Project
GFI1 WT vs 36n/n vs KD
Navigation
Downloads
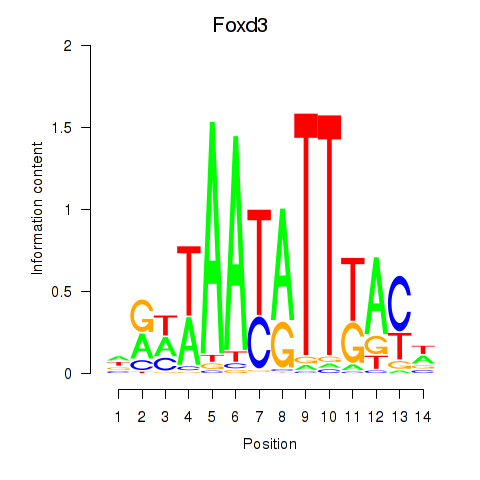
Results for Foxd3
Z-value: 0.45
Transcription factors associated with Foxd3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxd3
|
ENSMUSG00000067261.5 | forkhead box D3 |
Activity profile of Foxd3 motif
Sorted Z-values of Foxd3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_23715220 | 1.14 |
ENSMUST00000102972.6
|
H4c8
|
H4 clustered histone 8 |
| chr9_+_37995368 | 0.56 |
ENSMUST00000212502.4
|
Olfr887
|
olfactory receptor 887 |
| chr1_+_107456731 | 0.53 |
ENSMUST00000182198.8
|
Serpinb10
|
serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr2_-_172782089 | 0.46 |
ENSMUST00000009143.8
|
Bmp7
|
bone morphogenetic protein 7 |
| chr7_+_28834276 | 0.42 |
ENSMUST00000161522.8
ENSMUST00000204845.3 ENSMUST00000205027.3 ENSMUST00000204194.3 ENSMUST00000203070.3 ENSMUST00000203380.3 |
Rasgrp4
|
RAS guanyl releasing protein 4 |
| chr2_-_111779785 | 0.36 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307 |
| chrX_-_156198282 | 0.35 |
ENSMUST00000079945.11
ENSMUST00000138396.3 |
Phex
|
phosphate regulating endopeptidase homolog, X-linked |
| chr4_+_115594951 | 0.35 |
ENSMUST00000106522.9
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr10_+_66932235 | 0.33 |
ENSMUST00000174317.8
|
Jmjd1c
|
jumonji domain containing 1C |
| chr16_-_4698148 | 0.33 |
ENSMUST00000037843.7
|
Ubald1
|
UBA-like domain containing 1 |
| chr5_-_23889607 | 0.33 |
ENSMUST00000197985.5
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chr10_+_93324624 | 0.31 |
ENSMUST00000129421.8
|
Hal
|
histidine ammonia lyase |
| chr12_-_103597663 | 0.31 |
ENSMUST00000121625.2
ENSMUST00000044231.12 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr10_+_129320621 | 0.29 |
ENSMUST00000213236.2
ENSMUST00000213992.2 |
Olfr789
|
olfactory receptor 789 |
| chr4_+_115594918 | 0.25 |
ENSMUST00000106525.9
|
Efcab14
|
EF-hand calcium binding domain 14 |
| chr10_-_89457115 | 0.24 |
ENSMUST00000020102.14
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chr6_-_131224305 | 0.24 |
ENSMUST00000032306.15
ENSMUST00000088867.7 |
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr2_-_45000389 | 0.24 |
ENSMUST00000201804.4
ENSMUST00000028229.13 ENSMUST00000202187.4 ENSMUST00000153561.6 ENSMUST00000201490.2 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr16_-_16345197 | 0.21 |
ENSMUST00000069284.14
|
Fgd4
|
FYVE, RhoGEF and PH domain containing 4 |
| chr13_+_41040657 | 0.17 |
ENSMUST00000069958.15
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr11_+_29413734 | 0.15 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr11_-_34048538 | 0.13 |
ENSMUST00000223852.2
|
4930469K13Rik
|
RIKEN cDNA 4930469K13 gene |
| chr2_-_18397547 | 0.13 |
ENSMUST00000091418.12
ENSMUST00000166495.8 |
Dnajc1
|
DnaJ heat shock protein family (Hsp40) member C1 |
| chr2_-_45001141 | 0.12 |
ENSMUST00000201969.4
ENSMUST00000201623.4 |
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr6_-_137548004 | 0.12 |
ENSMUST00000100841.9
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr15_+_8997480 | 0.10 |
ENSMUST00000227191.3
|
Ranbp3l
|
RAN binding protein 3-like |
| chr7_+_97480125 | 0.08 |
ENSMUST00000206351.2
|
Pak1
|
p21 (RAC1) activated kinase 1 |
| chr12_-_110807330 | 0.08 |
ENSMUST00000177224.2
ENSMUST00000084974.11 ENSMUST00000070565.15 |
Mok
|
MOK protein kinase |
| chr10_-_128579879 | 0.07 |
ENSMUST00000026414.9
|
Dgka
|
diacylglycerol kinase, alpha |
| chr11_-_75330302 | 0.07 |
ENSMUST00000043696.9
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chr10_-_95678786 | 0.06 |
ENSMUST00000211096.2
|
Gm33543
|
predicted gene, 33543 |
| chr15_-_98505508 | 0.06 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr10_-_95678748 | 0.06 |
ENSMUST00000210336.2
|
Gm33543
|
predicted gene, 33543 |
| chr11_+_83193495 | 0.06 |
ENSMUST00000176430.8
ENSMUST00000065692.14 ENSMUST00000142680.2 |
Ap2b1
|
adaptor-related protein complex 2, beta 1 subunit |
| chr5_-_86475595 | 0.06 |
ENSMUST00000122377.2
|
Tmprss11d
|
transmembrane protease, serine 11d |
| chr2_+_51928017 | 0.06 |
ENSMUST00000065927.6
|
Tnfaip6
|
tumor necrosis factor alpha induced protein 6 |
| chr11_+_58549642 | 0.05 |
ENSMUST00000214392.2
|
Olfr322
|
olfactory receptor 322 |
| chr7_-_99629637 | 0.05 |
ENSMUST00000080817.6
|
Rnf169
|
ring finger protein 169 |
| chr11_-_75330415 | 0.05 |
ENSMUST00000128330.8
|
Serpinf2
|
serine (or cysteine) peptidase inhibitor, clade F, member 2 |
| chrY_-_70026134 | 0.05 |
ENSMUST00000188554.7
ENSMUST00000191151.2 |
Gm28079
|
predicted gene 28079 |
| chr14_-_70867588 | 0.05 |
ENSMUST00000228009.2
|
Dmtn
|
dematin actin binding protein |
| chr2_+_131792774 | 0.04 |
ENSMUST00000110170.8
ENSMUST00000110172.9 ENSMUST00000110171.9 |
Prnd
|
prion like protein doppel |
| chr5_-_23889591 | 0.03 |
ENSMUST00000198549.2
|
Srpk2
|
serine/arginine-rich protein specific kinase 2 |
| chrX_-_49517375 | 0.03 |
ENSMUST00000215270.2
|
Olfr1324
|
olfactory receptor 1324 |
| chr12_-_101924407 | 0.02 |
ENSMUST00000159883.2
ENSMUST00000160251.8 ENSMUST00000161011.8 ENSMUST00000021606.12 |
Atxn3
|
ataxin 3 |
| chr18_+_68470616 | 0.02 |
ENSMUST00000172148.4
ENSMUST00000239469.2 |
Mc5r
|
melanocortin 5 receptor |
| chr10_+_57521930 | 0.02 |
ENSMUST00000177325.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr10_+_42736771 | 0.02 |
ENSMUST00000105494.8
|
Scml4
|
Scm polycomb group protein like 4 |
| chr10_+_57521958 | 0.02 |
ENSMUST00000177473.8
|
Pkib
|
protein kinase inhibitor beta, cAMP dependent, testis specific |
| chr18_+_89224219 | 0.01 |
ENSMUST00000236835.2
|
Cd226
|
CD226 antigen |
| chr16_+_43056218 | 0.01 |
ENSMUST00000146708.8
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_+_38895902 | 0.01 |
ENSMUST00000003017.13
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr15_-_6904450 | 0.01 |
ENSMUST00000022746.13
ENSMUST00000176826.2 |
Osmr
|
oncostatin M receptor |
| chr1_+_88334678 | 0.01 |
ENSMUST00000027518.12
|
Spp2
|
secreted phosphoprotein 2 |
| chr5_+_148202117 | 0.01 |
ENSMUST00000110514.8
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr5_+_148202075 | 0.00 |
ENSMUST00000071878.12
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr7_-_142223662 | 0.00 |
ENSMUST00000228850.2
|
Gm49394
|
predicted gene, 49394 |
| chr6_-_21851827 | 0.00 |
ENSMUST00000202353.2
ENSMUST00000134635.2 ENSMUST00000123116.8 ENSMUST00000120965.8 ENSMUST00000143531.2 |
Tspan12
|
tetraspanin 12 |
| chr1_+_6805048 | 0.00 |
ENSMUST00000139838.8
|
St18
|
suppression of tumorigenicity 18 |
Network of associatons between targets according to the STRING database.
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:1900104 | hyaluranon cable assembly(GO:0036118) nephrogenic mesenchyme development(GO:0072076) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.1 | 0.3 | GO:0006548 | histidine catabolic process(GO:0006548) |
| 0.1 | 0.4 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.4 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.1 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) cellular response to vasopressin(GO:1904117) |
| 0.0 | 0.1 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:0002034 | regulation of blood vessel size by renin-angiotensin(GO:0002034) renal control of peripheral vascular resistance involved in regulation of systemic arterial blood pressure(GO:0003072) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0016841 | ammonia-lyase activity(GO:0016841) |
| 0.0 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.5 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ALK2 PATHWAY | ALK2 signaling events |