7 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.7 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 2.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 1.7 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.1 | GO:0032129 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.2 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0045322 | unmethylated CpG binding(GO:0045322) |
| 0.0 | 0.3 | GO:0070008 | serine-type carboxypeptidase activity(GO:0004185) serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.5 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0047276 | N-acetyllactosaminide 3-alpha-galactosyltransferase activity(GO:0047276) |
| 0.0 | 0.1 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.0 | 1.0 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.2 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.0 | 0.3 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 1.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 1.3 | GO:0008138 | protein tyrosine/serine/threonine phosphatase activity(GO:0008138) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.6 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0016743 | carboxyl- or carbamoyltransferase activity(GO:0016743) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.8 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.2 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.5 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.7 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.8 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 1.7 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 1.7 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 4.8 | GO:0003924 | GTPase activity(GO:0003924) |
| 0.0 | 0.4 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 1.5 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.5 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 6.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.4 | GO:0001786 | phosphatidylserine binding(GO:0001786) |
| 0.0 | 0.6 | GO:0008146 | sulfotransferase activity(GO:0008146) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.1 | 4.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 1.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.1 | 0.8 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 1.0 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.7 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 2.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.8 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 2.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 1.1 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 1.0 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.8 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.8 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.9 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 2.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.9 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.9 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.5 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 2.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 1.0 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 1.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.4 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.5 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.4 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.6 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.2 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 4.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 2.0 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 2.1 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.1 | 1.7 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.1 | 0.6 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.6 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.9 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.1 | 1.9 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.1 | 1.9 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.1 | 1.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.1 | 0.6 | REACTOME OPSINS | Genes involved in Opsins |
| 0.1 | 2.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 1.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.9 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.7 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 1.6 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 2.5 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 2.0 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 5.2 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 0.8 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 1.0 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 0.9 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.6 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.5 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 5.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 1.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.0 | 0.9 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.8 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.6 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.7 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.6 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.5 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.9 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.3 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.0 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.7 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 2.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.3 | REACTOME RESOLUTION OF AP SITES VIA THE MULTIPLE NUCLEOTIDE PATCH REPLACEMENT PATHWAY | Genes involved in Resolution of AP sites via the multiple-nucleotide patch replacement pathway |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.5 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.3 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.0 | 0.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |