Project
GFI1 WT vs 36n/n vs KD
Navigation
Downloads
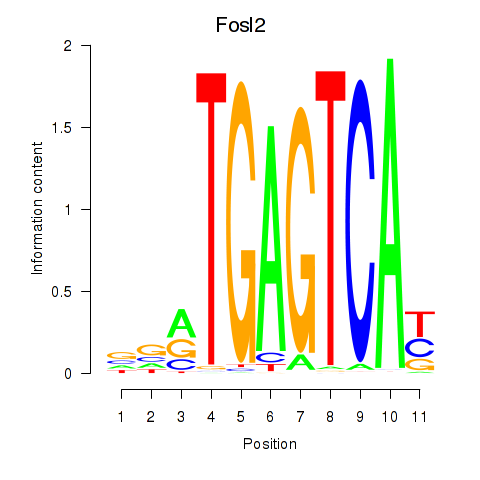
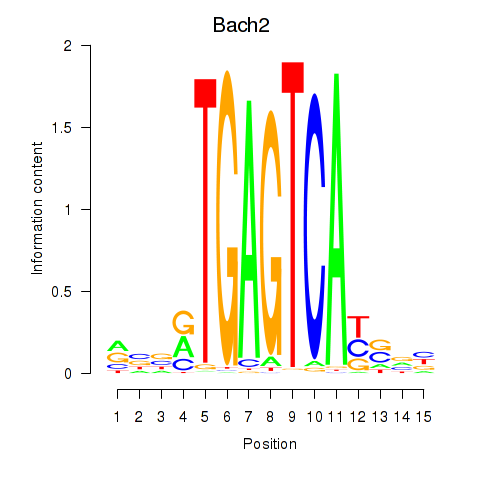
Results for Fosl2_Bach2
Z-value: 0.66
Transcription factors associated with Fosl2_Bach2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Fosl2
|
ENSMUSG00000029135.11 | fos-like antigen 2 |
|
Bach2
|
ENSMUSG00000040270.17 | BTB and CNC homology, basic leucine zipper transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bach2 | mm39_v1_chr4_+_32238950_32238964 | -0.41 | 4.9e-01 | Click! |
| Fosl2 | mm39_v1_chr5_+_32293145_32293201 | 0.15 | 8.1e-01 | Click! |
Activity profile of Fosl2_Bach2 motif
Sorted Z-values of Fosl2_Bach2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_90563510 | 0.76 |
ENSMUST00000014777.9
ENSMUST00000064391.12 |
Cpne8
|
copine VIII |
| chr3_-_88410495 | 0.66 |
ENSMUST00000120377.8
ENSMUST00000029699.13 |
Lmna
|
lamin A |
| chr3_+_142326363 | 0.62 |
ENSMUST00000165774.8
|
Gbp2
|
guanylate binding protein 2 |
| chr11_-_102255999 | 0.61 |
ENSMUST00000006749.10
|
Slc4a1
|
solute carrier family 4 (anion exchanger), member 1 |
| chr4_+_152093260 | 0.60 |
ENSMUST00000097773.4
|
Klhl21
|
kelch-like 21 |
| chr9_-_119812042 | 0.56 |
ENSMUST00000214058.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr18_-_38131766 | 0.51 |
ENSMUST00000236588.2
ENSMUST00000237272.2 ENSMUST00000236134.2 |
Arap3
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 |
| chr9_+_69361348 | 0.49 |
ENSMUST00000134907.8
|
Anxa2
|
annexin A2 |
| chr11_-_115915315 | 0.46 |
ENSMUST00000016703.8
|
H3f3b
|
H3.3 histone B |
| chrX_-_7999009 | 0.45 |
ENSMUST00000130832.8
ENSMUST00000033506.13 ENSMUST00000115623.8 ENSMUST00000153839.2 |
Wdr13
|
WD repeat domain 13 |
| chr1_+_75213044 | 0.44 |
ENSMUST00000188931.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chrX_+_73468140 | 0.44 |
ENSMUST00000135165.8
ENSMUST00000114128.8 ENSMUST00000004330.10 ENSMUST00000114133.9 ENSMUST00000114129.9 ENSMUST00000132749.2 |
Ikbkg
|
inhibitor of kappaB kinase gamma |
| chr1_+_170104889 | 0.43 |
ENSMUST00000179976.3
|
Sh2d1b1
|
SH2 domain containing 1B1 |
| chr7_-_126398165 | 0.42 |
ENSMUST00000205890.2
ENSMUST00000205336.2 ENSMUST00000087566.11 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr18_-_43610829 | 0.40 |
ENSMUST00000057110.11
|
Eif3j2
|
eukaryotic translation initiation factor 3, subunit J2 |
| chr8_-_61407760 | 0.39 |
ENSMUST00000110302.8
|
Clcn3
|
chloride channel, voltage-sensitive 3 |
| chr17_-_45883421 | 0.39 |
ENSMUST00000130406.2
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr5_-_24597009 | 0.38 |
ENSMUST00000059401.7
|
Atg9b
|
autophagy related 9B |
| chr4_-_126215462 | 0.38 |
ENSMUST00000102617.5
|
Adprs
|
ADP-ribosylserine hydrolase |
| chr4_-_6454262 | 0.38 |
ENSMUST00000029910.12
|
Nsmaf
|
neutral sphingomyelinase (N-SMase) activation associated factor |
| chr11_+_95733489 | 0.37 |
ENSMUST00000100532.10
ENSMUST00000036088.11 |
Gngt2
|
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr7_+_4463686 | 0.37 |
ENSMUST00000167298.2
ENSMUST00000171445.8 |
Eps8l1
|
EPS8-like 1 |
| chr19_-_32080496 | 0.36 |
ENSMUST00000235213.2
ENSMUST00000236504.2 |
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr1_+_75213082 | 0.36 |
ENSMUST00000055223.14
ENSMUST00000082158.13 ENSMUST00000188346.7 |
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr8_+_94954280 | 0.35 |
ENSMUST00000109547.2
|
Nup93
|
nucleoporin 93 |
| chr11_-_69906171 | 0.34 |
ENSMUST00000018718.8
ENSMUST00000102574.10 |
Acadvl
|
acyl-Coenzyme A dehydrogenase, very long chain |
| chr9_+_69360902 | 0.33 |
ENSMUST00000034756.15
ENSMUST00000123470.8 |
Anxa2
|
annexin A2 |
| chr14_-_52257113 | 0.33 |
ENSMUST00000166169.4
ENSMUST00000226605.2 |
Zfp219
|
zinc finger protein 219 |
| chr15_-_38518406 | 0.33 |
ENSMUST00000151319.8
|
Azin1
|
antizyme inhibitor 1 |
| chr14_-_56499690 | 0.32 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr16_+_20470402 | 0.32 |
ENSMUST00000007212.9
ENSMUST00000232629.2 |
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr2_+_121859025 | 0.29 |
ENSMUST00000028668.8
|
Eif3j1
|
eukaryotic translation initiation factor 3, subunit J1 |
| chr9_+_7272514 | 0.29 |
ENSMUST00000015394.10
|
Mmp13
|
matrix metallopeptidase 13 |
| chr11_-_95733235 | 0.29 |
ENSMUST00000059026.10
|
Abi3
|
ABI family member 3 |
| chr1_-_52271455 | 0.28 |
ENSMUST00000114512.8
|
Gls
|
glutaminase |
| chr6_+_17693941 | 0.27 |
ENSMUST00000115420.8
ENSMUST00000115419.8 |
St7
|
suppression of tumorigenicity 7 |
| chr11_+_50101717 | 0.26 |
ENSMUST00000147468.8
|
Mgat4b
|
mannoside acetylglucosaminyltransferase 4, isoenzyme B |
| chr17_+_36152559 | 0.26 |
ENSMUST00000174124.2
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr11_-_3864664 | 0.26 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr2_-_25114660 | 0.26 |
ENSMUST00000043584.5
|
Tubb4b
|
tubulin, beta 4B class IVB |
| chr16_+_35590745 | 0.26 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr1_+_171386752 | 0.26 |
ENSMUST00000004829.13
|
Cd244a
|
CD244 molecule A |
| chr17_-_45903410 | 0.25 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr17_+_46471950 | 0.25 |
ENSMUST00000024748.14
ENSMUST00000172170.8 |
Gtpbp2
|
GTP binding protein 2 |
| chr18_-_38102799 | 0.24 |
ENSMUST00000166148.8
ENSMUST00000163131.8 ENSMUST00000043437.14 |
Fchsd1
|
FCH and double SH3 domains 1 |
| chr14_+_79753055 | 0.24 |
ENSMUST00000110835.3
ENSMUST00000227192.2 |
Elf1
|
E74-like factor 1 |
| chr2_+_151947444 | 0.24 |
ENSMUST00000041500.8
|
Srxn1
|
sulfiredoxin 1 homolog (S. cerevisiae) |
| chr7_-_112946481 | 0.23 |
ENSMUST00000117577.8
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr14_-_36857202 | 0.23 |
ENSMUST00000165649.4
ENSMUST00000224769.2 |
Ghitm
|
growth hormone inducible transmembrane protein |
| chr4_-_58912678 | 0.23 |
ENSMUST00000144512.8
ENSMUST00000102889.10 ENSMUST00000055822.15 |
Ecpas
|
Ecm29 proteasome adaptor and scaffold |
| chr5_+_117501557 | 0.22 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chr13_-_8921732 | 0.22 |
ENSMUST00000054251.13
ENSMUST00000176813.8 ENSMUST00000175958.2 |
Wdr37
|
WD repeat domain 37 |
| chr1_+_75213114 | 0.22 |
ENSMUST00000188290.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr10_+_80765900 | 0.22 |
ENSMUST00000015456.10
ENSMUST00000220246.2 |
Gadd45b
|
growth arrest and DNA-damage-inducible 45 beta |
| chr1_+_174000304 | 0.22 |
ENSMUST00000027817.8
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr3_+_109481223 | 0.22 |
ENSMUST00000106576.3
|
Vav3
|
vav 3 oncogene |
| chrX_+_7588505 | 0.21 |
ENSMUST00000207675.2
ENSMUST00000116633.9 ENSMUST00000208996.2 ENSMUST00000144148.4 ENSMUST00000125991.9 ENSMUST00000148624.8 |
Wdr45
|
WD repeat domain 45 |
| chr2_+_30331839 | 0.21 |
ENSMUST00000131476.8
|
Ptpa
|
protein phosphatase 2 protein activator |
| chr4_-_140393185 | 0.21 |
ENSMUST00000069623.12
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr12_-_85335193 | 0.21 |
ENSMUST00000121930.2
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr5_-_148489593 | 0.20 |
ENSMUST00000201595.4
ENSMUST00000164904.2 |
Ubl3
|
ubiquitin-like 3 |
| chr8_-_110419867 | 0.20 |
ENSMUST00000034164.6
|
Ist1
|
increased sodium tolerance 1 homolog (yeast) |
| chr10_-_121462219 | 0.20 |
ENSMUST00000039810.8
ENSMUST00000218004.2 |
Xpot
|
exportin, tRNA (nuclear export receptor for tRNAs) |
| chr6_+_129510117 | 0.20 |
ENSMUST00000032264.9
|
Gabarapl1
|
gamma-aminobutyric acid (GABA) A receptor-associated protein-like 1 |
| chr11_-_99045894 | 0.20 |
ENSMUST00000103134.4
|
Ccr7
|
chemokine (C-C motif) receptor 7 |
| chr2_+_131751848 | 0.20 |
ENSMUST00000091288.13
ENSMUST00000124100.8 ENSMUST00000136783.2 |
Prnp
Prn
|
prion protein prion protein readthrough transcript |
| chr11_+_94218810 | 0.19 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr7_+_126843446 | 0.19 |
ENSMUST00000052509.6
|
Zfp771
|
zinc finger protein 771 |
| chr15_-_74650673 | 0.19 |
ENSMUST00000186014.2
ENSMUST00000191407.2 |
Ly6g6g
|
lymphocyte antigen 6 complex, locus G6G |
| chr11_-_69771797 | 0.18 |
ENSMUST00000238978.2
|
Kctd11
|
potassium channel tetramerisation domain containing 11 |
| chrY_-_1286623 | 0.18 |
ENSMUST00000091190.12
|
Ddx3y
|
DEAD box helicase 3, Y-linked |
| chr2_+_84891281 | 0.18 |
ENSMUST00000238769.2
|
Tnks1bp1
|
tankyrase 1 binding protein 1 |
| chr1_-_37575313 | 0.18 |
ENSMUST00000042161.15
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr13_+_30844025 | 0.18 |
ENSMUST00000110310.9
ENSMUST00000095914.7 |
Dusp22
|
dual specificity phosphatase 22 |
| chr11_-_51541610 | 0.18 |
ENSMUST00000142721.2
ENSMUST00000156835.8 ENSMUST00000001080.16 |
N4bp3
|
NEDD4 binding protein 3 |
| chr1_-_30988772 | 0.18 |
ENSMUST00000238874.2
ENSMUST00000027232.15 ENSMUST00000076587.6 ENSMUST00000233506.2 |
Ptp4a1
|
protein tyrosine phosphatase 4a1 |
| chr2_-_38534099 | 0.17 |
ENSMUST00000028083.6
|
Psmb7
|
proteasome (prosome, macropain) subunit, beta type 7 |
| chr14_+_51181956 | 0.17 |
ENSMUST00000178092.2
ENSMUST00000227052.2 |
Pnp
Gm49342
|
purine-nucleoside phosphorylase predicted gene, 49342 |
| chr18_-_35087355 | 0.17 |
ENSMUST00000025217.11
|
Hspa9
|
heat shock protein 9 |
| chr5_-_90487583 | 0.17 |
ENSMUST00000197021.2
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr11_+_117123107 | 0.17 |
ENSMUST00000106354.9
|
Septin9
|
septin 9 |
| chr10_+_128247598 | 0.17 |
ENSMUST00000096386.13
|
Rnf41
|
ring finger protein 41 |
| chr8_+_34222058 | 0.17 |
ENSMUST00000167264.8
ENSMUST00000187392.7 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr6_-_56878854 | 0.16 |
ENSMUST00000101367.9
|
Nt5c3
|
5'-nucleotidase, cytosolic III |
| chr18_+_38088597 | 0.16 |
ENSMUST00000070709.9
ENSMUST00000177058.8 ENSMUST00000169360.9 ENSMUST00000163591.9 ENSMUST00000091932.12 |
Rell2
|
RELT-like 2 |
| chr17_-_30831576 | 0.16 |
ENSMUST00000235171.2
ENSMUST00000236335.2 ENSMUST00000167624.2 |
Glo1
|
glyoxalase 1 |
| chr8_+_34222266 | 0.16 |
ENSMUST00000190675.2
ENSMUST00000171010.8 |
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr1_+_75213171 | 0.16 |
ENSMUST00000187058.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr13_-_58276353 | 0.16 |
ENSMUST00000007980.7
|
Hnrnpa0
|
heterogeneous nuclear ribonucleoprotein A0 |
| chr11_-_109364424 | 0.16 |
ENSMUST00000070152.12
|
Slc16a6
|
solute carrier family 16 (monocarboxylic acid transporters), member 6 |
| chr5_-_146731810 | 0.16 |
ENSMUST00000085614.6
|
Usp12
|
ubiquitin specific peptidase 12 |
| chr2_+_69210775 | 0.15 |
ENSMUST00000063690.4
|
Dhrs9
|
dehydrogenase/reductase (SDR family) member 9 |
| chr7_-_19005721 | 0.15 |
ENSMUST00000032561.9
|
Vasp
|
vasodilator-stimulated phosphoprotein |
| chr6_+_41515152 | 0.15 |
ENSMUST00000103291.2
ENSMUST00000192856.6 |
Trbc1
|
T cell receptor beta, constant region 1 |
| chr6_-_115735935 | 0.15 |
ENSMUST00000072933.13
|
Tmem40
|
transmembrane protein 40 |
| chr12_+_51737775 | 0.15 |
ENSMUST00000218820.2
ENSMUST00000021338.10 |
Ap4s1
|
adaptor-related protein complex AP-4, sigma 1 |
| chr10_+_128626772 | 0.15 |
ENSMUST00000219404.2
ENSMUST00000026411.8 |
Mmp19
|
matrix metallopeptidase 19 |
| chr8_+_70275079 | 0.15 |
ENSMUST00000164890.8
ENSMUST00000034325.6 ENSMUST00000238452.2 |
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr11_-_48762170 | 0.15 |
ENSMUST00000049519.4
ENSMUST00000097271.4 |
Irgm1
|
immunity-related GTPase family M member 1 |
| chr3_-_10273628 | 0.15 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte |
| chr14_-_36857083 | 0.15 |
ENSMUST00000042564.17
|
Ghitm
|
growth hormone inducible transmembrane protein |
| chr9_-_44253630 | 0.14 |
ENSMUST00000097558.5
|
Hmbs
|
hydroxymethylbilane synthase |
| chr11_+_94219046 | 0.14 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr18_+_35251912 | 0.14 |
ENSMUST00000042345.8
ENSMUST00000236037.2 |
Ctnna1
|
catenin (cadherin associated protein), alpha 1 |
| chr6_+_95094721 | 0.14 |
ENSMUST00000032107.10
ENSMUST00000119582.3 |
Kbtbd8
|
kelch repeat and BTB (POZ) domain containing 8 |
| chr15_-_97665524 | 0.14 |
ENSMUST00000128775.9
ENSMUST00000134885.3 |
Rapgef3
|
Rap guanine nucleotide exchange factor (GEF) 3 |
| chr10_-_76562002 | 0.14 |
ENSMUST00000001147.5
|
Col6a1
|
collagen, type VI, alpha 1 |
| chr10_+_128745214 | 0.14 |
ENSMUST00000220308.2
|
Cd63
|
CD63 antigen |
| chr9_+_120762466 | 0.14 |
ENSMUST00000007130.15
ENSMUST00000178812.9 |
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr4_-_140376047 | 0.14 |
ENSMUST00000105799.8
ENSMUST00000039204.10 ENSMUST00000097820.9 |
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr7_+_127773115 | 0.13 |
ENSMUST00000033051.16
ENSMUST00000177111.8 |
Itgad
|
integrin, alpha D |
| chr13_-_99027544 | 0.13 |
ENSMUST00000109399.9
|
Tnpo1
|
transportin 1 |
| chr14_+_66205932 | 0.13 |
ENSMUST00000022616.14
|
Clu
|
clusterin |
| chrX_-_133483849 | 0.13 |
ENSMUST00000113213.2
ENSMUST00000033617.13 |
Btk
|
Bruton agammaglobulinemia tyrosine kinase |
| chr10_+_115979787 | 0.13 |
ENSMUST00000105271.9
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr17_-_56440817 | 0.13 |
ENSMUST00000167545.3
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr11_+_77692116 | 0.13 |
ENSMUST00000100794.10
ENSMUST00000151373.4 ENSMUST00000130305.9 ENSMUST00000172303.10 ENSMUST00000092884.11 ENSMUST00000164334.8 |
Myo18a
|
myosin XVIIIA |
| chr9_+_108367801 | 0.12 |
ENSMUST00000006854.13
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr9_-_78388080 | 0.12 |
ENSMUST00000156988.2
|
Eef1a1
|
eukaryotic translation elongation factor 1 alpha 1 |
| chr11_-_43638790 | 0.12 |
ENSMUST00000048578.3
ENSMUST00000109278.8 |
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr12_+_16860931 | 0.12 |
ENSMUST00000020908.9
|
E2f6
|
E2F transcription factor 6 |
| chr4_+_116544509 | 0.12 |
ENSMUST00000030454.6
|
Prdx1
|
peroxiredoxin 1 |
| chr8_-_23184070 | 0.12 |
ENSMUST00000131767.2
|
Ikbkb
|
inhibitor of kappaB kinase beta |
| chr7_+_27186335 | 0.12 |
ENSMUST00000008528.8
|
Sertad1
|
SERTA domain containing 1 |
| chr5_-_92496730 | 0.12 |
ENSMUST00000038816.13
ENSMUST00000118006.3 |
Cxcl10
|
chemokine (C-X-C motif) ligand 10 |
| chr18_+_82929037 | 0.12 |
ENSMUST00000236858.2
|
Zfp516
|
zinc finger protein 516 |
| chr1_+_171265103 | 0.12 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr8_-_25581354 | 0.12 |
ENSMUST00000125466.2
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr17_+_25381414 | 0.12 |
ENSMUST00000073277.12
ENSMUST00000182621.8 |
Ccdc154
|
coiled-coil domain containing 154 |
| chr15_-_11399666 | 0.12 |
ENSMUST00000022849.7
|
Tars
|
threonyl-tRNA synthetase |
| chr15_-_38518458 | 0.11 |
ENSMUST00000127848.2
|
Azin1
|
antizyme inhibitor 1 |
| chr17_-_45884179 | 0.11 |
ENSMUST00000165127.8
ENSMUST00000166469.8 ENSMUST00000024739.14 |
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chrX_+_161684221 | 0.11 |
ENSMUST00000101095.9
|
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr6_+_17694003 | 0.11 |
ENSMUST00000052113.12
ENSMUST00000081635.13 |
St7
|
suppression of tumorigenicity 7 |
| chr16_+_20367327 | 0.11 |
ENSMUST00000003319.6
ENSMUST00000232680.2 ENSMUST00000232490.2 |
Abcf3
|
ATP-binding cassette, sub-family F (GCN20), member 3 |
| chr17_+_29170924 | 0.11 |
ENSMUST00000153462.8
|
Kctd20
|
potassium channel tetramerisation domain containing 20 |
| chr2_+_11710523 | 0.11 |
ENSMUST00000138856.2
ENSMUST00000078834.12 ENSMUST00000114834.10 ENSMUST00000114833.10 ENSMUST00000114831.9 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr13_+_93441307 | 0.11 |
ENSMUST00000080127.12
|
Homer1
|
homer scaffolding protein 1 |
| chr13_+_33187205 | 0.11 |
ENSMUST00000063191.14
|
Serpinb9
|
serine (or cysteine) peptidase inhibitor, clade B, member 9 |
| chr17_-_45903188 | 0.11 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr8_+_34221861 | 0.10 |
ENSMUST00000170705.8
|
Gtf2e2
|
general transcription factor II E, polypeptide 2 (beta subunit) |
| chr3_+_95836558 | 0.10 |
ENSMUST00000165307.8
ENSMUST00000015893.13 ENSMUST00000169426.8 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr9_-_72018933 | 0.10 |
ENSMUST00000185117.8
|
Tcf12
|
transcription factor 12 |
| chr14_+_34032684 | 0.10 |
ENSMUST00000022322.17
|
Glud1
|
glutamate dehydrogenase 1 |
| chr19_-_53020531 | 0.10 |
ENSMUST00000236008.2
ENSMUST00000237294.2 |
Xpnpep1
|
X-prolyl aminopeptidase (aminopeptidase P) 1, soluble |
| chr10_+_87926932 | 0.10 |
ENSMUST00000048621.8
|
Pmch
|
pro-melanin-concentrating hormone |
| chr3_-_95646856 | 0.10 |
ENSMUST00000153026.8
ENSMUST00000123143.8 ENSMUST00000137912.8 ENSMUST00000029753.14 ENSMUST00000131376.8 ENSMUST00000117507.10 ENSMUST00000128885.8 ENSMUST00000147217.2 |
Ecm1
|
extracellular matrix protein 1 |
| chr7_-_127545896 | 0.10 |
ENSMUST00000118755.8
ENSMUST00000094026.10 |
Prss36
|
protease, serine 36 |
| chr5_+_8848138 | 0.10 |
ENSMUST00000009058.10
|
Abcb1b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1B |
| chr4_-_40279382 | 0.10 |
ENSMUST00000108108.3
ENSMUST00000095128.10 |
Ndufb6
|
NADH:ubiquinone oxidoreductase subunit B6 |
| chr19_-_4978238 | 0.10 |
ENSMUST00000237394.2
ENSMUST00000025851.4 |
Dpp3
|
dipeptidylpeptidase 3 |
| chr6_+_67838100 | 0.10 |
ENSMUST00000200586.2
ENSMUST00000103309.3 |
Igkv17-127
|
immunoglobulin kappa variable 17-127 |
| chr7_+_24310171 | 0.10 |
ENSMUST00000206422.2
|
Phldb3
|
pleckstrin homology like domain, family B, member 3 |
| chr6_-_143045493 | 0.09 |
ENSMUST00000204655.3
ENSMUST00000111758.9 |
C2cd5
|
C2 calcium-dependent domain containing 5 |
| chr11_-_93859064 | 0.09 |
ENSMUST00000107844.3
ENSMUST00000170303.2 |
Nme1
Gm20390
|
NME/NM23 nucleoside diphosphate kinase 1 predicted gene 20390 |
| chr8_+_106099894 | 0.09 |
ENSMUST00000160650.2
|
Plekhg4
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 4 |
| chr19_+_5100475 | 0.09 |
ENSMUST00000225427.2
|
Rin1
|
Ras and Rab interactor 1 |
| chr7_+_30463175 | 0.09 |
ENSMUST00000165887.8
ENSMUST00000085691.11 ENSMUST00000054427.13 ENSMUST00000085688.11 |
Dmkn
|
dermokine |
| chr9_+_108782646 | 0.09 |
ENSMUST00000112070.8
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr12_+_111725282 | 0.09 |
ENSMUST00000239017.2
ENSMUST00000084941.12 |
Klc1
|
kinesin light chain 1 |
| chr7_-_7124478 | 0.09 |
ENSMUST00000056246.8
|
Zfp954
|
zinc finger protein 954 |
| chr1_+_172328768 | 0.09 |
ENSMUST00000111228.2
|
Tagln2
|
transgelin 2 |
| chr1_-_180021039 | 0.09 |
ENSMUST00000160482.8
ENSMUST00000170472.8 |
Coq8a
|
coenzyme Q8A |
| chr10_+_127126643 | 0.09 |
ENSMUST00000026475.15
ENSMUST00000139091.2 ENSMUST00000230446.2 |
Ddit3
Ddit3
|
DNA-damage inducible transcript 3 DNA-damage inducible transcript 3 |
| chr5_+_35156389 | 0.09 |
ENSMUST00000114281.8
ENSMUST00000114280.8 |
Rgs12
|
regulator of G-protein signaling 12 |
| chr2_-_20948230 | 0.09 |
ENSMUST00000140230.2
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr9_-_75448979 | 0.09 |
ENSMUST00000214171.2
|
Tmod3
|
tropomodulin 3 |
| chrX_-_10083157 | 0.09 |
ENSMUST00000072393.9
ENSMUST00000044598.13 ENSMUST00000073392.11 ENSMUST00000115533.8 ENSMUST00000115532.2 |
Rpgr
|
retinitis pigmentosa GTPase regulator |
| chr8_+_108020092 | 0.08 |
ENSMUST00000169453.8
|
Nfat5
|
nuclear factor of activated T cells 5 |
| chr10_+_86614864 | 0.08 |
ENSMUST00000099396.3
|
Nt5dc3
|
5'-nucleotidase domain containing 3 |
| chr7_-_120269462 | 0.08 |
ENSMUST00000127845.2
ENSMUST00000208635.2 ENSMUST00000033178.4 |
Pdzd9
|
PDZ domain containing 9 |
| chr2_+_3425159 | 0.08 |
ENSMUST00000100463.10
ENSMUST00000061852.12 ENSMUST00000102988.10 ENSMUST00000115066.8 |
Dclre1c
|
DNA cross-link repair 1C |
| chr1_+_153625243 | 0.08 |
ENSMUST00000182722.8
|
Rnasel
|
ribonuclease L (2', 5'-oligoisoadenylate synthetase-dependent) |
| chr7_+_29515485 | 0.08 |
ENSMUST00000178162.3
ENSMUST00000032796.14 |
Zfp790
|
zinc finger protein 790 |
| chr19_+_5100815 | 0.08 |
ENSMUST00000224178.2
ENSMUST00000225799.3 ENSMUST00000025818.8 |
Rin1
|
Ras and Rab interactor 1 |
| chr5_+_104194383 | 0.08 |
ENSMUST00000150226.8
|
Nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr16_+_58548273 | 0.08 |
ENSMUST00000023426.12
ENSMUST00000162057.8 ENSMUST00000162191.2 |
Cldnd1
|
claudin domain containing 1 |
| chr14_-_86986541 | 0.08 |
ENSMUST00000226254.2
|
Diaph3
|
diaphanous related formin 3 |
| chr7_-_126183392 | 0.08 |
ENSMUST00000128970.8
ENSMUST00000116269.9 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr1_-_135040044 | 0.08 |
ENSMUST00000086432.7
|
Ptprv
|
protein tyrosine phosphatase, receptor type, V |
| chr7_+_55491819 | 0.08 |
ENSMUST00000032629.16
ENSMUST00000173783.8 ENSMUST00000085255.11 ENSMUST00000163845.4 |
Cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr14_+_76714350 | 0.08 |
ENSMUST00000140251.9
|
Tsc22d1
|
TSC22 domain family, member 1 |
| chr8_-_108315024 | 0.08 |
ENSMUST00000044106.6
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr5_+_104194930 | 0.08 |
ENSMUST00000134313.8
|
Nudt9
|
nudix (nucleoside diphosphate linked moiety X)-type motif 9 |
| chr11_+_78079243 | 0.08 |
ENSMUST00000002128.14
ENSMUST00000150941.8 |
Rab34
|
RAB34, member RAS oncogene family |
| chr12_+_111725357 | 0.08 |
ENSMUST00000118471.8
ENSMUST00000122300.8 |
Klc1
|
kinesin light chain 1 |
| chr1_-_149836974 | 0.08 |
ENSMUST00000190507.2
ENSMUST00000070200.15 |
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr2_+_29968596 | 0.08 |
ENSMUST00000045246.8
|
Pkn3
|
protein kinase N3 |
| chr19_-_41252370 | 0.08 |
ENSMUST00000237871.2
ENSMUST00000025989.10 |
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr4_+_151012375 | 0.07 |
ENSMUST00000139826.8
ENSMUST00000116257.8 |
Tnfrsf9
|
tumor necrosis factor receptor superfamily, member 9 |
| chr5_+_124045238 | 0.07 |
ENSMUST00000023869.15
|
Denr
|
density-regulated protein |
| chr14_-_36690636 | 0.07 |
ENSMUST00000067700.13
|
Ccser2
|
coiled-coil serine rich 2 |
| chr1_+_165130192 | 0.07 |
ENSMUST00000111450.3
|
Gpr161
|
G protein-coupled receptor 161 |
| chr5_-_65248927 | 0.07 |
ENSMUST00000043352.8
|
Tmem156
|
transmembrane protein 156 |
| chr9_+_108368032 | 0.07 |
ENSMUST00000166103.9
ENSMUST00000085044.14 ENSMUST00000193678.6 ENSMUST00000178075.8 |
Usp19
|
ubiquitin specific peptidase 19 |
| chr5_+_25452342 | 0.07 |
ENSMUST00000114950.2
|
Galnt11
|
polypeptide N-acetylgalactosaminyltransferase 11 |
| chr3_+_84081411 | 0.07 |
ENSMUST00000193882.2
|
Gm6525
|
predicted pseudogene 6525 |
| chr4_+_48124902 | 0.07 |
ENSMUST00000107721.8
ENSMUST00000153502.8 ENSMUST00000107720.9 ENSMUST00000064765.6 |
Stx17
|
syntaxin 17 |
| chr9_+_108782664 | 0.07 |
ENSMUST00000026740.6
|
Col7a1
|
collagen, type VII, alpha 1 |
| chr7_+_27307422 | 0.07 |
ENSMUST00000142365.8
|
Akt2
|
thymoma viral proto-oncogene 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Fosl2_Bach2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.2 | 0.5 | GO:1903659 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.4 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.1 | 0.8 | GO:0044362 | modulation of molecular function in other organism(GO:0044359) negative regulation of molecular function in other organism(GO:0044362) negative regulation of molecular function in other organism involved in symbiotic interaction(GO:0052204) modulation of molecular function in other organism involved in symbiotic interaction(GO:0052205) negative regulation by host of symbiont molecular function(GO:0052405) modification by host of symbiont molecular function(GO:0052428) |
| 0.1 | 0.7 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.1 | 0.4 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.1 | 0.4 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.1 | 0.5 | GO:1902340 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.1 | 0.3 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.2 | GO:0030472 | mitotic spindle organization in nucleus(GO:0030472) |
| 0.1 | 0.2 | GO:0018160 | peptidyl-pyrromethane cofactor linkage(GO:0018160) |
| 0.1 | 0.2 | GO:2000547 | lymphocyte migration into lymphoid organs(GO:0097021) positive regulation of thymocyte migration(GO:2000412) regulation of dendritic cell dendrite assembly(GO:2000547) |
| 0.1 | 0.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 0.2 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.1 | 0.2 | GO:0046032 | ADP catabolic process(GO:0046032) |
| 0.1 | 0.2 | GO:0009838 | abscission(GO:0009838) |
| 0.1 | 0.4 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.2 | GO:0006409 | tRNA export from nucleus(GO:0006409) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.0 | 0.4 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.4 | GO:0044805 | late nucleophagy(GO:0044805) |
| 0.0 | 0.1 | GO:1904499 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.1 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.2 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 | 0.2 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.1 | GO:1903027 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.3 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.2 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.2 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.5 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
| 0.0 | 0.1 | GO:1901740 | negative regulation of myoblast fusion(GO:1901740) |
| 0.0 | 0.3 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.4 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.6 | GO:0044406 | adhesion of symbiont to host(GO:0044406) |
| 0.0 | 0.1 | GO:0038032 | termination of G-protein coupled receptor signaling pathway(GO:0038032) |
| 0.0 | 0.1 | GO:1901098 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.6 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) negative regulation of determination of dorsal identity(GO:2000016) |
| 0.0 | 0.1 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.1 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.1 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.4 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.0 | 0.3 | GO:0034497 | protein localization to pre-autophagosomal structure(GO:0034497) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.2 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.0 | 0.3 | GO:0071501 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0018214 | protein carboxylation(GO:0018214) |
| 0.0 | 0.2 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0006669 | sphinganine-1-phosphate biosynthetic process(GO:0006669) |
| 0.0 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.0 | 0.0 | GO:0006218 | uridine catabolic process(GO:0006218) uridine metabolic process(GO:0046108) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.0 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.6 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.3 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.1 | GO:0002266 | follicular dendritic cell activation(GO:0002266) follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.2 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.2 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.4 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.1 | 0.4 | GO:0005673 | transcription factor TFIIE complex(GO:0005673) |
| 0.1 | 0.5 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.1 | 0.7 | GO:0005638 | lamin filament(GO:0005638) |
| 0.1 | 0.6 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.6 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.2 | GO:0097574 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) |
| 0.0 | 0.7 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.6 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.5 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.3 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.4 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0070992 | translation initiation complex(GO:0070992) |
| 0.0 | 0.2 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.4 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.3 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.0 | 1.2 | GO:0000502 | proteasome complex(GO:0000502) |
| 0.0 | 0.2 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.1 | 0.4 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.4 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 0.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.5 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.2 | GO:0004418 | hydroxymethylbilane synthase activity(GO:0004418) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.2 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.1 | 0.2 | GO:0008160 | protein tyrosine phosphatase activator activity(GO:0008160) |
| 0.1 | 0.2 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.0 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 1.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.4 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:1903135 | cupric ion binding(GO:1903135) |
| 0.0 | 0.2 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.7 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.6 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.3 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.4 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.8 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.5 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.0 | 0.0 | GO:0031800 | type 3 metabotropic glutamate receptor binding(GO:0031800) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.5 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.7 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |