Project
GFI1 WT vs 36n/n vs KD
Navigation
Downloads
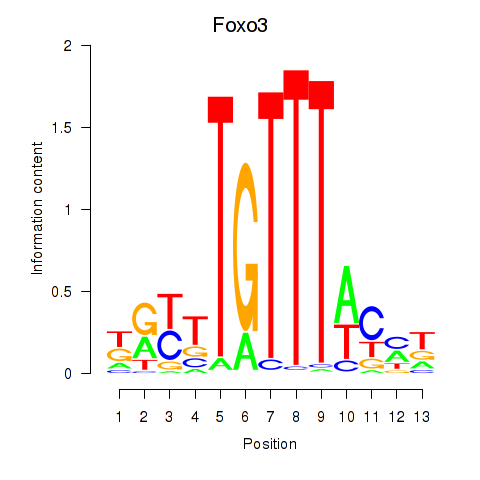
Results for Foxo3
Z-value: 0.56
Transcription factors associated with Foxo3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Foxo3
|
ENSMUSG00000048756.12 | forkhead box O3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Foxo3 | mm39_v1_chr10_-_42152684_42152759 | 0.84 | 7.2e-02 | Click! |
Activity profile of Foxo3 motif
Sorted Z-values of Foxo3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_64659004 | 0.75 |
ENSMUST00000066166.6
|
Tll1
|
tolloid-like |
| chr11_+_45946800 | 0.74 |
ENSMUST00000011400.8
|
Adam19
|
a disintegrin and metallopeptidase domain 19 (meltrin beta) |
| chr2_+_3514071 | 0.48 |
ENSMUST00000036350.3
|
Cdnf
|
cerebral dopamine neurotrophic factor |
| chr1_+_172525613 | 0.47 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related |
| chr15_+_98390278 | 0.44 |
ENSMUST00000205772.3
ENSMUST00000216822.2 |
Olfr279
|
olfactory receptor 279 |
| chr3_+_63203235 | 0.42 |
ENSMUST00000194134.6
|
Mme
|
membrane metallo endopeptidase |
| chr11_+_60244132 | 0.36 |
ENSMUST00000070805.13
ENSMUST00000094140.9 ENSMUST00000108723.9 ENSMUST00000108722.5 |
Drc3
|
dynein regulatory complex subunit 3 |
| chr3_+_53396120 | 0.33 |
ENSMUST00000029307.4
|
Stoml3
|
stomatin (Epb7.2)-like 3 |
| chr2_-_104324035 | 0.32 |
ENSMUST00000111124.8
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr18_-_80751327 | 0.28 |
ENSMUST00000236310.2
ENSMUST00000167977.8 ENSMUST00000035800.8 |
Nfatc1
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 1 |
| chr2_-_111400026 | 0.28 |
ENSMUST00000217772.2
ENSMUST00000207283.3 |
Olfr1295
|
olfactory receptor 1295 |
| chr14_+_51333816 | 0.28 |
ENSMUST00000169895.3
|
Rnase4
|
ribonuclease, RNase A family 4 |
| chr1_+_17672117 | 0.27 |
ENSMUST00000088476.4
|
Pi15
|
peptidase inhibitor 15 |
| chr4_-_119272690 | 0.27 |
ENSMUST00000238287.2
ENSMUST00000238759.2 ENSMUST00000063642.10 |
Ccdc30
|
coiled-coil domain containing 30 |
| chr11_-_65636651 | 0.26 |
ENSMUST00000138093.2
|
Map2k4
|
mitogen-activated protein kinase kinase 4 |
| chr11_-_60243695 | 0.25 |
ENSMUST00000095254.12
ENSMUST00000102683.11 ENSMUST00000093048.13 ENSMUST00000093046.13 ENSMUST00000064019.15 ENSMUST00000102682.5 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr1_+_171052623 | 0.24 |
ENSMUST00000111321.8
ENSMUST00000005824.12 ENSMUST00000111320.8 ENSMUST00000111319.2 |
Apoa2
|
apolipoprotein A-II |
| chr10_+_128540049 | 0.23 |
ENSMUST00000217836.2
|
Pmel
|
premelanosome protein |
| chr2_+_124452194 | 0.22 |
ENSMUST00000051419.15
ENSMUST00000076335.12 ENSMUST00000078621.12 ENSMUST00000077847.12 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr7_-_30259253 | 0.22 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans) |
| chr2_-_51039112 | 0.22 |
ENSMUST00000154545.2
ENSMUST00000017288.9 |
Rnd3
|
Rho family GTPase 3 |
| chr17_+_41121979 | 0.21 |
ENSMUST00000024721.8
ENSMUST00000233740.2 |
Rhag
|
Rhesus blood group-associated A glycoprotein |
| chr19_-_27988393 | 0.21 |
ENSMUST00000172907.8
ENSMUST00000046898.17 |
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr12_+_71063431 | 0.21 |
ENSMUST00000125125.2
|
Arid4a
|
AT rich interactive domain 4A (RBP1-like) |
| chr4_+_105014536 | 0.21 |
ENSMUST00000064139.8
|
Plpp3
|
phospholipid phosphatase 3 |
| chr12_-_31549538 | 0.21 |
ENSMUST00000064240.14
ENSMUST00000185739.8 ENSMUST00000188326.3 ENSMUST00000101499.10 ENSMUST00000085487.12 |
Cbll1
|
Casitas B-lineage lymphoma-like 1 |
| chr11_+_29413734 | 0.20 |
ENSMUST00000155854.8
|
Ccdc88a
|
coiled coil domain containing 88A |
| chr14_+_45457168 | 0.18 |
ENSMUST00000227086.2
ENSMUST00000147957.2 |
Gpr137c
|
G protein-coupled receptor 137C |
| chr11_-_86884507 | 0.18 |
ENSMUST00000018571.5
|
Ypel2
|
yippee like 2 |
| chr13_+_83723743 | 0.16 |
ENSMUST00000198217.5
ENSMUST00000199210.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr17_+_17669082 | 0.16 |
ENSMUST00000140134.2
|
Lix1
|
limb and CNS expressed 1 |
| chr11_+_85243970 | 0.16 |
ENSMUST00000108056.8
ENSMUST00000108061.8 ENSMUST00000108062.8 ENSMUST00000138423.8 ENSMUST00000092821.10 ENSMUST00000074875.11 |
Bcas3
|
breast carcinoma amplified sequence 3 |
| chrX_+_7744535 | 0.16 |
ENSMUST00000033495.15
|
Pim2
|
proviral integration site 2 |
| chr19_-_27988534 | 0.15 |
ENSMUST00000174850.8
|
Rfx3
|
regulatory factor X, 3 (influences HLA class II expression) |
| chr7_-_103463120 | 0.15 |
ENSMUST00000098192.4
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr9_+_40092216 | 0.15 |
ENSMUST00000218134.2
ENSMUST00000216720.2 ENSMUST00000214763.2 |
Olfr986
|
olfactory receptor 986 |
| chr2_+_91541245 | 0.14 |
ENSMUST00000142692.2
ENSMUST00000090608.6 |
Harbi1
|
harbinger transposase derived 1 |
| chr19_+_5927876 | 0.14 |
ENSMUST00000235340.2
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr11_-_94133527 | 0.13 |
ENSMUST00000061469.4
|
Wfikkn2
|
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
| chr14_+_30601157 | 0.13 |
ENSMUST00000040715.8
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr7_-_119461027 | 0.13 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr13_+_83720457 | 0.13 |
ENSMUST00000196730.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr11_+_69657275 | 0.12 |
ENSMUST00000132528.8
ENSMUST00000153943.2 |
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr18_-_39622932 | 0.12 |
ENSMUST00000152853.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr17_+_43700327 | 0.12 |
ENSMUST00000113599.2
ENSMUST00000224278.2 ENSMUST00000225466.2 |
Adgrf5
|
adhesion G protein-coupled receptor F5 |
| chrX_-_47297746 | 0.11 |
ENSMUST00000088935.4
|
Zdhhc9
|
zinc finger, DHHC domain containing 9 |
| chr15_-_82796308 | 0.10 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr1_+_63655127 | 0.09 |
ENSMUST00000226288.2
|
Gm39653
|
predicted gene, 39653 |
| chr4_-_44710408 | 0.09 |
ENSMUST00000134968.9
ENSMUST00000173821.8 ENSMUST00000174319.8 ENSMUST00000173733.8 ENSMUST00000172866.8 ENSMUST00000165417.9 ENSMUST00000107825.9 ENSMUST00000102932.10 ENSMUST00000107827.9 ENSMUST00000107826.9 ENSMUST00000014174.14 |
Pax5
|
paired box 5 |
| chr2_+_84670543 | 0.09 |
ENSMUST00000111624.8
|
Slc43a1
|
solute carrier family 43, member 1 |
| chr19_-_19088543 | 0.09 |
ENSMUST00000112832.8
|
Rorb
|
RAR-related orphan receptor beta |
| chr13_+_83720484 | 0.09 |
ENSMUST00000196207.5
|
Mef2c
|
myocyte enhancer factor 2C |
| chr4_-_155430153 | 0.08 |
ENSMUST00000103178.11
|
Prkcz
|
protein kinase C, zeta |
| chr7_+_24607042 | 0.08 |
ENSMUST00000151121.2
|
Arhgef1
|
Rho guanine nucleotide exchange factor (GEF) 1 |
| chr7_-_109215754 | 0.08 |
ENSMUST00000084738.5
|
Denn2b
|
DENN domain containing 2B |
| chr9_-_37627519 | 0.08 |
ENSMUST00000215727.2
ENSMUST00000211952.3 |
Olfr160
|
olfactory receptor 160 |
| chr9_+_65268304 | 0.07 |
ENSMUST00000147185.3
|
Ubap1l
|
ubiquitin-associated protein 1-like |
| chr7_-_65020655 | 0.06 |
ENSMUST00000032729.8
|
Tjp1
|
tight junction protein 1 |
| chr13_+_60749995 | 0.06 |
ENSMUST00000044083.9
|
Dapk1
|
death associated protein kinase 1 |
| chr7_-_65020955 | 0.06 |
ENSMUST00000102592.10
|
Tjp1
|
tight junction protein 1 |
| chr18_-_39622295 | 0.06 |
ENSMUST00000131885.2
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr13_+_83723255 | 0.06 |
ENSMUST00000199167.5
ENSMUST00000195904.5 |
Mef2c
|
myocyte enhancer factor 2C |
| chr10_+_25308466 | 0.05 |
ENSMUST00000219224.2
ENSMUST00000219166.2 |
Epb41l2
|
erythrocyte membrane protein band 4.1 like 2 |
| chr12_-_85317359 | 0.05 |
ENSMUST00000166821.8
ENSMUST00000019378.8 ENSMUST00000220854.2 |
Mlh3
|
mutL homolog 3 |
| chr1_+_75119472 | 0.05 |
ENSMUST00000189650.7
|
Retreg2
|
reticulophagy regulator family member 2 |
| chr19_+_34268071 | 0.05 |
ENSMUST00000112472.4
ENSMUST00000235232.2 |
Fas
|
Fas (TNF receptor superfamily member 6) |
| chr1_-_127782735 | 0.05 |
ENSMUST00000208183.3
|
Map3k19
|
mitogen-activated protein kinase kinase kinase 19 |
| chr2_+_124452493 | 0.05 |
ENSMUST00000103239.10
ENSMUST00000103240.9 |
Sema6d
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
| chr7_+_29991101 | 0.05 |
ENSMUST00000150892.2
ENSMUST00000126216.2 ENSMUST00000014065.16 |
Clip3
|
CAP-GLY domain containing linker protein 3 |
| chr6_-_69741999 | 0.04 |
ENSMUST00000103365.3
|
Igkv12-46
|
immunoglobulin kappa variable 12-46 |
| chr2_-_85675827 | 0.04 |
ENSMUST00000213515.2
|
Olfr1019
|
olfactory receptor 1019 |
| chr6_-_69835868 | 0.04 |
ENSMUST00000103369.2
|
Igkv12-41
|
immunoglobulin kappa chain variable 12-41 |
| chr8_-_41469332 | 0.04 |
ENSMUST00000131965.2
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr9_+_78020554 | 0.04 |
ENSMUST00000009972.6
ENSMUST00000117330.8 ENSMUST00000044551.8 |
Cilk1
|
ciliogenesis associated kinase 1 |
| chr4_-_119272667 | 0.04 |
ENSMUST00000238609.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr5_-_18093739 | 0.04 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr19_-_34856853 | 0.03 |
ENSMUST00000036584.13
|
Pank1
|
pantothenate kinase 1 |
| chr8_-_26275182 | 0.03 |
ENSMUST00000038498.10
|
Bag4
|
BCL2-associated athanogene 4 |
| chr10_+_128139227 | 0.03 |
ENSMUST00000218315.2
ENSMUST00000219721.2 |
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chr2_-_118558852 | 0.03 |
ENSMUST00000102524.8
|
Plcb2
|
phospholipase C, beta 2 |
| chr16_+_35590745 | 0.03 |
ENSMUST00000231579.2
|
Hspbap1
|
Hspb associated protein 1 |
| chr10_+_40225272 | 0.03 |
ENSMUST00000044672.11
ENSMUST00000095743.4 |
Cdk19
|
cyclin-dependent kinase 19 |
| chr18_-_39622829 | 0.03 |
ENSMUST00000025300.13
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr7_-_70010341 | 0.02 |
ENSMUST00000032768.15
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr10_+_128139191 | 0.02 |
ENSMUST00000005825.8
|
Pan2
|
PAN2 poly(A) specific ribonuclease subunit |
| chrX_+_158480304 | 0.02 |
ENSMUST00000123433.8
|
Sh3kbp1
|
SH3-domain kinase binding protein 1 |
| chr4_-_58206596 | 0.02 |
ENSMUST00000042850.9
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr4_+_134042423 | 0.02 |
ENSMUST00000105875.8
ENSMUST00000030638.7 |
Trim63
|
tripartite motif-containing 63 |
| chr7_+_113365235 | 0.02 |
ENSMUST00000046687.16
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chr2_-_7086066 | 0.02 |
ENSMUST00000183209.8
|
Celf2
|
CUGBP, Elav-like family member 2 |
| chr3_-_145355725 | 0.01 |
ENSMUST00000029846.5
|
Ccn1
|
cellular communication network factor 1 |
| chr10_+_103203552 | 0.01 |
ENSMUST00000179636.3
ENSMUST00000217905.2 ENSMUST00000074204.12 |
Slc6a15
|
solute carrier family 6 (neurotransmitter transporter), member 15 |
| chr1_-_154692678 | 0.01 |
ENSMUST00000238369.2
|
Cacna1e
|
calcium channel, voltage-dependent, R type, alpha 1E subunit |
| chr3_+_107137924 | 0.01 |
ENSMUST00000179399.3
|
A630076J17Rik
|
RIKEN cDNA A630076J17 gene |
| chr7_+_113365356 | 0.01 |
ENSMUST00000084696.6
|
Spon1
|
spondin 1, (f-spondin) extracellular matrix protein |
| chr8_-_41469786 | 0.01 |
ENSMUST00000117735.8
|
Mtus1
|
mitochondrial tumor suppressor 1 |
| chr11_-_103844870 | 0.01 |
ENSMUST00000103075.11
|
Nsf
|
N-ethylmaleimide sensitive fusion protein |
| chr15_+_79232591 | 0.01 |
ENSMUST00000229285.2
|
Maff
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein F (avian) |
| chr12_+_30934320 | 0.01 |
ENSMUST00000067087.7
|
Alkal2
|
ALK and LTK ligand 2 |
| chr7_-_106423873 | 0.01 |
ENSMUST00000208895.3
|
Olfr702
|
olfactory receptor 702 |
| chr19_+_5927821 | 0.00 |
ENSMUST00000145200.8
ENSMUST00000025732.14 ENSMUST00000125114.8 ENSMUST00000155697.8 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr11_+_19874354 | 0.00 |
ENSMUST00000093299.13
|
Spred2
|
sprouty-related EVH1 domain containing 2 |
| chr11_+_74150634 | 0.00 |
ENSMUST00000217016.2
ENSMUST00000214303.2 |
Olfr406
|
olfactory receptor 406 |
| chr8_-_33374282 | 0.00 |
ENSMUST00000209107.4
ENSMUST00000209022.3 |
Nrg1
|
neuregulin 1 |
| chr5_+_8106527 | 0.00 |
ENSMUST00000148633.4
|
Sri
|
sorcin |
| chr3_+_63203516 | 0.00 |
ENSMUST00000029400.7
|
Mme
|
membrane metallo endopeptidase |
| chr10_+_102348076 | 0.00 |
ENSMUST00000219445.2
|
Rassf9
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Foxo3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 0.4 | GO:0002071 | glandular epithelial cell maturation(GO:0002071) |
| 0.1 | 0.4 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.1 | 0.2 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.5 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.2 | GO:0072488 | ammonium transmembrane transport(GO:0072488) |
| 0.0 | 0.2 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) |
| 0.0 | 0.4 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.0 | 0.2 | GO:1903566 | positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.3 | GO:2000672 | negative regulation of motor neuron apoptotic process(GO:2000672) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.0 | GO:0031104 | dendrite regeneration(GO:0031104) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.0 | 0.2 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0018230 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.1 | GO:2000667 | positive regulation of interleukin-5 secretion(GO:2000664) positive regulation of interleukin-13 secretion(GO:2000667) |
| 0.0 | 0.0 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.2 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.1 | GO:0005712 | chiasma(GO:0005712) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.1 | 0.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 0.3 | GO:0001225 | RNA polymerase II transcription coactivator binding(GO:0001225) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0031721 | hemoglobin alpha binding(GO:0031721) |
| 0.0 | 0.3 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0010428 | methyl-CpNpG binding(GO:0010428) |
| 0.0 | 0.2 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.4 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |