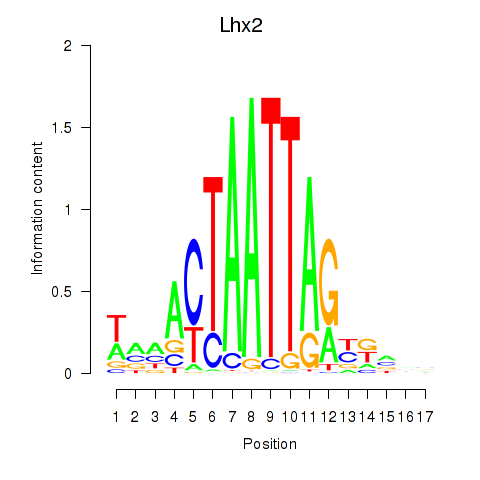
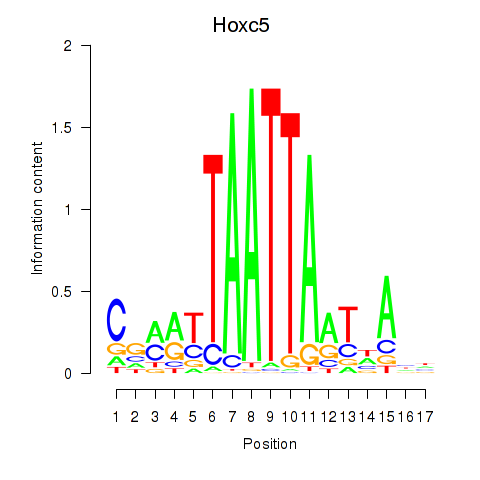
|
chr13_+_21901791
Show fit
|
3.96 |
ENSMUST00000188775.2
|
H3c10
|
H3 clustered histone 10
|
|
chr13_+_21995906
Show fit
|
3.85 |
ENSMUST00000104941.4
|
H4c17
|
H4 clustered histone 17
|
|
chr2_+_87610895
Show fit
|
3.12 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152
|
|
chr9_+_118892497
Show fit
|
2.95 |
ENSMUST00000141185.8
ENSMUST00000126251.8
ENSMUST00000136561.2
|
Vill
|
villin-like
|
|
chrM_+_9870
Show fit
|
2.91 |
ENSMUST00000084013.1
|
mt-Nd4l
|
mitochondrially encoded NADH dehydrogenase 4L
|
|
chr13_-_23929490
Show fit
|
2.33 |
ENSMUST00000091752.5
|
H3c3
|
H3 clustered histone 3
|
|
chr19_-_12313274
Show fit
|
1.77 |
ENSMUST00000208398.3
|
Olfr1438-ps1
|
olfactory receptor 1438, pseudogene 1
|
|
chr13_-_21900313
Show fit
|
1.75 |
ENSMUST00000091756.2
|
H2bc13
|
H2B clustered histone 13
|
|
chrX_+_16485937
Show fit
|
1.71 |
ENSMUST00000026013.6
|
Maoa
|
monoamine oxidase A
|
|
chr2_-_111820618
Show fit
|
1.68 |
ENSMUST00000216948.2
ENSMUST00000214935.2
ENSMUST00000217452.2
ENSMUST00000215045.2
|
Olfr1309
|
olfactory receptor 1309
|
|
chr17_+_33483650
Show fit
|
1.61 |
ENSMUST00000217023.3
|
Olfr63
|
olfactory receptor 63
|
|
chr2_-_85966272
Show fit
|
1.60 |
ENSMUST00000216566.3
ENSMUST00000214364.2
|
Olfr1039
|
olfactory receptor 1039
|
|
chr13_+_23728222
Show fit
|
1.55 |
ENSMUST00000075558.5
|
H3c7
|
H3 clustered histone 7
|
|
chrM_+_2743
Show fit
|
1.36 |
ENSMUST00000082392.1
|
mt-Nd1
|
mitochondrially encoded NADH dehydrogenase 1
|
|
chr13_+_23930717
Show fit
|
1.35 |
ENSMUST00000099703.5
|
H2bc3
|
H2B clustered histone 3
|
|
chrM_+_10167
Show fit
|
1.34 |
ENSMUST00000082414.1
|
mt-Nd4
|
mitochondrially encoded NADH dehydrogenase 4
|
|
chr9_+_123195986
Show fit
|
1.31 |
ENSMUST00000038863.9
ENSMUST00000216843.2
|
Lars2
|
leucyl-tRNA synthetase, mitochondrial
|
|
chr7_-_11414074
Show fit
|
1.28 |
ENSMUST00000227010.2
ENSMUST00000209638.3
|
Vmn1r72
|
vomeronasal 1 receptor 72
|
|
chr7_+_23752679
Show fit
|
1.24 |
ENSMUST00000236959.2
|
Vmn1r183
|
vomeronasal 1 receptor 183
|
|
chr2_-_111843053
Show fit
|
1.23 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310
|
|
chr7_+_130375799
Show fit
|
1.10 |
ENSMUST00000048453.7
ENSMUST00000208593.2
|
Btbd16
|
BTB (POZ) domain containing 16
|
|
chr7_-_10292412
Show fit
|
1.10 |
ENSMUST00000236246.2
|
Vmn1r68
|
vomeronasal 1 receptor 68
|
|
chr2_+_88217406
Show fit
|
1.07 |
ENSMUST00000214040.3
|
Olfr1178
|
olfactory receptor 1178
|
|
chr13_+_21900554
Show fit
|
1.05 |
ENSMUST00000070124.5
|
H2ac13
|
H2A clustered histone 13
|
|
chr17_-_59320257
Show fit
|
1.05 |
ENSMUST00000174122.2
ENSMUST00000025065.12
|
Nudt12
|
nudix (nucleoside diphosphate linked moiety X)-type motif 12
|
|
chr11_+_73851643
Show fit
|
1.04 |
ENSMUST00000213134.2
ENSMUST00000216291.2
|
Olfr397
|
olfactory receptor 397
|
|
chr14_-_8146867
Show fit
|
1.01 |
ENSMUST00000217035.2
ENSMUST00000206009.3
|
Olfr31
|
olfactory receptor 31
|
|
chr7_-_107633196
Show fit
|
1.00 |
ENSMUST00000210173.3
|
Olfr478
|
olfactory receptor 478
|
|
chr7_-_41708447
Show fit
|
0.99 |
ENSMUST00000168489.3
ENSMUST00000233456.2
|
Vmn2r59
|
vomeronasal 2, receptor 59
|
|
chr2_-_87868043
Show fit
|
0.98 |
ENSMUST00000129056.3
|
Olfr73
|
olfactory receptor 73
|
|
chr4_-_131802606
Show fit
|
0.98 |
ENSMUST00000146021.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr6_+_57180275
Show fit
|
0.97 |
ENSMUST00000226892.2
ENSMUST00000227421.2
|
Vmn1r13
|
vomeronasal 1 receptor 13
|
|
chrM_-_14061
Show fit
|
0.97 |
ENSMUST00000082419.1
|
mt-Nd6
|
mitochondrially encoded NADH dehydrogenase 6
|
|
chrX_-_100838004
Show fit
|
0.96 |
ENSMUST00000147742.9
|
Gm4779
|
predicted gene 4779
|
|
chr15_-_77840856
Show fit
|
0.96 |
ENSMUST00000117725.2
ENSMUST00000016696.13
|
Foxred2
|
FAD-dependent oxidoreductase domain containing 2
|
|
chr3_-_64473251
Show fit
|
0.92 |
ENSMUST00000176481.9
|
Vmn2r6
|
vomeronasal 2, receptor 6
|
|
chr6_-_57340712
Show fit
|
0.91 |
ENSMUST00000228156.2
ENSMUST00000227186.2
ENSMUST00000228294.2
ENSMUST00000228342.2
|
Vmn1r17
|
vomeronasal 1 receptor 17
|
|
chrX_+_102400061
Show fit
|
0.89 |
ENSMUST00000116547.3
|
Chic1
|
cysteine-rich hydrophobic domain 1
|
|
chr2_-_73284262
Show fit
|
0.88 |
ENSMUST00000102679.8
|
Wipf1
|
WAS/WASL interacting protein family, member 1
|
|
chr9_-_19275301
Show fit
|
0.86 |
ENSMUST00000214810.2
|
Olfr846
|
olfactory receptor 846
|
|
chr2_-_89855921
Show fit
|
0.85 |
ENSMUST00000216616.3
|
Olfr1264
|
olfactory receptor 1264
|
|
chr9_-_39618413
Show fit
|
0.84 |
ENSMUST00000215192.2
|
Olfr149
|
olfactory receptor 149
|
|
chr7_-_12829100
Show fit
|
0.80 |
ENSMUST00000209822.3
ENSMUST00000235753.2
|
Vmn1r85
|
vomeronasal 1 receptor 85
|
|
chr3_-_129834788
Show fit
|
0.80 |
ENSMUST00000168644.3
|
Sec24b
|
Sec24 related gene family, member B (S. cerevisiae)
|
|
chr8_+_66838927
Show fit
|
0.79 |
ENSMUST00000039540.12
ENSMUST00000110253.3
|
Marchf1
|
membrane associated ring-CH-type finger 1
|
|
chr2_+_181632922
Show fit
|
0.78 |
ENSMUST00000071760.8
ENSMUST00000236373.2
ENSMUST00000184507.3
|
Gm14496
|
predicted gene 14496
|
|
chr6_-_57306479
Show fit
|
0.78 |
ENSMUST00000227283.2
ENSMUST00000228356.2
|
Vmn1r16
|
vomeronasal 1 receptor 16
|
|
chr1_+_40478787
Show fit
|
0.76 |
ENSMUST00000097772.10
|
Il1rl1
|
interleukin 1 receptor-like 1
|
|
chr7_-_126574959
Show fit
|
0.76 |
ENSMUST00000206296.2
ENSMUST00000205437.3
|
Cdiptos
|
CDIP transferase, opposite strand
|
|
chr2_-_87798643
Show fit
|
0.75 |
ENSMUST00000099841.4
|
Olfr1157
|
olfactory receptor 1157
|
|
chr1_+_16758629
Show fit
|
0.73 |
ENSMUST00000026881.11
|
Ly96
|
lymphocyte antigen 96
|
|
chr11_-_73382303
Show fit
|
0.72 |
ENSMUST00000119863.2
ENSMUST00000215358.2
ENSMUST00000214623.2
|
Olfr381
|
olfactory receptor 381
|
|
chr7_+_28869770
Show fit
|
0.72 |
ENSMUST00000033886.8
ENSMUST00000209019.2
ENSMUST00000208330.2
|
Ggn
|
gametogenetin
|
|
chr10_+_129072073
Show fit
|
0.72 |
ENSMUST00000203248.3
|
Olfr774
|
olfactory receptor 774
|
|
chr17_-_47998953
Show fit
|
0.71 |
ENSMUST00000113301.2
ENSMUST00000113302.10
|
Tomm6
|
translocase of outer mitochondrial membrane 6
|
|
chr11_+_49410475
Show fit
|
0.71 |
ENSMUST00000204706.3
|
Olfr1383
|
olfactory receptor 1383
|
|
chr2_-_111253457
Show fit
|
0.69 |
ENSMUST00000213210.2
ENSMUST00000184954.4
|
Olfr1286
|
olfactory receptor 1286
|
|
chr2_-_111104451
Show fit
|
0.69 |
ENSMUST00000214760.2
|
Olfr1277
|
olfactory receptor 1277
|
|
chr17_+_21060706
Show fit
|
0.68 |
ENSMUST00000232909.2
ENSMUST00000233670.2
ENSMUST00000233939.2
|
Vmn1r230
|
vomeronasal 1 receptor 230
|
|
chr10_-_75946790
Show fit
|
0.67 |
ENSMUST00000120757.2
|
Slc5a4b
|
solute carrier family 5 (neutral amino acid transporters, system A), member 4b
|
|
chr7_-_12002196
Show fit
|
0.67 |
ENSMUST00000227973.2
ENSMUST00000228764.2
ENSMUST00000228482.2
ENSMUST00000227080.2
|
Vmn1r81
|
vomeronasal 1 receptor 81
|
|
chr16_-_58898368
Show fit
|
0.67 |
ENSMUST00000216495.3
|
Olfr190
|
olfactory receptor 190
|
|
chr7_+_107585900
Show fit
|
0.67 |
ENSMUST00000214677.2
|
Olfr477
|
olfactory receptor 477
|
|
chr7_-_30259025
Show fit
|
0.66 |
ENSMUST00000043975.11
ENSMUST00000156241.2
|
Lin37
|
lin-37 homolog (C. elegans)
|
|
chr2_-_111880531
Show fit
|
0.66 |
ENSMUST00000213582.2
ENSMUST00000213961.3
ENSMUST00000215531.2
|
Olfr1312
|
olfactory receptor 1312
|
|
chr17_+_20634408
Show fit
|
0.66 |
ENSMUST00000233980.2
ENSMUST00000233642.2
ENSMUST00000233743.2
ENSMUST00000233318.2
ENSMUST00000233161.2
ENSMUST00000233891.2
ENSMUST00000233600.2
ENSMUST00000233863.2
|
Vmn1r224
|
vomeronasal 1 receptor 224
|
|
chr5_-_125119855
Show fit
|
0.66 |
ENSMUST00000125053.8
|
Ncor2
|
nuclear receptor co-repressor 2
|
|
chr5_-_84565218
Show fit
|
0.65 |
ENSMUST00000113401.4
|
Epha5
|
Eph receptor A5
|
|
chr7_+_108265625
Show fit
|
0.63 |
ENSMUST00000213979.3
ENSMUST00000216331.2
ENSMUST00000217170.2
|
Olfr510
|
olfactory receptor 510
|
|
chr7_+_23650057
Show fit
|
0.63 |
ENSMUST00000173816.5
|
Vmn1r180
|
vomeronasal 1 receptor 180
|
|
chr6_-_58195806
Show fit
|
0.63 |
ENSMUST00000228530.2
ENSMUST00000226666.2
|
Vmn1r27
|
vomeronasal 1 receptor 27
|
|
chr16_+_51851948
Show fit
|
0.62 |
ENSMUST00000226593.2
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chrM_+_3906
Show fit
|
0.60 |
ENSMUST00000082396.1
|
mt-Nd2
|
mitochondrially encoded NADH dehydrogenase 2
|
|
chr7_-_5152669
Show fit
|
0.60 |
ENSMUST00000236378.2
|
Vmn1r55
|
vomeronasal 1 receptor 55
|
|
chr3_-_15902583
Show fit
|
0.60 |
ENSMUST00000108354.8
ENSMUST00000108349.2
ENSMUST00000108352.9
ENSMUST00000108350.8
ENSMUST00000050623.11
|
Sirpb1c
|
signal-regulatory protein beta 1C
|
|
chr14_-_50586329
Show fit
|
0.59 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735
|
|
chr13_-_110493665
Show fit
|
0.59 |
ENSMUST00000058806.7
ENSMUST00000224534.2
|
Gapt
|
Grb2-binding adaptor, transmembrane
|
|
chr8_+_114362181
Show fit
|
0.59 |
ENSMUST00000179926.9
|
Mon1b
|
MON1 homolog B, secretory traffciking associated
|
|
chr3_-_66204228
Show fit
|
0.58 |
ENSMUST00000029419.8
|
Veph1
|
ventricular zone expressed PH domain-containing 1
|
|
chr11_-_73742280
Show fit
|
0.57 |
ENSMUST00000213365.2
|
Olfr393
|
olfactory receptor 393
|
|
chr14_-_50564218
Show fit
|
0.57 |
ENSMUST00000217152.2
|
Olfr734
|
olfactory receptor 734
|
|
chr8_-_70963202
Show fit
|
0.57 |
ENSMUST00000125184.8
|
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1
|
|
chr15_-_66985760
Show fit
|
0.57 |
ENSMUST00000092640.6
|
St3gal1
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 1
|
|
chr16_+_51851917
Show fit
|
0.57 |
ENSMUST00000227062.2
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chr16_+_65317389
Show fit
|
0.56 |
ENSMUST00000176330.8
ENSMUST00000004964.15
ENSMUST00000176038.8
|
Pou1f1
|
POU domain, class 1, transcription factor 1
|
|
chr19_+_12364643
Show fit
|
0.56 |
ENSMUST00000217062.3
ENSMUST00000216145.2
ENSMUST00000213657.2
|
Olfr1440
|
olfactory receptor 1440
|
|
chr2_-_86528739
Show fit
|
0.56 |
ENSMUST00000214141.2
|
Olfr1087
|
olfactory receptor 1087
|
|
chr4_-_155729865
Show fit
|
0.56 |
ENSMUST00000115821.3
|
Gm10563
|
predicted gene 10563
|
|
chr16_-_48592372
Show fit
|
0.56 |
ENSMUST00000231701.3
|
Trat1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr16_-_59138611
Show fit
|
0.55 |
ENSMUST00000216261.2
|
Olfr204
|
olfactory receptor 204
|
|
chrM_+_14138
Show fit
|
0.55 |
ENSMUST00000082421.1
|
mt-Cytb
|
mitochondrially encoded cytochrome b
|
|
chr14_+_51366512
Show fit
|
0.55 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6
|
|
chr2_-_86257093
Show fit
|
0.54 |
ENSMUST00000217481.2
|
Olfr1062
|
olfactory receptor 1062
|
|
chr1_-_92412835
Show fit
|
0.54 |
ENSMUST00000214928.3
|
Olfr1416
|
olfactory receptor 1416
|
|
chr7_-_103113358
Show fit
|
0.54 |
ENSMUST00000214347.2
|
Olfr607
|
olfactory receptor 607
|
|
chr7_+_5221492
Show fit
|
0.54 |
ENSMUST00000228062.2
ENSMUST00000227798.2
|
Vmn1r57
|
vomeronasal 1 receptor 57
|
|
chr19_+_13339600
Show fit
|
0.52 |
ENSMUST00000215096.2
|
Olfr1467
|
olfactory receptor 1467
|
|
chr9_+_38374440
Show fit
|
0.52 |
ENSMUST00000216724.3
|
Olfr904
|
olfactory receptor 904
|
|
chr7_-_103701873
Show fit
|
0.52 |
ENSMUST00000214299.2
|
Olfr642
|
olfactory receptor 642
|
|
chr6_+_57133904
Show fit
|
0.51 |
ENSMUST00000226866.2
ENSMUST00000227581.2
|
Vmn1r12
|
vomeronasal 1 receptor 12
|
|
chr8_+_72860884
Show fit
|
0.51 |
ENSMUST00000210435.4
|
Olfr374
|
olfactory receptor 374
|
|
chr17_+_56920389
Show fit
|
0.50 |
ENSMUST00000080492.7
|
Rpl36
|
ribosomal protein L36
|
|
chr2_-_34990689
Show fit
|
0.50 |
ENSMUST00000226631.2
ENSMUST00000045776.5
ENSMUST00000226972.2
|
AI182371
|
expressed sequence AI182371
|
|
chr17_-_37974666
Show fit
|
0.50 |
ENSMUST00000215414.2
ENSMUST00000213638.3
|
Olfr117
|
olfactory receptor 117
|
|
chr11_+_58839716
Show fit
|
0.50 |
ENSMUST00000078267.5
|
H2bu2
|
H2B.U histone 2
|
|
chr9_+_38491474
Show fit
|
0.49 |
ENSMUST00000217160.3
|
Olfr912
|
olfactory receptor 912
|
|
chr7_+_5023375
Show fit
|
0.48 |
ENSMUST00000076251.7
|
Zfp865
|
zinc finger protein 865
|
|
chr7_+_5023552
Show fit
|
0.48 |
ENSMUST00000208728.2
ENSMUST00000085427.6
|
Ccdc106
Zfp865
|
coiled-coil domain containing 106
zinc finger protein 865
|
|
chr17_-_37523969
Show fit
|
0.48 |
ENSMUST00000060728.7
ENSMUST00000216318.2
|
Olfr95
|
olfactory receptor 95
|
|
chr14_+_99536111
Show fit
|
0.48 |
ENSMUST00000005279.8
|
Klf5
|
Kruppel-like factor 5
|
|
chr10_-_81243475
Show fit
|
0.47 |
ENSMUST00000140916.8
|
Nfic
|
nuclear factor I/C
|
|
chr2_-_88745501
Show fit
|
0.47 |
ENSMUST00000213136.2
|
Olfr1209
|
olfactory receptor 1209
|
|
chr7_+_28869629
Show fit
|
0.46 |
ENSMUST00000098609.4
|
Ggn
|
gametogenetin
|
|
chr16_+_45044678
Show fit
|
0.46 |
ENSMUST00000102802.10
ENSMUST00000063654.6
|
Btla
|
B and T lymphocyte associated
|
|
chr19_-_12302465
Show fit
|
0.46 |
ENSMUST00000207241.3
|
Olfr1437
|
olfactory receptor 1437
|
|
chr1_-_150341911
Show fit
|
0.45 |
ENSMUST00000162367.8
ENSMUST00000161611.8
ENSMUST00000161320.8
ENSMUST00000159035.2
|
Prg4
|
proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein)
|
|
chr14_-_50538979
Show fit
|
0.45 |
ENSMUST00000216195.2
ENSMUST00000214372.2
ENSMUST00000214756.2
|
Olfr733
|
olfactory receptor 733
|
|
chr6_+_57405915
Show fit
|
0.45 |
ENSMUST00000227909.2
|
Vmn1r20
|
vomeronasal 1 receptor 20
|
|
chr6_+_129022843
Show fit
|
0.44 |
ENSMUST00000032257.10
ENSMUST00000204320.2
|
Klrb1f
|
killer cell lectin-like receptor subfamily B member 1F
|
|
chr17_+_38456172
Show fit
|
0.43 |
ENSMUST00000215078.3
ENSMUST00000215549.3
ENSMUST00000173610.2
|
Olfr133
|
olfactory receptor 133
|
|
chr1_-_54596754
Show fit
|
0.43 |
ENSMUST00000097739.5
|
Pgap1
|
post-GPI attachment to proteins 1
|
|
chr16_-_58695131
Show fit
|
0.42 |
ENSMUST00000217377.2
|
Olfr177
|
olfactory receptor 177
|
|
chr11_-_50183129
Show fit
|
0.42 |
ENSMUST00000059458.5
|
Maml1
|
mastermind like transcriptional coactivator 1
|
|
chr7_-_106531426
Show fit
|
0.42 |
ENSMUST00000215468.2
|
Olfr709-ps1
|
olfactory receptor 709, pseudogene 1
|
|
chrY_-_1245685
Show fit
|
0.42 |
ENSMUST00000143286.8
ENSMUST00000137048.8
ENSMUST00000069309.14
ENSMUST00000139365.8
ENSMUST00000154004.8
|
Uty
|
ubiquitously transcribed tetratricopeptide repeat containing, Y-linked
|
|
chr8_+_114362419
Show fit
|
0.42 |
ENSMUST00000035777.10
|
Mon1b
|
MON1 homolog B, secretory traffciking associated
|
|
chr2_-_88157559
Show fit
|
0.42 |
ENSMUST00000214207.2
|
Olfr1175
|
olfactory receptor 1175
|
|
chr2_-_89423470
Show fit
|
0.41 |
ENSMUST00000217254.2
ENSMUST00000217192.2
ENSMUST00000213221.2
|
Olfr1246
|
olfactory receptor 1246
|
|
chr14_+_8282925
Show fit
|
0.41 |
ENSMUST00000217642.2
|
Olfr720
|
olfactory receptor 720
|
|
chr11_-_96859484
Show fit
|
0.41 |
ENSMUST00000107623.8
|
Sp2
|
Sp2 transcription factor
|
|
chr4_-_148236516
Show fit
|
0.41 |
ENSMUST00000056965.12
ENSMUST00000168503.8
ENSMUST00000152098.8
|
Fbxo6
|
F-box protein 6
|
|
chr4_-_131802561
Show fit
|
0.41 |
ENSMUST00000105970.8
ENSMUST00000105975.8
|
Epb41
|
erythrocyte membrane protein band 4.1
|
|
chr4_-_119217079
Show fit
|
0.41 |
ENSMUST00000143494.3
ENSMUST00000154606.9
|
Ccdc30
|
coiled-coil domain containing 30
|
|
chr9_-_62895197
Show fit
|
0.41 |
ENSMUST00000216209.2
|
Pias1
|
protein inhibitor of activated STAT 1
|
|
chr18_-_60981981
Show fit
|
0.41 |
ENSMUST00000177172.8
ENSMUST00000175934.8
ENSMUST00000176630.8
|
Tcof1
|
treacle ribosome biogenesis factor 1
|
|
chr7_-_140596811
Show fit
|
0.40 |
ENSMUST00000081924.5
|
Ifitm6
|
interferon induced transmembrane protein 6
|
|
chr9_+_20193647
Show fit
|
0.40 |
ENSMUST00000071725.4
ENSMUST00000212983.3
|
Olfr39
|
olfactory receptor 39
|
|
chr7_-_30259253
Show fit
|
0.40 |
ENSMUST00000108164.8
|
Lin37
|
lin-37 homolog (C. elegans)
|
|
chr9_+_38398381
Show fit
|
0.39 |
ENSMUST00000214344.3
|
Olfr906
|
olfactory receptor 906
|
|
chr5_-_100521343
Show fit
|
0.39 |
ENSMUST00000182433.8
|
Sec31a
|
Sec31 homolog A (S. cerevisiae)
|
|
chr1_-_184464810
Show fit
|
0.39 |
ENSMUST00000048572.7
|
Hlx
|
H2.0-like homeobox
|
|
chr6_-_37419030
Show fit
|
0.39 |
ENSMUST00000041093.6
|
Creb3l2
|
cAMP responsive element binding protein 3-like 2
|
|
chr2_+_88470886
Show fit
|
0.38 |
ENSMUST00000217379.2
ENSMUST00000120598.3
|
Olfr1191-ps1
|
olfactory receptor 1191, pseudogene 1
|
|
chr16_+_11224481
Show fit
|
0.38 |
ENSMUST00000122168.8
|
Snx29
|
sorting nexin 29
|
|
chr7_-_140597465
Show fit
|
0.38 |
ENSMUST00000211330.2
|
Ifitm6
|
interferon induced transmembrane protein 6
|
|
chr16_+_51851588
Show fit
|
0.38 |
ENSMUST00000114471.3
|
Cblb
|
Casitas B-lineage lymphoma b
|
|
chr2_+_118877610
Show fit
|
0.38 |
ENSMUST00000153300.8
ENSMUST00000028799.12
|
Knl1
|
kinetochore scaffold 1
|
|
chr9_+_38259893
Show fit
|
0.37 |
ENSMUST00000216304.2
|
Olfr898
|
olfactory receptor 898
|
|
chr2_-_111779785
Show fit
|
0.37 |
ENSMUST00000099604.6
|
Olfr1307
|
olfactory receptor 1307
|
|
chr15_+_98350469
Show fit
|
0.37 |
ENSMUST00000217517.2
|
Olfr281
|
olfactory receptor 281
|
|
chr6_+_90078412
Show fit
|
0.37 |
ENSMUST00000089417.8
ENSMUST00000226577.2
|
Vmn1r50
|
vomeronasal 1 receptor 50
|
|
chr10_-_4382283
Show fit
|
0.37 |
ENSMUST00000155172.8
|
Rmnd1
|
required for meiotic nuclear division 1 homolog
|
|
chr6_-_36787096
Show fit
|
0.37 |
ENSMUST00000201321.2
ENSMUST00000101534.5
|
Ptn
|
pleiotrophin
|
|
chr16_+_44687460
Show fit
|
0.37 |
ENSMUST00000102805.4
|
Cd200r2
|
Cd200 receptor 2
|
|
chr5_-_116162415
Show fit
|
0.36 |
ENSMUST00000031486.14
ENSMUST00000111999.8
|
Prkab1
|
protein kinase, AMP-activated, beta 1 non-catalytic subunit
|
|
chr19_+_13208692
Show fit
|
0.36 |
ENSMUST00000207246.4
|
Olfr1463
|
olfactory receptor 1463
|
|
chr9_+_38119661
Show fit
|
0.36 |
ENSMUST00000211975.3
|
Olfr893
|
olfactory receptor 893
|
|
chr16_-_64591509
Show fit
|
0.36 |
ENSMUST00000076991.7
|
4930453N24Rik
|
RIKEN cDNA 4930453N24 gene
|
|
chr2_+_87260619
Show fit
|
0.36 |
ENSMUST00000215909.2
|
Olfr1124
|
olfactory receptor 1124
|
|
chr2_-_5680801
Show fit
|
0.36 |
ENSMUST00000114987.4
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID
|
|
chr7_+_19095111
Show fit
|
0.36 |
ENSMUST00000047621.14
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like
|
|
chr2_-_111815654
Show fit
|
0.35 |
ENSMUST00000214537.2
|
Olfr1309
|
olfactory receptor 1309
|
|
chr13_-_21652632
Show fit
|
0.35 |
ENSMUST00000068235.6
|
Nkapl
|
NFKB activating protein-like
|
|
chr10_+_78864575
Show fit
|
0.34 |
ENSMUST00000203906.3
|
Olfr57
|
olfactory receptor 57
|
|
chr10_-_128462616
Show fit
|
0.34 |
ENSMUST00000026420.7
|
Rps26
|
ribosomal protein S26
|
|
chr19_+_13594739
Show fit
|
0.34 |
ENSMUST00000217061.3
ENSMUST00000209005.4
ENSMUST00000208347.3
|
Olfr1487
|
olfactory receptor 1487
|
|
chr6_-_58452315
Show fit
|
0.34 |
ENSMUST00000177318.3
ENSMUST00000176147.9
ENSMUST00000176023.3
ENSMUST00000228586.2
|
Vmn1r31
|
vomeronasal 1 receptor 31
|
|
chr11_-_95966477
Show fit
|
0.34 |
ENSMUST00000090541.12
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9)
|
|
chr2_-_111400026
Show fit
|
0.33 |
ENSMUST00000217772.2
ENSMUST00000207283.3
|
Olfr1295
|
olfactory receptor 1295
|
|
chr4_+_145311722
Show fit
|
0.33 |
ENSMUST00000105739.8
|
Zfp268
|
zinc finger protein 268
|
|
chr11_-_99134885
Show fit
|
0.33 |
ENSMUST00000103132.10
ENSMUST00000038214.7
|
Krt222
|
keratin 222
|
|
chr18_-_46444941
Show fit
|
0.33 |
ENSMUST00000072835.7
|
Ccdc112
|
coiled-coil domain containing 112
|
|
chr4_+_146033882
Show fit
|
0.33 |
ENSMUST00000105730.2
ENSMUST00000091878.6
|
Zfp987
|
zinc finger protein 987
|
|
chr16_-_36695166
Show fit
|
0.33 |
ENSMUST00000075946.12
|
Eaf2
|
ELL associated factor 2
|
|
chr7_+_23400128
Show fit
|
0.33 |
ENSMUST00000226233.2
ENSMUST00000227987.2
|
Vmn1r173
|
vomeronasal 1 receptor 173
|
|
chr12_+_119356318
Show fit
|
0.32 |
ENSMUST00000221866.2
|
Macc1
|
metastasis associated in colon cancer 1
|
|
chrM_+_7006
Show fit
|
0.32 |
ENSMUST00000082405.1
|
mt-Co2
|
mitochondrially encoded cytochrome c oxidase II
|
|
chr7_-_107726656
Show fit
|
0.32 |
ENSMUST00000214722.2
|
Olfr484
|
olfactory receptor 484
|
|
chr4_-_41045381
Show fit
|
0.32 |
ENSMUST00000054945.8
|
Aqp7
|
aquaporin 7
|
|
chr1_-_183003077
Show fit
|
0.32 |
ENSMUST00000194033.2
|
Disp1
|
dispatched RND transporter family member 1
|
|
chr7_-_29894471
Show fit
|
0.31 |
ENSMUST00000126116.3
|
Capns1
|
calpain, small subunit 1
|
|
chr6_-_66757618
Show fit
|
0.31 |
ENSMUST00000227493.2
|
Vmn1r38
|
vomeronasal 1 receptor 38
|
|
chr3_-_14676309
Show fit
|
0.31 |
ENSMUST00000185423.2
ENSMUST00000186870.7
ENSMUST00000185384.7
|
Rbis
|
ribosomal biogenesis factor
|
|
chr6_+_136509922
Show fit
|
0.31 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein
|
|
chr7_+_104236232
Show fit
|
0.30 |
ENSMUST00000213984.2
|
Olfr654
|
olfactory receptor 654
|
|
chr17_-_38618461
Show fit
|
0.30 |
ENSMUST00000213505.2
|
Olfr137
|
olfactory receptor 137
|
|
chr2_+_111984716
Show fit
|
0.30 |
ENSMUST00000214063.2
ENSMUST00000217533.2
|
Olfr1318
|
olfactory receptor 1318
|
|
chr2_-_86926352
Show fit
|
0.30 |
ENSMUST00000217066.3
ENSMUST00000214636.2
|
Olfr1109
|
olfactory receptor 1109
|
|
chrM_+_5319
Show fit
|
0.30 |
ENSMUST00000082402.1
|
mt-Co1
|
mitochondrially encoded cytochrome c oxidase I
|
|
chr9_-_40004854
Show fit
|
0.30 |
ENSMUST00000050996.6
ENSMUST00000213087.4
|
Olfr983
|
olfactory receptor 983
|
|
chr13_+_44882998
Show fit
|
0.30 |
ENSMUST00000174068.8
|
Jarid2
|
jumonji, AT rich interactive domain 2
|
|
chr10_+_7708178
Show fit
|
0.29 |
ENSMUST00000039484.6
|
Zc3h12d
|
zinc finger CCCH type containing 12D
|
|
chr17_+_20903105
Show fit
|
0.29 |
ENSMUST00000232708.2
ENSMUST00000233808.2
ENSMUST00000232818.2
ENSMUST00000233159.2
ENSMUST00000233959.2
ENSMUST00000233722.2
|
Vmn1r226
|
vomeronasal 1 receptor 226
|
|
chr3_-_97329336
Show fit
|
0.29 |
ENSMUST00000199297.4
|
Olfr1402
|
olfactory receptor 1402
|
|
chr14_+_56813060
Show fit
|
0.29 |
ENSMUST00000161553.2
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4
|
|
chr10_+_129153986
Show fit
|
0.29 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780
|
|
chr16_+_75389732
Show fit
|
0.29 |
ENSMUST00000046378.14
ENSMUST00000114249.8
ENSMUST00000114253.2
|
Rbm11
|
RNA binding motif protein 11
|
|
chr5_-_3691453
Show fit
|
0.29 |
ENSMUST00000140871.2
|
Gatad1
|
GATA zinc finger domain containing 1
|
|
chr7_+_48608800
Show fit
|
0.28 |
ENSMUST00000183659.8
|
Nav2
|
neuron navigator 2
|
|
chr1_+_173093568
Show fit
|
0.28 |
ENSMUST00000213420.2
|
Olfr418
|
olfactory receptor 418
|