Project
GFI1 WT vs 36n/n vs KD
Navigation
Downloads
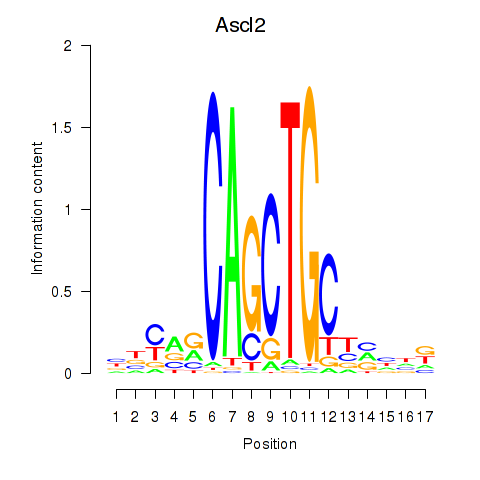
Results for Ascl2
Z-value: 0.45
Transcription factors associated with Ascl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ascl2
|
ENSMUSG00000009248.7 | achaete-scute family bHLH transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ascl2 | mm39_v1_chr7_-_142522454_142522514 | -0.91 | 3.2e-02 | Click! |
Activity profile of Ascl2 motif
Sorted Z-values of Ascl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_12342715 | 0.44 |
ENSMUST00000233892.2
|
Vmn2r53
|
vomeronasal 2, receptor 53 |
| chr2_-_5719302 | 0.43 |
ENSMUST00000044009.14
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr5_-_125201872 | 0.41 |
ENSMUST00000055256.14
|
Ncor2
|
nuclear receptor co-repressor 2 |
| chr7_-_12468931 | 0.34 |
ENSMUST00000233304.2
ENSMUST00000233373.2 ENSMUST00000233874.2 ENSMUST00000233902.2 |
Vmn2r56
|
vomeronasal 2, receptor 56 |
| chr7_+_3339059 | 0.30 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_+_3339077 | 0.30 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr4_+_140633646 | 0.30 |
ENSMUST00000030765.7
|
Padi2
|
peptidyl arginine deiminase, type II |
| chr15_-_101268036 | 0.26 |
ENSMUST00000077196.6
|
Krt80
|
keratin 80 |
| chr17_+_69071546 | 0.25 |
ENSMUST00000233625.2
|
L3mbtl4
|
L3MBTL4 histone methyl-lysine binding protein |
| chr6_-_25689781 | 0.24 |
ENSMUST00000200812.2
|
Gpr37
|
G protein-coupled receptor 37 |
| chr5_+_135197228 | 0.24 |
ENSMUST00000111187.10
ENSMUST00000111188.5 ENSMUST00000202606.3 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr10_-_127016448 | 0.24 |
ENSMUST00000222911.3
ENSMUST00000095270.3 |
Slc26a10
|
solute carrier family 26, member 10 |
| chr2_-_5947551 | 0.23 |
ENSMUST00000026924.7
ENSMUST00000095147.9 |
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr6_+_82379456 | 0.23 |
ENSMUST00000032122.11
|
Tacr1
|
tachykinin receptor 1 |
| chr17_+_56259617 | 0.22 |
ENSMUST00000003274.8
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr6_+_82379768 | 0.22 |
ENSMUST00000203775.2
|
Tacr1
|
tachykinin receptor 1 |
| chr13_+_93908138 | 0.22 |
ENSMUST00000091403.6
|
Arsb
|
arylsulfatase B |
| chr3_+_135143910 | 0.22 |
ENSMUST00000196446.5
ENSMUST00000106291.10 ENSMUST00000199613.5 |
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr7_-_114162125 | 0.22 |
ENSMUST00000211506.2
ENSMUST00000119712.8 ENSMUST00000032908.15 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr17_-_89099404 | 0.21 |
ENSMUST00000024916.7
|
Lhcgr
|
luteinizing hormone/choriogonadotropin receptor |
| chr8_+_66419809 | 0.21 |
ENSMUST00000072482.13
|
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr1_+_87254729 | 0.21 |
ENSMUST00000172794.8
ENSMUST00000164992.9 ENSMUST00000173173.8 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr1_-_88133472 | 0.21 |
ENSMUST00000119972.4
|
Dnajb3
|
DnaJ heat shock protein family (Hsp40) member B3 |
| chr8_+_66070661 | 0.21 |
ENSMUST00000110258.8
ENSMUST00000110256.8 ENSMUST00000110255.8 |
Marchf1
|
membrane associated ring-CH-type finger 1 |
| chr3_-_131096792 | 0.21 |
ENSMUST00000200236.2
ENSMUST00000106337.7 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr7_+_142014546 | 0.20 |
ENSMUST00000105968.8
ENSMUST00000018963.11 ENSMUST00000105967.8 |
Lsp1
|
lymphocyte specific 1 |
| chr3_-_129763801 | 0.20 |
ENSMUST00000029624.15
|
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr2_+_118610184 | 0.20 |
ENSMUST00000063975.10
ENSMUST00000037547.9 ENSMUST00000110846.8 ENSMUST00000110843.2 |
Disp2
|
dispatched RND tramsporter family member 2 |
| chr19_+_10502612 | 0.20 |
ENSMUST00000237321.2
ENSMUST00000038379.5 |
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr11_+_33913013 | 0.19 |
ENSMUST00000020362.3
|
Kcnmb1
|
potassium large conductance calcium-activated channel, subfamily M, beta member 1 |
| chr7_+_142025817 | 0.19 |
ENSMUST00000105966.2
|
Lsp1
|
lymphocyte specific 1 |
| chr15_-_74983786 | 0.19 |
ENSMUST00000191451.2
ENSMUST00000100542.10 |
Ly6c2
|
lymphocyte antigen 6 complex, locus C2 |
| chr7_-_106531426 | 0.18 |
ENSMUST00000215468.2
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1 |
| chr1_-_60605867 | 0.18 |
ENSMUST00000027168.12
ENSMUST00000090293.11 ENSMUST00000140485.8 |
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr9_-_57743989 | 0.18 |
ENSMUST00000164010.8
ENSMUST00000171444.8 ENSMUST00000098686.4 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr7_+_142025575 | 0.18 |
ENSMUST00000038946.9
|
Lsp1
|
lymphocyte specific 1 |
| chr15_-_76090508 | 0.17 |
ENSMUST00000073418.13
ENSMUST00000171634.8 ENSMUST00000076442.12 |
Plec
|
plectin |
| chr5_-_66330394 | 0.17 |
ENSMUST00000201544.4
|
Rbm47
|
RNA binding motif protein 47 |
| chr9_+_44893077 | 0.17 |
ENSMUST00000034602.9
|
Cd3d
|
CD3 antigen, delta polypeptide |
| chrX_-_166047275 | 0.17 |
ENSMUST00000112170.2
|
Tlr8
|
toll-like receptor 8 |
| chr16_+_51852435 | 0.17 |
ENSMUST00000227879.2
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr7_+_44465714 | 0.17 |
ENSMUST00000208172.2
|
Nup62
|
nucleoporin 62 |
| chr19_+_10502679 | 0.17 |
ENSMUST00000235674.2
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr5_-_125119855 | 0.17 |
ENSMUST00000125053.8
|
Ncor2
|
nuclear receptor co-repressor 2 |
| chr3_-_129763638 | 0.17 |
ENSMUST00000146340.2
ENSMUST00000153506.8 |
Mcub
|
mitochondrial calcium uniporter dominant negative beta subunit |
| chr8_-_32499513 | 0.16 |
ENSMUST00000208205.3
|
Nrg1
|
neuregulin 1 |
| chr17_-_32408431 | 0.16 |
ENSMUST00000087721.10
ENSMUST00000162117.3 |
Ephx3
|
epoxide hydrolase 3 |
| chr2_+_32253692 | 0.16 |
ENSMUST00000113331.8
ENSMUST00000113338.9 |
Ciz1
|
CDKN1A interacting zinc finger protein 1 |
| chr8_-_32499556 | 0.15 |
ENSMUST00000207470.3
ENSMUST00000207417.3 ENSMUST00000238826.2 |
Nrg1
|
neuregulin 1 |
| chr7_-_140590605 | 0.15 |
ENSMUST00000026565.7
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr1_+_177269845 | 0.15 |
ENSMUST00000195002.2
|
Zbtb18
|
zinc finger and BTB domain containing 18 |
| chr12_+_105302853 | 0.15 |
ENSMUST00000180458.9
|
Tunar
|
Tcl1 upstream neural differentiation associated RNA |
| chr9_-_62888156 | 0.15 |
ENSMUST00000098651.6
ENSMUST00000214830.2 |
Pias1
|
protein inhibitor of activated STAT 1 |
| chr8_+_106002772 | 0.15 |
ENSMUST00000014920.8
|
Nol3
|
nucleolar protein 3 (apoptosis repressor with CARD domain) |
| chr15_-_58078274 | 0.15 |
ENSMUST00000022986.8
|
Fbxo32
|
F-box protein 32 |
| chr1_-_130643471 | 0.14 |
ENSMUST00000066863.13
ENSMUST00000169659.8 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr5_+_135197137 | 0.14 |
ENSMUST00000031692.12
|
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr16_+_57173456 | 0.14 |
ENSMUST00000159816.8
|
Filip1l
|
filamin A interacting protein 1-like |
| chr11_+_69806866 | 0.14 |
ENSMUST00000134581.2
|
Gps2
|
G protein pathway suppressor 2 |
| chr4_-_156312961 | 0.14 |
ENSMUST00000217885.2
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr5_+_123280250 | 0.14 |
ENSMUST00000174836.8
ENSMUST00000163030.9 |
Setd1b
|
SET domain containing 1B |
| chr11_-_78950698 | 0.14 |
ENSMUST00000141409.8
|
Ksr1
|
kinase suppressor of ras 1 |
| chr19_+_37423198 | 0.14 |
ENSMUST00000025944.9
|
Hhex
|
hematopoietically expressed homeobox |
| chr4_-_156312996 | 0.13 |
ENSMUST00000105571.4
|
Plekhn1
|
pleckstrin homology domain containing, family N member 1 |
| chr10_-_7162196 | 0.13 |
ENSMUST00000015346.12
|
Cnksr3
|
Cnksr family member 3 |
| chr7_+_48895879 | 0.13 |
ENSMUST00000064395.13
|
Nav2
|
neuron navigator 2 |
| chr12_-_114752425 | 0.13 |
ENSMUST00000103510.2
|
Ighv1-26
|
immunoglobulin heavy variable 1-26 |
| chr3_+_131791042 | 0.13 |
ENSMUST00000029665.7
|
Dkk2
|
dickkopf WNT signaling pathway inhibitor 2 |
| chr15_+_101310283 | 0.13 |
ENSMUST00000068904.9
|
Krt7
|
keratin 7 |
| chr2_+_32617671 | 0.13 |
ENSMUST00000113242.5
|
Sh2d3c
|
SH2 domain containing 3C |
| chr14_-_104081119 | 0.13 |
ENSMUST00000227824.2
ENSMUST00000172237.2 |
Ednrb
|
endothelin receptor type B |
| chr7_-_84339156 | 0.12 |
ENSMUST00000209117.2
ENSMUST00000207975.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr2_-_103315483 | 0.12 |
ENSMUST00000028610.10
|
Cat
|
catalase |
| chr18_+_38809771 | 0.12 |
ENSMUST00000134388.2
ENSMUST00000148850.8 |
9630014M24Rik
Arhgap26
|
RIKEN cDNA 9630014M24 gene Rho GTPase activating protein 26 |
| chr6_-_34887530 | 0.12 |
ENSMUST00000149448.8
ENSMUST00000133336.8 |
Wdr91
|
WD repeat domain 91 |
| chr16_+_21613068 | 0.12 |
ENSMUST00000211443.2
ENSMUST00000231300.2 ENSMUST00000209449.2 ENSMUST00000181780.9 ENSMUST00000209728.2 ENSMUST00000181960.3 ENSMUST00000209429.2 ENSMUST00000180830.3 ENSMUST00000231988.2 |
1300002E11Rik
Map3k13
|
RIKEN cDNA 1300002E11 gene mitogen-activated protein kinase kinase kinase 13 |
| chr3_+_107803137 | 0.12 |
ENSMUST00000004134.11
|
Gstm5
|
glutathione S-transferase, mu 5 |
| chr12_+_17740831 | 0.12 |
ENSMUST00000071858.5
|
Hpcal1
|
hippocalcin-like 1 |
| chr1_-_167221344 | 0.11 |
ENSMUST00000028005.3
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr2_+_89804937 | 0.11 |
ENSMUST00000214630.2
ENSMUST00000111512.10 ENSMUST00000144710.3 ENSMUST00000216678.2 |
Olfr1260
|
olfactory receptor 1260 |
| chr4_-_82803384 | 0.11 |
ENSMUST00000048430.4
|
Cer1
|
cerberus 1, DAN family BMP antagonist |
| chr15_+_61858883 | 0.11 |
ENSMUST00000159338.2
|
Myc
|
myelocytomatosis oncogene |
| chr8_-_106198112 | 0.11 |
ENSMUST00000014990.13
|
Tppp3
|
tubulin polymerization-promoting protein family member 3 |
| chrX_-_97934387 | 0.11 |
ENSMUST00000113826.8
ENSMUST00000033560.9 ENSMUST00000142267.2 |
Ophn1
|
oligophrenin 1 |
| chr1_+_132119169 | 0.11 |
ENSMUST00000188169.7
ENSMUST00000112357.9 ENSMUST00000188175.2 |
Lemd1
Gm29695
|
LEM domain containing 1 predicted gene, 29695 |
| chr14_+_66378382 | 0.11 |
ENSMUST00000022620.11
|
Chrna2
|
cholinergic receptor, nicotinic, alpha polypeptide 2 (neuronal) |
| chr5_+_117979899 | 0.11 |
ENSMUST00000142742.9
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr4_-_155870321 | 0.11 |
ENSMUST00000097742.3
|
Tmem88b
|
transmembrane protein 88B |
| chr12_-_76224025 | 0.11 |
ENSMUST00000101291.11
ENSMUST00000218621.2 ENSMUST00000076634.5 |
Esr2
|
estrogen receptor 2 (beta) |
| chrX_+_7439839 | 0.11 |
ENSMUST00000144719.9
ENSMUST00000234896.2 |
Flicr
Foxp3
|
Foxp3 regulating long intergenic noncoding RNA forkhead box P3 |
| chr1_-_184615415 | 0.11 |
ENSMUST00000048308.6
|
C130074G19Rik
|
RIKEN cDNA C130074G19 gene |
| chr11_+_53410697 | 0.11 |
ENSMUST00000120878.9
ENSMUST00000147912.2 |
Septin8
|
septin 8 |
| chr13_+_95833359 | 0.11 |
ENSMUST00000022182.5
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr10_+_4432488 | 0.11 |
ENSMUST00000138112.8
|
Ccdc170
|
coiled-coil domain containing 170 |
| chr5_+_149335214 | 0.11 |
ENSMUST00000093110.12
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr15_-_103231921 | 0.10 |
ENSMUST00000229551.2
|
Zfp385a
|
zinc finger protein 385A |
| chr6_-_77956499 | 0.10 |
ENSMUST00000159626.8
ENSMUST00000075340.12 ENSMUST00000162273.2 |
Ctnna2
|
catenin (cadherin associated protein), alpha 2 |
| chr8_-_106427696 | 0.10 |
ENSMUST00000042608.8
|
Acd
|
adrenocortical dysplasia |
| chr4_+_150699670 | 0.10 |
ENSMUST00000219467.2
|
Rere
|
arginine glutamic acid dipeptide (RE) repeats |
| chr9_+_72892850 | 0.10 |
ENSMUST00000150826.9
ENSMUST00000085350.11 ENSMUST00000140675.8 |
Ccpg1
|
cell cycle progression 1 |
| chr11_+_83855245 | 0.10 |
ENSMUST00000049714.15
ENSMUST00000092834.12 ENSMUST00000183714.8 ENSMUST00000183456.8 |
Synrg
|
synergin, gamma |
| chr7_-_141241632 | 0.10 |
ENSMUST00000239500.1
|
ENSMUSG00000118661.1
|
mucin 6, gastric |
| chr9_-_21963306 | 0.10 |
ENSMUST00000003501.9
ENSMUST00000215901.2 |
Elavl3
|
ELAV like RNA binding protein 3 |
| chr5_-_136275407 | 0.10 |
ENSMUST00000196245.2
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr5_-_31065036 | 0.10 |
ENSMUST00000132034.5
ENSMUST00000132253.5 |
Ost4
|
oligosaccharyltransferase complex subunit 4 (non-catalytic) |
| chr7_+_75105282 | 0.10 |
ENSMUST00000207750.2
ENSMUST00000166315.7 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr4_-_66322695 | 0.10 |
ENSMUST00000084496.3
|
Astn2
|
astrotactin 2 |
| chr2_+_164782642 | 0.09 |
ENSMUST00000137626.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr16_+_14523696 | 0.09 |
ENSMUST00000023356.8
|
Snai2
|
snail family zinc finger 2 |
| chr7_+_48896560 | 0.09 |
ENSMUST00000184945.8
|
Nav2
|
neuron navigator 2 |
| chr3_+_135144202 | 0.09 |
ENSMUST00000166033.6
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_-_18397547 | 0.09 |
ENSMUST00000091418.12
ENSMUST00000166495.8 |
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr17_+_48571298 | 0.09 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr4_-_148236516 | 0.09 |
ENSMUST00000056965.12
ENSMUST00000168503.8 ENSMUST00000152098.8 |
Fbxo6
|
F-box protein 6 |
| chr4_+_101843823 | 0.09 |
ENSMUST00000106914.8
|
Gm12789
|
predicted gene 12789 |
| chr14_+_33645539 | 0.09 |
ENSMUST00000168727.3
|
Gdf10
|
growth differentiation factor 10 |
| chr4_-_46536088 | 0.09 |
ENSMUST00000102924.3
ENSMUST00000046897.13 |
Trim14
|
tripartite motif-containing 14 |
| chr16_+_14398085 | 0.09 |
ENSMUST00000147024.8
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr11_+_115054157 | 0.09 |
ENSMUST00000021077.4
|
Slc9a3r1
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 |
| chr11_-_69088635 | 0.09 |
ENSMUST00000094078.4
ENSMUST00000021262.10 |
Alox8
|
arachidonate 8-lipoxygenase |
| chr11_+_117700479 | 0.09 |
ENSMUST00000026649.14
ENSMUST00000177131.8 ENSMUST00000120928.2 ENSMUST00000175737.2 ENSMUST00000132298.2 |
Syngr2
Gm20708
|
synaptogyrin 2 predicted gene 20708 |
| chr6_-_125143425 | 0.09 |
ENSMUST00000117757.9
ENSMUST00000073605.15 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr17_-_17845293 | 0.09 |
ENSMUST00000041047.4
|
Lnpep
|
leucyl/cystinyl aminopeptidase |
| chr4_-_66322750 | 0.09 |
ENSMUST00000068214.11
|
Astn2
|
astrotactin 2 |
| chr1_+_89507952 | 0.09 |
ENSMUST00000074945.8
|
Agap1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
| chr6_+_48624295 | 0.09 |
ENSMUST00000078223.6
ENSMUST00000203509.2 |
Gimap8
|
GTPase, IMAP family member 8 |
| chr5_+_137639538 | 0.09 |
ENSMUST00000177466.8
ENSMUST00000166099.3 |
Sap25
|
sin3 associated polypeptide |
| chr5_-_36987917 | 0.09 |
ENSMUST00000031002.10
|
Man2b2
|
mannosidase 2, alpha B2 |
| chr4_-_148215135 | 0.09 |
ENSMUST00000030862.5
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr1_+_131671751 | 0.09 |
ENSMUST00000049027.10
|
Slc26a9
|
solute carrier family 26, member 9 |
| chr7_+_78563964 | 0.09 |
ENSMUST00000120331.4
|
Isg20
|
interferon-stimulated protein |
| chr1_+_120048890 | 0.09 |
ENSMUST00000027637.13
ENSMUST00000112644.9 ENSMUST00000056038.15 |
3110009E18Rik
|
RIKEN cDNA 3110009E18 gene |
| chr9_-_108338111 | 0.08 |
ENSMUST00000193895.6
|
Klhdc8b
|
kelch domain containing 8B |
| chrX_-_16777913 | 0.08 |
ENSMUST00000040134.8
|
Ndp
|
Norrie disease (pseudoglioma) (human) |
| chr3_+_27237143 | 0.08 |
ENSMUST00000091284.5
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr11_-_106679671 | 0.08 |
ENSMUST00000123339.2
|
Ddx5
|
DEAD box helicase 5 |
| chr9_-_21763898 | 0.08 |
ENSMUST00000217336.2
ENSMUST00000034728.9 |
Dock6
|
dedicator of cytokinesis 6 |
| chr10_+_122514669 | 0.08 |
ENSMUST00000161487.8
ENSMUST00000067918.12 |
Ppm1h
|
protein phosphatase 1H (PP2C domain containing) |
| chr2_-_150510116 | 0.08 |
ENSMUST00000028944.4
|
Acss1
|
acyl-CoA synthetase short-chain family member 1 |
| chr7_+_44866635 | 0.08 |
ENSMUST00000097216.5
ENSMUST00000209343.2 ENSMUST00000209678.2 |
Tead2
|
TEA domain family member 2 |
| chr6_+_125326662 | 0.08 |
ENSMUST00000032491.15
|
Tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr11_+_29323618 | 0.08 |
ENSMUST00000040182.13
ENSMUST00000109477.2 |
Ccdc88a
|
coiled coil domain containing 88A |
| chr7_-_28038129 | 0.08 |
ENSMUST00000209141.2
ENSMUST00000003527.10 |
Supt5
|
suppressor of Ty 5, DSIF elongation factor subunit |
| chr15_-_71599664 | 0.08 |
ENSMUST00000022953.10
|
Fam135b
|
family with sequence similarity 135, member B |
| chr9_-_81515865 | 0.07 |
ENSMUST00000183482.2
|
Htr1b
|
5-hydroxytryptamine (serotonin) receptor 1B |
| chr12_+_71062733 | 0.07 |
ENSMUST00000046305.12
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr3_-_100396635 | 0.07 |
ENSMUST00000061455.9
|
Tent5c
|
terminal nucleotidyltransferase 5C |
| chr6_-_52168675 | 0.07 |
ENSMUST00000101395.3
|
Hoxa4
|
homeobox A4 |
| chr6_+_3993774 | 0.07 |
ENSMUST00000031673.7
|
Gngt1
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr5_+_115681003 | 0.07 |
ENSMUST00000157050.8
|
Pxn
|
paxillin |
| chr13_+_16186410 | 0.07 |
ENSMUST00000042603.14
|
Inhba
|
inhibin beta-A |
| chr17_-_57554631 | 0.07 |
ENSMUST00000233568.2
ENSMUST00000005975.8 |
Gpr108
|
G protein-coupled receptor 108 |
| chr7_-_28001624 | 0.07 |
ENSMUST00000108315.4
|
Dll3
|
delta like canonical Notch ligand 3 |
| chr2_-_179976458 | 0.07 |
ENSMUST00000015771.3
|
Gata5
|
GATA binding protein 5 |
| chr2_+_167380112 | 0.07 |
ENSMUST00000052631.8
|
Snai1
|
snail family zinc finger 1 |
| chr9_+_64718708 | 0.07 |
ENSMUST00000213926.2
|
Dennd4a
|
DENN/MADD domain containing 4A |
| chr5_-_106606032 | 0.06 |
ENSMUST00000086795.8
|
Barhl2
|
BarH like homeobox 2 |
| chr3_+_27237114 | 0.06 |
ENSMUST00000046515.15
|
Nceh1
|
neutral cholesterol ester hydrolase 1 |
| chr4_-_57301791 | 0.06 |
ENSMUST00000075637.11
|
Ptpn3
|
protein tyrosine phosphatase, non-receptor type 3 |
| chr4_+_42916666 | 0.06 |
ENSMUST00000132173.8
ENSMUST00000107975.8 |
Phf24
|
PHD finger protein 24 |
| chr8_-_85500010 | 0.06 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X |
| chr1_-_60606237 | 0.06 |
ENSMUST00000142258.3
|
Raph1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
| chr4_-_141660390 | 0.06 |
ENSMUST00000036701.8
|
Fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr11_-_46057224 | 0.06 |
ENSMUST00000020679.3
|
Nipal4
|
NIPA-like domain containing 4 |
| chr3_-_108117754 | 0.06 |
ENSMUST00000117784.8
ENSMUST00000119650.8 ENSMUST00000117409.8 |
Atxn7l2
|
ataxin 7-like 2 |
| chr11_-_106679983 | 0.06 |
ENSMUST00000129585.8
|
Ddx5
|
DEAD box helicase 5 |
| chr14_-_36770898 | 0.06 |
ENSMUST00000225070.2
ENSMUST00000022338.7 |
Rgr
|
retinal G protein coupled receptor |
| chr14_+_34395845 | 0.06 |
ENSMUST00000048263.14
|
Wapl
|
WAPL cohesin release factor |
| chr6_+_114620054 | 0.06 |
ENSMUST00000032457.17
|
Atg7
|
autophagy related 7 |
| chr18_+_65022035 | 0.06 |
ENSMUST00000224385.3
ENSMUST00000163516.9 |
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr12_-_70394074 | 0.06 |
ENSMUST00000223160.2
ENSMUST00000222316.2 ENSMUST00000167755.3 ENSMUST00000110520.10 ENSMUST00000110522.10 ENSMUST00000221041.2 ENSMUST00000222603.3 |
Trim9
|
tripartite motif-containing 9 |
| chr1_+_87603952 | 0.06 |
ENSMUST00000170300.8
ENSMUST00000167032.2 |
Inpp5d
|
inositol polyphosphate-5-phosphatase D |
| chr12_+_110816687 | 0.06 |
ENSMUST00000221549.2
ENSMUST00000170060.4 |
Zfp839
|
zinc finger protein 839 |
| chr11_+_112673041 | 0.06 |
ENSMUST00000000579.3
|
Sox9
|
SRY (sex determining region Y)-box 9 |
| chr2_-_77000936 | 0.06 |
ENSMUST00000164114.9
ENSMUST00000049544.14 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr8_+_95703506 | 0.06 |
ENSMUST00000212581.2
|
Adgrg1
|
adhesion G protein-coupled receptor G1 |
| chr4_-_8239034 | 0.06 |
ENSMUST00000066674.8
|
Car8
|
carbonic anhydrase 8 |
| chr11_-_75329726 | 0.06 |
ENSMUST00000108437.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr7_-_43906802 | 0.06 |
ENSMUST00000107945.8
ENSMUST00000118216.8 |
Acp4
|
acid phosphatase 4 |
| chr5_+_150597204 | 0.06 |
ENSMUST00000202170.4
ENSMUST00000016569.11 |
Pds5b
|
PDS5 cohesin associated factor B |
| chr2_+_31360219 | 0.06 |
ENSMUST00000102840.5
|
Ass1
|
argininosuccinate synthetase 1 |
| chr2_-_93164812 | 0.06 |
ENSMUST00000111265.9
|
Tspan18
|
tetraspanin 18 |
| chr3_-_40856935 | 0.06 |
ENSMUST00000099123.5
|
1700034I23Rik
|
RIKEN cDNA 1700034I23 gene |
| chr2_+_27599259 | 0.06 |
ENSMUST00000100251.9
|
Rxra
|
retinoid X receptor alpha |
| chr9_+_107784065 | 0.06 |
ENSMUST00000035203.9
|
Mst1r
|
macrophage stimulating 1 receptor (c-met-related tyrosine kinase) |
| chr2_-_33321306 | 0.06 |
ENSMUST00000113158.8
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr7_-_28246530 | 0.06 |
ENSMUST00000239002.2
ENSMUST00000057974.4 |
Nccrp1
|
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chr11_+_106680062 | 0.06 |
ENSMUST00000103068.10
ENSMUST00000018516.11 |
Cep95
|
centrosomal protein 95 |
| chr12_+_73333553 | 0.05 |
ENSMUST00000140523.8
ENSMUST00000126488.8 |
Slc38a6
|
solute carrier family 38, member 6 |
| chr5_+_123153072 | 0.05 |
ENSMUST00000051016.5
ENSMUST00000121652.8 |
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr13_+_47196975 | 0.05 |
ENSMUST00000037025.16
ENSMUST00000143868.2 |
Kdm1b
|
lysine (K)-specific demethylase 1B |
| chr15_+_102052797 | 0.05 |
ENSMUST00000023807.7
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr7_-_80037622 | 0.05 |
ENSMUST00000206698.2
|
Fes
|
feline sarcoma oncogene |
| chr6_+_114620067 | 0.05 |
ENSMUST00000169310.10
ENSMUST00000182169.8 ENSMUST00000183165.8 ENSMUST00000182098.8 ENSMUST00000182793.8 ENSMUST00000182902.8 ENSMUST00000182428.8 ENSMUST00000182035.8 |
Atg7
|
autophagy related 7 |
| chr8_-_37200051 | 0.05 |
ENSMUST00000098826.10
|
Dlc1
|
deleted in liver cancer 1 |
| chr1_-_120048788 | 0.05 |
ENSMUST00000027634.13
|
Dbi
|
diazepam binding inhibitor |
| chr2_-_10053381 | 0.05 |
ENSMUST00000026888.11
|
Taf3
|
TATA-box binding protein associated factor 3 |
| chrX_+_95757092 | 0.05 |
ENSMUST00000033554.5
|
Gpr165
|
G protein-coupled receptor 165 |
| chr1_+_87254719 | 0.05 |
ENSMUST00000027475.15
|
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr17_-_34170401 | 0.05 |
ENSMUST00000087543.5
|
B3galt4
|
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ascl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0072365 | regulation of cellular ketone metabolic process by negative regulation of transcription from RNA polymerase II promoter(GO:0072365) |
| 0.1 | 0.5 | GO:0046878 | positive regulation of saliva secretion(GO:0046878) |
| 0.1 | 0.3 | GO:0070100 | negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 0.6 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.2 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.1 | 0.2 | GO:1903413 | response to bile acid(GO:1903412) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0071550 | death-inducing signaling complex assembly(GO:0071550) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.4 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:0032298 | positive regulation of DNA-dependent DNA replication initiation(GO:0032298) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:0002851 | positive regulation of tolerance induction dependent upon immune response(GO:0002654) regulation of peripheral tolerance induction(GO:0002658) positive regulation of peripheral tolerance induction(GO:0002660) regulation of peripheral T cell tolerance induction(GO:0002849) positive regulation of peripheral T cell tolerance induction(GO:0002851) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.3 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0098583 | mastication(GO:0071626) learned vocalization behavior(GO:0098583) |
| 0.0 | 0.2 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0072186 | metanephric cap development(GO:0072185) metanephric cap morphogenesis(GO:0072186) metanephric cap mesenchymal cell proliferation involved in metanephros development(GO:0090094) |
| 0.0 | 0.2 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0035582 | sequestering of BMP in extracellular matrix(GO:0035582) |
| 0.0 | 0.1 | GO:0039521 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) amino acid homeostasis(GO:0080144) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.0 | 0.2 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.1 | GO:0061017 | hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.1 | GO:1901204 | regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.0 | 0.1 | GO:0019413 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate biosynthetic process(GO:0019542) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.2 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.2 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.0 | 0.1 | GO:0070458 | detoxification of nitrogen compound(GO:0051410) cellular detoxification of nitrogen compound(GO:0070458) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.1 | GO:0072034 | intrahepatic bile duct development(GO:0035622) response to heparin(GO:0071503) cellular response to heparin(GO:0071504) renal vesicle induction(GO:0072034) |
| 0.0 | 0.1 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.1 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0086023 | adrenergic receptor signaling pathway involved in heart process(GO:0086023) |
| 0.0 | 0.3 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.1 | GO:0045659 | negative regulation of neutrophil differentiation(GO:0045659) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0010046 | response to mycotoxin(GO:0010046) |
| 0.0 | 0.2 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.1 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.0 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.1 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.0 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.1 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0006447 | regulation of translational initiation by iron(GO:0006447) positive regulation of translational initiation by iron(GO:0045994) |
| 0.0 | 0.0 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.1 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.4 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.1 | GO:0043512 | inhibin A complex(GO:0043512) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
| 0.0 | 0.3 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.0 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.0 | 0.1 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.4 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.3 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.1 | GO:0004962 | endothelin receptor activity(GO:0004962) |
| 0.0 | 0.2 | GO:0004096 | catalase activity(GO:0004096) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0038052 | RNA polymerase II transcription factor activity, estrogen-activated sequence-specific DNA binding(GO:0038052) |
| 0.0 | 0.2 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0019779 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.4 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.1 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.3 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.7 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0051435 | BH4 domain binding(GO:0051435) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.1 | GO:0034647 | histone demethylase activity (H3-trimethyl-K4 specific)(GO:0034647) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0019002 | GMP binding(GO:0019002) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |