Project
GFI1 WT vs 36n/n vs KD
Navigation
Downloads
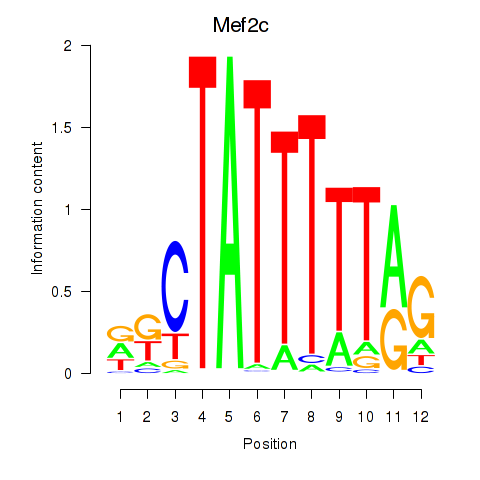
Results for Mef2c
Z-value: 0.52
Transcription factors associated with Mef2c
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Mef2c
|
ENSMUSG00000005583.17 | myocyte enhancer factor 2C |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Mef2c | mm39_v1_chr13_+_83720484_83720509 | -0.65 | 2.4e-01 | Click! |
Activity profile of Mef2c motif
Sorted Z-values of Mef2c motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_34578842 | 0.58 |
ENSMUST00000110819.4
|
Hdac9
|
histone deacetylase 9 |
| chr11_+_3280401 | 0.54 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr11_+_120839879 | 0.45 |
ENSMUST00000154187.8
ENSMUST00000100130.10 ENSMUST00000129473.8 ENSMUST00000168579.8 |
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr11_+_23234644 | 0.41 |
ENSMUST00000150750.3
|
Xpo1
|
exportin 1 |
| chr4_-_94940425 | 0.34 |
ENSMUST00000107094.2
|
Jun
|
jun proto-oncogene |
| chr15_+_102391614 | 0.34 |
ENSMUST00000229432.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr7_-_126399208 | 0.31 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_126399778 | 0.28 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr1_-_13061333 | 0.27 |
ENSMUST00000115403.9
ENSMUST00000136197.8 ENSMUST00000115402.8 |
Slco5a1
|
solute carrier organic anion transporter family, member 5A1 |
| chr11_-_120538928 | 0.27 |
ENSMUST00000239158.2
ENSMUST00000026134.3 |
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr15_+_102379503 | 0.26 |
ENSMUST00000229222.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr2_-_32584132 | 0.25 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr4_-_9643636 | 0.24 |
ENSMUST00000108333.8
ENSMUST00000108334.8 ENSMUST00000108335.8 ENSMUST00000152526.8 ENSMUST00000103004.10 |
Asph
|
aspartate-beta-hydroxylase |
| chr2_-_164675357 | 0.22 |
ENSMUST00000042775.5
|
Neurl2
|
neuralized E3 ubiquitin protein ligase 2 |
| chr5_-_88675700 | 0.21 |
ENSMUST00000087033.6
|
Jchain
|
immunoglobulin joining chain |
| chr15_-_89310060 | 0.21 |
ENSMUST00000109313.9
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr11_+_94218810 | 0.20 |
ENSMUST00000107818.9
ENSMUST00000051221.13 |
Ankrd40
|
ankyrin repeat domain 40 |
| chr11_+_70548022 | 0.20 |
ENSMUST00000157027.8
ENSMUST00000072841.12 ENSMUST00000108548.8 ENSMUST00000126241.8 |
Eno3
|
enolase 3, beta muscle |
| chr7_-_126399574 | 0.19 |
ENSMUST00000106348.8
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr3_+_96552895 | 0.19 |
ENSMUST00000119365.8
ENSMUST00000029744.6 |
Itga10
|
integrin, alpha 10 |
| chr5_-_113044216 | 0.19 |
ENSMUST00000086617.11
|
Myo18b
|
myosin XVIIIb |
| chr2_+_120294046 | 0.19 |
ENSMUST00000028749.15
ENSMUST00000110721.9 ENSMUST00000239364.2 |
Capn3
|
calpain 3 |
| chr2_+_154633265 | 0.18 |
ENSMUST00000140713.3
ENSMUST00000137333.8 |
Raly
a
|
hnRNP-associated with lethal yellow nonagouti |
| chr7_+_141995545 | 0.18 |
ENSMUST00000105971.8
ENSMUST00000145287.8 |
Tnni2
|
troponin I, skeletal, fast 2 |
| chr7_-_127805518 | 0.18 |
ENSMUST00000033049.9
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr12_+_117621644 | 0.18 |
ENSMUST00000222105.2
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr7_-_84328553 | 0.17 |
ENSMUST00000069537.3
ENSMUST00000207865.2 ENSMUST00000178385.9 ENSMUST00000208782.2 |
Zfand6
|
zinc finger, AN1-type domain 6 |
| chr1_+_180678677 | 0.17 |
ENSMUST00000038091.8
|
Sde2
|
SDE2 telomere maintenance homolog (S. pombe) |
| chr4_+_144619696 | 0.17 |
ENSMUST00000142808.8
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr15_+_73620213 | 0.16 |
ENSMUST00000053232.8
|
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr11_+_77692116 | 0.16 |
ENSMUST00000100794.10
ENSMUST00000151373.4 ENSMUST00000130305.9 ENSMUST00000172303.10 ENSMUST00000092884.11 ENSMUST00000164334.8 |
Myo18a
|
myosin XVIIIA |
| chr17_-_71305003 | 0.16 |
ENSMUST00000024846.13
ENSMUST00000232766.2 |
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_112586501 | 0.16 |
ENSMUST00000180015.9
|
Adssl1
|
adenylosuccinate synthetase like 1 |
| chr4_-_141391406 | 0.16 |
ENSMUST00000084203.11
|
Plekhm2
|
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
| chr9_-_35030479 | 0.16 |
ENSMUST00000213526.2
ENSMUST00000215089.2 |
St3gal4
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr2_-_48839218 | 0.16 |
ENSMUST00000090976.10
ENSMUST00000149679.8 |
Orc4
|
origin recognition complex, subunit 4 |
| chr7_+_46495521 | 0.16 |
ENSMUST00000133062.2
|
Ldha
|
lactate dehydrogenase A |
| chr12_-_84447702 | 0.16 |
ENSMUST00000122194.8
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr8_-_72124359 | 0.16 |
ENSMUST00000177517.8
ENSMUST00000030170.15 |
Unc13a
|
unc-13 homolog A |
| chr11_+_120123727 | 0.16 |
ENSMUST00000122148.8
ENSMUST00000044985.14 |
Bahcc1
|
BAH domain and coiled-coil containing 1 |
| chr17_+_73433422 | 0.16 |
ENSMUST00000232703.2
|
Lclat1
|
lysocardiolipin acyltransferase 1 |
| chr11_+_94219046 | 0.15 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr10_+_127337541 | 0.15 |
ENSMUST00000160019.8
ENSMUST00000160610.2 ENSMUST00000035839.3 |
Stac3
|
SH3 and cysteine rich domain 3 |
| chr17_-_66079681 | 0.15 |
ENSMUST00000070673.9
|
Rab31
|
RAB31, member RAS oncogene family |
| chr9_-_42368880 | 0.15 |
ENSMUST00000125995.8
|
Tbcel
|
tubulin folding cofactor E-like |
| chr19_-_6065415 | 0.14 |
ENSMUST00000237519.2
|
Capn1
|
calpain 1 |
| chr5_+_64317550 | 0.14 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr6_-_25809209 | 0.14 |
ENSMUST00000115330.8
|
Pot1a
|
protection of telomeres 1A |
| chr17_-_45903410 | 0.14 |
ENSMUST00000166119.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr12_-_84447625 | 0.13 |
ENSMUST00000117286.2
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr7_-_4517367 | 0.13 |
ENSMUST00000166161.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr18_-_35631914 | 0.13 |
ENSMUST00000236007.2
ENSMUST00000237896.2 ENSMUST00000235778.2 ENSMUST00000235524.2 ENSMUST00000235691.2 ENSMUST00000235619.2 ENSMUST00000025215.10 |
Sil1
|
endoplasmic reticulum chaperone SIL1 homolog (S. cerevisiae) |
| chr10_-_79744726 | 0.13 |
ENSMUST00000165684.8
ENSMUST00000164705.8 ENSMUST00000105378.9 ENSMUST00000170409.2 |
Med16
|
mediator complex subunit 16 |
| chr7_-_100164007 | 0.13 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr2_-_48839276 | 0.13 |
ENSMUST00000028098.11
|
Orc4
|
origin recognition complex, subunit 4 |
| chr11_+_95925711 | 0.13 |
ENSMUST00000006217.10
ENSMUST00000107700.4 |
Snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr1_+_75213044 | 0.12 |
ENSMUST00000188931.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr3_-_65865656 | 0.12 |
ENSMUST00000029416.14
|
Ccnl1
|
cyclin L1 |
| chr4_+_156300325 | 0.12 |
ENSMUST00000105572.3
|
Perm1
|
PPARGC1 and ESRR induced regulator, muscle 1 |
| chr19_-_6065799 | 0.11 |
ENSMUST00000235138.2
|
Capn1
|
calpain 1 |
| chrX_-_92875712 | 0.11 |
ENSMUST00000045748.7
|
Pdk3
|
pyruvate dehydrogenase kinase, isoenzyme 3 |
| chr17_+_57556449 | 0.11 |
ENSMUST00000224947.2
ENSMUST00000019631.11 ENSMUST00000224885.2 ENSMUST00000224152.2 |
Trip10
|
thyroid hormone receptor interactor 10 |
| chr2_+_163535925 | 0.11 |
ENSMUST00000109400.3
|
Pkig
|
protein kinase inhibitor, gamma |
| chr11_-_120521382 | 0.11 |
ENSMUST00000106181.8
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr11_-_102120953 | 0.11 |
ENSMUST00000107150.8
ENSMUST00000156337.2 ENSMUST00000107151.9 ENSMUST00000107152.9 |
Hdac5
|
histone deacetylase 5 |
| chr15_+_102379621 | 0.11 |
ENSMUST00000229918.2
|
Pcbp2
|
poly(rC) binding protein 2 |
| chr17_-_79076487 | 0.11 |
ENSMUST00000233850.2
|
Heatr5b
|
HEAT repeat containing 5B |
| chr17_+_46989695 | 0.11 |
ENSMUST00000233032.2
|
Mea1
|
male enhanced antigen 1 |
| chr5_+_149202157 | 0.10 |
ENSMUST00000200806.4
|
Alox5ap
|
arachidonate 5-lipoxygenase activating protein |
| chr3_+_68491487 | 0.10 |
ENSMUST00000182997.3
|
Schip1
|
schwannomin interacting protein 1 |
| chr16_-_56533179 | 0.10 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr13_+_44994167 | 0.10 |
ENSMUST00000173906.3
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chrX_+_158086253 | 0.10 |
ENSMUST00000112491.2
|
Rps6ka3
|
ribosomal protein S6 kinase polypeptide 3 |
| chr18_+_37898633 | 0.10 |
ENSMUST00000044851.8
|
Pcdhga12
|
protocadherin gamma subfamily A, 12 |
| chr10_+_69048914 | 0.10 |
ENSMUST00000163497.8
ENSMUST00000164212.8 ENSMUST00000067908.14 |
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr3_+_116356592 | 0.09 |
ENSMUST00000029573.8
|
Lrrc39
|
leucine rich repeat containing 39 |
| chr11_-_102109748 | 0.09 |
ENSMUST00000131254.2
|
Hdac5
|
histone deacetylase 5 |
| chr5_-_5564730 | 0.09 |
ENSMUST00000115445.8
ENSMUST00000179804.8 ENSMUST00000125110.2 ENSMUST00000115446.8 |
Cldn12
|
claudin 12 |
| chr9_-_66421868 | 0.09 |
ENSMUST00000056890.10
|
Fbxl22
|
F-box and leucine-rich repeat protein 22 |
| chr10_-_24588030 | 0.09 |
ENSMUST00000105520.8
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr4_-_134099840 | 0.09 |
ENSMUST00000030643.3
|
Extl1
|
exostosin-like glycosyltransferase 1 |
| chr7_+_141996067 | 0.09 |
ENSMUST00000149529.8
|
Tnni2
|
troponin I, skeletal, fast 2 |
| chr1_+_75213082 | 0.09 |
ENSMUST00000055223.14
ENSMUST00000082158.13 ENSMUST00000188346.7 |
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr11_-_102120917 | 0.09 |
ENSMUST00000008999.12
|
Hdac5
|
histone deacetylase 5 |
| chr16_-_35589726 | 0.09 |
ENSMUST00000023554.9
|
Slc49a4
|
solute carrier family 49 member 4 |
| chr5_-_25047577 | 0.09 |
ENSMUST00000030787.9
|
Rheb
|
Ras homolog enriched in brain |
| chr6_+_58808733 | 0.09 |
ENSMUST00000126292.8
ENSMUST00000031823.12 |
Herc3
|
hect domain and RLD 3 |
| chr5_+_25964985 | 0.08 |
ENSMUST00000128727.8
ENSMUST00000088244.6 |
Actr3b
|
ARP3 actin-related protein 3B |
| chr13_+_38388904 | 0.08 |
ENSMUST00000091641.13
ENSMUST00000178564.2 |
Snrnp48
|
small nuclear ribonucleoprotein 48 (U11/U12) |
| chr7_+_130467564 | 0.08 |
ENSMUST00000075181.11
ENSMUST00000151119.9 ENSMUST00000048180.12 |
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr11_-_120520954 | 0.07 |
ENSMUST00000106180.2
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr5_+_33176160 | 0.07 |
ENSMUST00000019109.8
|
Ywhah
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide |
| chr5_-_129907878 | 0.07 |
ENSMUST00000026617.13
|
Phkg1
|
phosphorylase kinase gamma 1 |
| chr3_-_137837117 | 0.07 |
ENSMUST00000029805.13
|
Mttp
|
microsomal triglyceride transfer protein |
| chr7_+_80707328 | 0.07 |
ENSMUST00000107348.2
|
Alpk3
|
alpha-kinase 3 |
| chrX_+_9138995 | 0.07 |
ENSMUST00000015486.7
|
Xk
|
X-linked Kx blood group |
| chr15_-_91075933 | 0.07 |
ENSMUST00000069511.8
|
Abcd2
|
ATP-binding cassette, sub-family D (ALD), member 2 |
| chr8_+_110432210 | 0.07 |
ENSMUST00000212192.2
|
Zfp821
|
zinc finger protein 821 |
| chr1_+_179936757 | 0.07 |
ENSMUST00000143176.8
ENSMUST00000135056.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr19_+_6434416 | 0.07 |
ENSMUST00000035269.15
ENSMUST00000113483.2 |
Pygm
|
muscle glycogen phosphorylase |
| chr17_-_45903188 | 0.06 |
ENSMUST00000164769.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr1_+_75213114 | 0.06 |
ENSMUST00000188290.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr19_+_4053290 | 0.06 |
ENSMUST00000025806.5
|
Doc2g
|
double C2, gamma |
| chr6_-_70313491 | 0.06 |
ENSMUST00000103388.4
|
Igkv6-20
|
immunoglobulin kappa variable 6-20 |
| chr3_-_51248032 | 0.06 |
ENSMUST00000062009.14
ENSMUST00000194641.6 |
Elf2
|
E74-like factor 2 |
| chr6_+_55313409 | 0.06 |
ENSMUST00000004774.4
|
Aqp1
|
aquaporin 1 |
| chr7_+_107264518 | 0.06 |
ENSMUST00000239294.2
|
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr7_-_101749433 | 0.06 |
ENSMUST00000106937.8
|
Art5
|
ADP-ribosyltransferase 5 |
| chr11_+_73051228 | 0.05 |
ENSMUST00000006104.10
ENSMUST00000135202.8 ENSMUST00000136894.3 |
P2rx5
|
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr19_+_4053367 | 0.05 |
ENSMUST00000236482.2
|
Doc2g
|
double C2, gamma |
| chr9_-_79884920 | 0.05 |
ENSMUST00000239133.2
|
Filip1
|
filamin A interacting protein 1 |
| chr11_+_70548513 | 0.05 |
ENSMUST00000134087.8
|
Eno3
|
enolase 3, beta muscle |
| chr4_+_11486002 | 0.05 |
ENSMUST00000108307.3
|
Virma
|
vir like m6A methyltransferase associated |
| chr1_+_75213171 | 0.05 |
ENSMUST00000187058.7
|
Dnajb2
|
DnaJ heat shock protein family (Hsp40) member B2 |
| chr11_+_20597149 | 0.05 |
ENSMUST00000109585.2
|
Sertad2
|
SERTA domain containing 2 |
| chr14_-_32407203 | 0.05 |
ENSMUST00000096038.4
|
3425401B19Rik
|
RIKEN cDNA 3425401B19 gene |
| chr5_+_107645626 | 0.05 |
ENSMUST00000152474.8
ENSMUST00000060553.8 |
Btbd8
|
BTB (POZ) domain containing 8 |
| chr1_+_131755715 | 0.04 |
ENSMUST00000086559.7
|
Slc41a1
|
solute carrier family 41, member 1 |
| chrX_+_105626790 | 0.04 |
ENSMUST00000101296.9
ENSMUST00000101297.4 |
Gm5127
|
predicted gene 5127 |
| chr11_+_102159558 | 0.04 |
ENSMUST00000036467.5
|
Asb16
|
ankyrin repeat and SOCS box-containing 16 |
| chr17_-_45903494 | 0.04 |
ENSMUST00000163492.8
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr13_-_74512111 | 0.04 |
ENSMUST00000022064.5
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr19_-_6065181 | 0.04 |
ENSMUST00000236537.2
ENSMUST00000025891.11 |
Capn1
|
calpain 1 |
| chr7_-_4517608 | 0.04 |
ENSMUST00000166959.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr10_+_32959472 | 0.04 |
ENSMUST00000095762.5
ENSMUST00000218281.2 ENSMUST00000217779.2 ENSMUST00000219665.2 ENSMUST00000219931.2 |
Trdn
|
triadin |
| chr11_+_75546989 | 0.04 |
ENSMUST00000136935.2
|
Myo1c
|
myosin IC |
| chr9_-_110571645 | 0.04 |
ENSMUST00000006005.12
|
Pth1r
|
parathyroid hormone 1 receptor |
| chr11_+_75546671 | 0.04 |
ENSMUST00000108431.3
|
Myo1c
|
myosin IC |
| chr2_+_90948481 | 0.04 |
ENSMUST00000137942.8
ENSMUST00000111430.10 ENSMUST00000169776.2 |
Mybpc3
|
myosin binding protein C, cardiac |
| chr10_+_69048464 | 0.04 |
ENSMUST00000020101.12
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr6_-_116050081 | 0.04 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr9_-_79885063 | 0.04 |
ENSMUST00000093811.11
|
Filip1
|
filamin A interacting protein 1 |
| chr4_+_123095297 | 0.04 |
ENSMUST00000068262.6
|
Nt5c1a
|
5'-nucleotidase, cytosolic IA |
| chr1_-_75195127 | 0.04 |
ENSMUST00000079464.13
|
Tuba4a
|
tubulin, alpha 4A |
| chr3_-_129548954 | 0.03 |
ENSMUST00000029653.7
|
Egf
|
epidermal growth factor |
| chr12_+_59177780 | 0.03 |
ENSMUST00000177460.8
|
Mia2
|
MIA SH3 domain ER export factor 2 |
| chr19_-_6065872 | 0.03 |
ENSMUST00000164843.10
|
Capn1
|
calpain 1 |
| chr7_+_130467486 | 0.03 |
ENSMUST00000120441.8
|
Plekha1
|
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
| chr7_+_87927293 | 0.03 |
ENSMUST00000032779.12
ENSMUST00000131108.9 ENSMUST00000128791.2 |
Ctsc
|
cathepsin C |
| chrX_+_55825033 | 0.03 |
ENSMUST00000114772.9
ENSMUST00000114768.10 ENSMUST00000155882.8 |
Fhl1
|
four and a half LIM domains 1 |
| chr8_+_15107646 | 0.03 |
ENSMUST00000033842.4
|
Myom2
|
myomesin 2 |
| chr5_+_135916764 | 0.03 |
ENSMUST00000005077.7
|
Hspb1
|
heat shock protein 1 |
| chr5_+_135916847 | 0.03 |
ENSMUST00000111155.2
|
Hspb1
|
heat shock protein 1 |
| chr9_-_122016671 | 0.03 |
ENSMUST00000216738.2
|
Ano10
|
anoctamin 10 |
| chr13_+_38010203 | 0.03 |
ENSMUST00000128570.9
|
Rreb1
|
ras responsive element binding protein 1 |
| chr9_+_62249177 | 0.03 |
ENSMUST00000128636.8
|
Anp32a
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
| chr1_+_171265103 | 0.03 |
ENSMUST00000043839.5
|
F11r
|
F11 receptor |
| chr10_-_24587884 | 0.03 |
ENSMUST00000135846.2
|
Enpp1
|
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr5_+_75312939 | 0.03 |
ENSMUST00000202681.4
ENSMUST00000000476.15 |
Pdgfra
|
platelet derived growth factor receptor, alpha polypeptide |
| chr2_+_69500444 | 0.03 |
ENSMUST00000100050.4
|
Klhl41
|
kelch-like 41 |
| chr4_+_144619397 | 0.03 |
ENSMUST00000105744.8
ENSMUST00000171001.8 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr15_-_100952206 | 0.03 |
ENSMUST00000178140.2
|
Fignl2
|
fidgetin-like 2 |
| chr15_-_89309998 | 0.03 |
ENSMUST00000168376.2
|
Cpt1b
|
carnitine palmitoyltransferase 1b, muscle |
| chr6_-_70116066 | 0.03 |
ENSMUST00000103379.3
ENSMUST00000197371.2 |
Igkv6-29
|
immunoglobulin kappa chain variable 6-29 |
| chr10_+_80465481 | 0.02 |
ENSMUST00000085435.7
|
Csnk1g2
|
casein kinase 1, gamma 2 |
| chr5_-_18093739 | 0.02 |
ENSMUST00000169095.6
ENSMUST00000197574.2 |
Cd36
|
CD36 molecule |
| chr11_+_60066681 | 0.02 |
ENSMUST00000102688.2
|
Rai1
|
retinoic acid induced 1 |
| chr14_+_55813074 | 0.02 |
ENSMUST00000022826.7
|
Fitm1
|
fat storage-inducing transmembrane protein 1 |
| chr15_-_98705791 | 0.02 |
ENSMUST00000075444.8
|
Ddn
|
dendrin |
| chr3_-_20209220 | 0.02 |
ENSMUST00000184552.2
|
Gyg
|
glycogenin |
| chr3_-_94380898 | 0.02 |
ENSMUST00000029785.4
|
Riiad1
|
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 |
| chr3_+_69914946 | 0.02 |
ENSMUST00000053013.6
|
Otol1
|
otolin 1 |
| chr5_-_5564873 | 0.02 |
ENSMUST00000060947.14
|
Cldn12
|
claudin 12 |
| chr13_-_113800172 | 0.02 |
ENSMUST00000054650.5
|
Hspb3
|
heat shock protein 3 |
| chr6_-_142647985 | 0.02 |
ENSMUST00000205202.3
ENSMUST00000073173.12 ENSMUST00000111771.8 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr15_+_80507671 | 0.02 |
ENSMUST00000043149.9
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr17_-_68257900 | 0.02 |
ENSMUST00000164647.8
|
Arhgap28
|
Rho GTPase activating protein 28 |
| chr16_+_29884153 | 0.02 |
ENSMUST00000023171.8
|
Hes1
|
hes family bHLH transcription factor 1 |
| chr11_-_78950641 | 0.02 |
ENSMUST00000226282.2
|
Ksr1
|
kinase suppressor of ras 1 |
| chr14_-_52151537 | 0.02 |
ENSMUST00000227402.2
ENSMUST00000227237.2 |
Ndrg2
|
N-myc downstream regulated gene 2 |
| chr18_-_3280999 | 0.02 |
ENSMUST00000049942.13
|
Crem
|
cAMP responsive element modulator |
| chr4_-_120672900 | 0.02 |
ENSMUST00000120779.8
|
Nfyc
|
nuclear transcription factor-Y gamma |
| chr18_-_3281089 | 0.02 |
ENSMUST00000139537.2
ENSMUST00000124747.8 |
Crem
|
cAMP responsive element modulator |
| chr10_-_88440996 | 0.01 |
ENSMUST00000121629.8
|
Mybpc1
|
myosin binding protein C, slow-type |
| chr16_-_43836681 | 0.01 |
ENSMUST00000036174.10
|
Gramd1c
|
GRAM domain containing 1C |
| chr10_+_69048506 | 0.01 |
ENSMUST00000167384.8
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr9_-_7872970 | 0.01 |
ENSMUST00000115672.2
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr1_+_130947580 | 0.01 |
ENSMUST00000016673.6
|
Il10
|
interleukin 10 |
| chr6_+_108190050 | 0.01 |
ENSMUST00000032192.9
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
| chr2_-_86640362 | 0.01 |
ENSMUST00000216117.2
|
Olfr141
|
olfactory receptor 141 |
| chr11_+_98274637 | 0.01 |
ENSMUST00000008021.3
|
Tcap
|
titin-cap |
| chr14_+_26616514 | 0.01 |
ENSMUST00000238987.2
ENSMUST00000239004.2 ENSMUST00000165929.4 ENSMUST00000090337.12 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr1_+_128069716 | 0.01 |
ENSMUST00000187557.2
|
R3hdm1
|
R3H domain containing 1 |
| chr3_+_93301003 | 0.01 |
ENSMUST00000045912.3
|
Rptn
|
repetin |
| chrX_+_55824797 | 0.01 |
ENSMUST00000114773.10
|
Fhl1
|
four and a half LIM domains 1 |
| chr7_-_4517559 | 0.01 |
ENSMUST00000163538.8
|
Tnnt1
|
troponin T1, skeletal, slow |
| chr16_+_11131676 | 0.01 |
ENSMUST00000023140.6
|
Tnfrsf17
|
tumor necrosis factor receptor superfamily, member 17 |
| chr2_-_77533596 | 0.01 |
ENSMUST00000171063.8
|
Zfp385b
|
zinc finger protein 385B |
| chr10_+_90412827 | 0.01 |
ENSMUST00000182550.8
ENSMUST00000099364.12 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr8_+_55024446 | 0.01 |
ENSMUST00000239166.2
ENSMUST00000239106.2 ENSMUST00000239152.2 |
Asb5
|
ankyrin repeat and SOCs box-containing 5 |
| chr3_-_154760978 | 0.01 |
ENSMUST00000064076.6
|
Tnni3k
|
TNNI3 interacting kinase |
| chr4_+_11485945 | 0.01 |
ENSMUST00000055372.14
ENSMUST00000059914.13 |
Virma
|
vir like m6A methyltransferase associated |
| chrX_+_163221035 | 0.01 |
ENSMUST00000033755.6
|
Asb11
|
ankyrin repeat and SOCS box-containing 11 |
| chr17_-_14914484 | 0.01 |
ENSMUST00000170872.3
|
Thbs2
|
thrombospondin 2 |
| chr12_-_103304573 | 0.01 |
ENSMUST00000149431.2
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr3_-_49711706 | 0.01 |
ENSMUST00000191794.2
|
Pcdh18
|
protocadherin 18 |
| chr1_-_172047282 | 0.01 |
ENSMUST00000170700.2
ENSMUST00000003554.11 |
Casq1
|
calsequestrin 1 |
| chrX_+_163156359 | 0.01 |
ENSMUST00000033751.8
|
Vegfd
|
vascular endothelial growth factor D |
| chr10_-_88440869 | 0.01 |
ENSMUST00000119185.8
ENSMUST00000238199.2 |
Mybpc1
|
myosin binding protein C, slow-type |
| chr6_+_108190163 | 0.01 |
ENSMUST00000203615.3
|
Itpr1
|
inositol 1,4,5-trisphosphate receptor 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Mef2c
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.2 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.1 | 0.2 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.1 | 0.3 | GO:0035026 | leading edge cell differentiation(GO:0035026) cellular response to potassium ion starvation(GO:0051365) |
| 0.1 | 0.2 | GO:1903028 | regulation of opsonization(GO:1903027) positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.5 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.0 | 0.8 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.4 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.2 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.4 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0060382 | regulation of DNA strand elongation(GO:0060382) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.7 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.2 | GO:0099525 | presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.1 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.0 | 0.2 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.2 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:0019660 | glucose catabolic process to lactate(GO:0019659) glycolytic fermentation(GO:0019660) glucose catabolic process to lactate via pyruvate(GO:0019661) |
| 0.0 | 0.3 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.1 | GO:0045719 | negative regulation of glycogen biosynthetic process(GO:0045719) |
| 0.0 | 0.3 | GO:0051197 | positive regulation of glycolytic process(GO:0045821) positive regulation of cofactor metabolic process(GO:0051194) positive regulation of coenzyme metabolic process(GO:0051197) |
| 0.0 | 0.1 | GO:1901409 | positive regulation of phosphorylation of RNA polymerase II C-terminal domain(GO:1901409) |
| 0.0 | 0.2 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.1 | GO:0038089 | positive regulation of cell migration by vascular endothelial growth factor signaling pathway(GO:0038089) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.1 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.9 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.4 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.3 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0098831 | calyx of Held(GO:0044305) presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.3 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.3 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0070187 | telosome(GO:0070187) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.3 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.1 | 0.2 | GO:0004019 | adenylosuccinate synthase activity(GO:0004019) |
| 0.0 | 0.7 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.3 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.5 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.2 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.2 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.4 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.5 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.8 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.0 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.7 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |