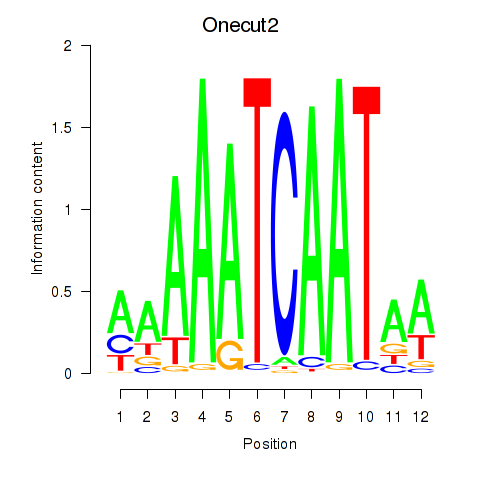
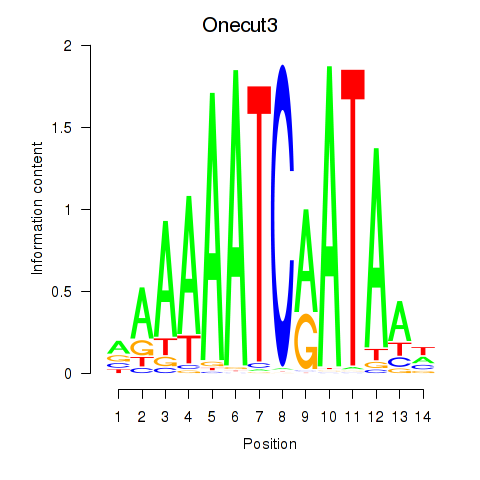
|
chr11_-_73382303
Show fit
|
2.27 |
ENSMUST00000119863.2
ENSMUST00000215358.2
ENSMUST00000214623.2
|
Olfr381
|
olfactory receptor 381
|
|
chr14_+_51366512
Show fit
|
1.97 |
ENSMUST00000095923.4
|
Rnase6
|
ribonuclease, RNase A family, 6
|
|
chr15_+_10224052
Show fit
|
1.85 |
ENSMUST00000128450.8
ENSMUST00000148257.8
ENSMUST00000128921.8
|
Prlr
|
prolactin receptor
|
|
chr13_-_22913799
Show fit
|
1.68 |
ENSMUST00000237024.2
ENSMUST00000236800.2
|
Vmn1r207
|
vomeronasal 1 receptor 207
|
|
chr1_+_172525613
Show fit
|
1.51 |
ENSMUST00000038495.5
|
Crp
|
C-reactive protein, pentraxin-related
|
|
chr7_+_6459167
Show fit
|
1.48 |
ENSMUST00000214301.3
|
Olfr1336
|
olfactory receptor 1336
|
|
chr19_+_13608985
Show fit
|
1.44 |
ENSMUST00000217182.3
|
Olfr1489
|
olfactory receptor 1489
|
|
chr3_-_15902583
Show fit
|
1.44 |
ENSMUST00000108354.8
ENSMUST00000108349.2
ENSMUST00000108352.9
ENSMUST00000108350.8
ENSMUST00000050623.11
|
Sirpb1c
|
signal-regulatory protein beta 1C
|
|
chr10_-_128505096
Show fit
|
1.34 |
ENSMUST00000238610.2
ENSMUST00000238712.2
|
Ikzf4
|
IKAROS family zinc finger 4
|
|
chr2_+_87610895
Show fit
|
1.32 |
ENSMUST00000215394.2
|
Olfr152
|
olfactory receptor 152
|
|
chr13_-_22993852
Show fit
|
1.22 |
ENSMUST00000227038.2
ENSMUST00000227265.2
|
Vmn1r209
|
vomeronasal 1 receptor 209
|
|
chr16_-_58620631
Show fit
|
1.17 |
ENSMUST00000206205.3
|
Olfr173
|
olfactory receptor 173
|
|
chr14_+_51366306
Show fit
|
1.10 |
ENSMUST00000226210.2
|
Rnase6
|
ribonuclease, RNase A family, 6
|
|
chr2_+_111205867
Show fit
|
1.08 |
ENSMUST00000062407.6
|
Olfr1284
|
olfactory receptor 1284
|
|
chr11_+_74721733
Show fit
|
1.04 |
ENSMUST00000000291.9
|
Mnt
|
max binding protein
|
|
chr2_-_111843053
Show fit
|
1.00 |
ENSMUST00000213559.3
|
Olfr1310
|
olfactory receptor 1310
|
|
chr10_+_129153986
Show fit
|
0.99 |
ENSMUST00000215503.2
|
Olfr780
|
olfactory receptor 780
|
|
chr2_-_88534814
Show fit
|
0.97 |
ENSMUST00000216928.2
ENSMUST00000216977.2
|
Olfr1196
|
olfactory receptor 1196
|
|
chr10_-_41894360
Show fit
|
0.94 |
ENSMUST00000162405.8
ENSMUST00000095729.11
ENSMUST00000161081.2
ENSMUST00000160262.9
|
Armc2
|
armadillo repeat containing 2
|
|
chrX_-_48823936
Show fit
|
0.91 |
ENSMUST00000215373.3
|
Olfr1321
|
olfactory receptor 1321
|
|
chr19_+_38252984
Show fit
|
0.90 |
ENSMUST00000198518.5
ENSMUST00000199812.5
|
Lgi1
|
leucine-rich repeat LGI family, member 1
|
|
chr13_+_49658249
Show fit
|
0.87 |
ENSMUST00000051504.8
|
Ecm2
|
extracellular matrix protein 2, female organ and adipocyte specific
|
|
chr7_+_4795873
Show fit
|
0.86 |
ENSMUST00000032597.12
ENSMUST00000078432.5
|
Rpl28
|
ribosomal protein L28
|
|
chr3_-_15640045
Show fit
|
0.83 |
ENSMUST00000192382.6
ENSMUST00000195778.3
ENSMUST00000091319.7
|
Sirpb1b
|
signal-regulatory protein beta 1B
|
|
chr7_+_104681905
Show fit
|
0.79 |
ENSMUST00000219602.2
|
Olfr676
|
olfactory receptor 676
|
|
chr2_+_37080286
Show fit
|
0.78 |
ENSMUST00000218602.2
|
Olfr365
|
olfactory receptor 365
|
|
chr8_+_123103348
Show fit
|
0.77 |
ENSMUST00000017622.12
ENSMUST00000093073.12
ENSMUST00000176699.2
|
Zc3h18
|
zinc finger CCCH-type containing 18
|
|
chr7_-_106730290
Show fit
|
0.76 |
ENSMUST00000214919.2
|
Olfr715
|
olfactory receptor 715
|
|
chr9_-_21710029
Show fit
|
0.75 |
ENSMUST00000034717.7
|
Kank2
|
KN motif and ankyrin repeat domains 2
|
|
chr2_-_89491811
Show fit
|
0.73 |
ENSMUST00000215730.3
|
Olfr1250
|
olfactory receptor 1250
|
|
chr11_+_49313894
Show fit
|
0.67 |
ENSMUST00000216641.2
ENSMUST00000217595.2
|
Olfr1389
|
olfactory receptor 1389
|
|
chr1_-_173569301
Show fit
|
0.65 |
ENSMUST00000042610.15
ENSMUST00000127730.2
|
Ifi207
|
interferon activated gene 207
|
|
chr1_-_5140504
Show fit
|
0.64 |
ENSMUST00000147158.2
ENSMUST00000118000.8
|
Rgs20
|
regulator of G-protein signaling 20
|
|
chr2_-_89491560
Show fit
|
0.63 |
ENSMUST00000111527.4
|
Olfr1250
|
olfactory receptor 1250
|
|
chr6_+_56947258
Show fit
|
0.63 |
ENSMUST00000226130.2
ENSMUST00000228276.2
|
Vmn1r5
|
vomeronasal 1 receptor 5
|
|
chr11_+_46126274
Show fit
|
0.62 |
ENSMUST00000060185.3
|
Fndc9
|
fibronectin type III domain containing 9
|
|
chr3_-_58792633
Show fit
|
0.61 |
ENSMUST00000055636.13
ENSMUST00000072551.7
ENSMUST00000051408.8
|
Clrn1
|
clarin 1
|
|
chr6_+_124001527
Show fit
|
0.59 |
ENSMUST00000032238.5
|
Vmn2r26
|
vomeronasal 2, receptor 26
|
|
chr17_+_12338161
Show fit
|
0.58 |
ENSMUST00000024594.9
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta)
|
|
chr2_-_104680104
Show fit
|
0.58 |
ENSMUST00000028593.11
|
Prrg4
|
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane)
|
|
chr2_-_111253457
Show fit
|
0.58 |
ENSMUST00000213210.2
ENSMUST00000184954.4
|
Olfr1286
|
olfactory receptor 1286
|
|
chr10_-_76304948
Show fit
|
0.58 |
ENSMUST00000049185.6
|
Ybey
|
ybeY metallopeptidase
|
|
chr19_-_13126896
Show fit
|
0.58 |
ENSMUST00000213493.2
|
Olfr1459
|
olfactory receptor 1459
|
|
chr15_-_79212323
Show fit
|
0.57 |
ENSMUST00000166977.9
|
Pla2g6
|
phospholipase A2, group VI
|
|
chr11_-_79414542
Show fit
|
0.56 |
ENSMUST00000179322.2
|
Evi2b
|
ecotropic viral integration site 2b
|
|
chr4_+_147390131
Show fit
|
0.55 |
ENSMUST00000148762.4
|
Zfp988
|
zinc finger protein 988
|
|
chr15_+_54975713
Show fit
|
0.54 |
ENSMUST00000096433.10
|
Deptor
|
DEP domain containing MTOR-interacting protein
|
|
chr13_-_23015518
Show fit
|
0.54 |
ENSMUST00000226294.2
ENSMUST00000226180.2
|
Vmn1r210
|
vomeronasal 1 receptor 210
|
|
chr8_+_67150055
Show fit
|
0.53 |
ENSMUST00000039303.7
|
Npy1r
|
neuropeptide Y receptor Y1
|
|
chr16_-_10131804
Show fit
|
0.53 |
ENSMUST00000078357.5
|
Emp2
|
epithelial membrane protein 2
|
|
chr4_+_145237329
Show fit
|
0.52 |
ENSMUST00000105742.8
ENSMUST00000136309.8
|
Zfp990
|
zinc finger protein 990
|
|
chr4_+_109200225
Show fit
|
0.51 |
ENSMUST00000030281.12
|
Eps15
|
epidermal growth factor receptor pathway substrate 15
|
|
chr9_+_55234197
Show fit
|
0.50 |
ENSMUST00000085754.10
ENSMUST00000034862.5
|
Tmem266
|
transmembrane protein 266
|
|
chr19_-_4109446
Show fit
|
0.50 |
ENSMUST00000189808.7
|
Gstp3
|
glutathione S-transferase pi 3
|
|
chr6_-_122833109
Show fit
|
0.50 |
ENSMUST00000042081.9
|
C3ar1
|
complement component 3a receptor 1
|
|
chr10_-_129715720
Show fit
|
0.49 |
ENSMUST00000216966.2
ENSMUST00000213742.2
|
Olfr814
|
olfactory receptor 814
|
|
chr1_+_131566044
Show fit
|
0.48 |
ENSMUST00000073350.13
|
Ctse
|
cathepsin E
|
|
chr1_-_158785937
Show fit
|
0.48 |
ENSMUST00000159861.9
|
Pappa2
|
pappalysin 2
|
|
chr14_+_20732804
Show fit
|
0.47 |
ENSMUST00000228545.2
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae)
|
|
chr1_-_64160557
Show fit
|
0.46 |
ENSMUST00000055001.10
ENSMUST00000114086.8
|
Klf7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr6_+_89946295
Show fit
|
0.45 |
ENSMUST00000205088.3
ENSMUST00000226715.2
ENSMUST00000228401.2
|
Vmn1r46
|
vomeronasal 1 receptor 46
|
|
chr4_+_147637714
Show fit
|
0.45 |
ENSMUST00000139784.8
ENSMUST00000143885.8
ENSMUST00000081742.7
|
Zfp985
|
zinc finger protein 985
|
|
chr8_+_67149815
Show fit
|
0.44 |
ENSMUST00000212588.2
|
Npy1r
|
neuropeptide Y receptor Y1
|
|
chr7_-_127048280
Show fit
|
0.44 |
ENSMUST00000053392.11
|
Zfp689
|
zinc finger protein 689
|
|
chr2_+_111156865
Show fit
|
0.44 |
ENSMUST00000208176.3
|
Olfr1281
|
olfactory receptor 1281
|
|
chr9_-_108991088
Show fit
|
0.44 |
ENSMUST00000199540.2
ENSMUST00000198076.5
ENSMUST00000054925.13
|
Fbxw21
|
F-box and WD-40 domain protein 21
|
|
chr19_-_12093187
Show fit
|
0.43 |
ENSMUST00000208391.3
ENSMUST00000214103.2
|
Olfr1428
|
olfactory receptor 1428
|
|
chr7_+_23330147
Show fit
|
0.42 |
ENSMUST00000227774.2
ENSMUST00000226771.2
ENSMUST00000228681.2
ENSMUST00000228559.2
ENSMUST00000228674.2
ENSMUST00000227866.2
ENSMUST00000227386.2
ENSMUST00000228484.2
ENSMUST00000226321.2
ENSMUST00000226128.2
ENSMUST00000226733.2
ENSMUST00000228228.2
|
Vmn1r171
|
vomeronasal 1 receptor 171
|
|
chr8_-_5155347
Show fit
|
0.42 |
ENSMUST00000023835.3
|
Slc10a2
|
solute carrier family 10, member 2
|
|
chr13_-_22961570
Show fit
|
0.42 |
ENSMUST00000227136.2
|
Vmn1r208
|
vomeronasal 1 receptor 208
|
|
chr1_+_92545510
Show fit
|
0.42 |
ENSMUST00000213247.2
|
Olfr12
|
olfactory receptor 12
|
|
chr9_-_19799300
Show fit
|
0.40 |
ENSMUST00000079660.5
|
Olfr862
|
olfactory receptor 862
|
|
chr13_-_22225527
Show fit
|
0.39 |
ENSMUST00000102977.4
|
H4c9
|
H4 clustered histone 9
|
|
chr2_-_94236991
Show fit
|
0.38 |
ENSMUST00000111237.9
ENSMUST00000094801.5
ENSMUST00000111238.8
|
Ttc17
|
tetratricopeptide repeat domain 17
|
|
chr11_+_29323618
Show fit
|
0.38 |
ENSMUST00000040182.13
ENSMUST00000109477.2
|
Ccdc88a
|
coiled coil domain containing 88A
|
|
chr17_+_18269686
Show fit
|
0.37 |
ENSMUST00000176802.3
|
Vmn2r124
|
vomeronasal 2, receptor 124
|
|
chr3_+_20011201
Show fit
|
0.37 |
ENSMUST00000091309.12
ENSMUST00000108329.8
ENSMUST00000003714.13
|
Cp
|
ceruloplasmin
|
|
chr7_-_30234422
Show fit
|
0.37 |
ENSMUST00000208522.2
ENSMUST00000207860.2
ENSMUST00000208538.2
|
Arhgap33
|
Rho GTPase activating protein 33
|
|
chr11_+_29324348
Show fit
|
0.36 |
ENSMUST00000239407.2
|
Ccdc88a
|
coiled coil domain containing 88A
|
|
chr7_+_18853778
Show fit
|
0.34 |
ENSMUST00000053109.5
|
Fbxo46
|
F-box protein 46
|
|
chr1_+_131566223
Show fit
|
0.34 |
ENSMUST00000112411.2
|
Ctse
|
cathepsin E
|
|
chr12_+_59178072
Show fit
|
0.34 |
ENSMUST00000176464.8
ENSMUST00000170992.9
ENSMUST00000176322.8
|
Mia2
|
MIA SH3 domain ER export factor 2
|
|
chr5_-_138153956
Show fit
|
0.34 |
ENSMUST00000132318.2
ENSMUST00000049393.15
|
Zfp113
|
zinc finger protein 113
|
|
chr8_-_85500998
Show fit
|
0.34 |
ENSMUST00000109762.8
|
Nfix
|
nuclear factor I/X
|
|
chr11_+_97206542
Show fit
|
0.33 |
ENSMUST00000019026.10
ENSMUST00000132168.2
|
Mrpl45
|
mitochondrial ribosomal protein L45
|
|
chr3_+_101917455
Show fit
|
0.33 |
ENSMUST00000066187.6
ENSMUST00000198675.2
|
Nhlh2
|
nescient helix loop helix 2
|
|
chr8_-_32408380
Show fit
|
0.32 |
ENSMUST00000208497.3
ENSMUST00000207584.3
|
Nrg1
|
neuregulin 1
|
|
chr13_+_60749735
Show fit
|
0.32 |
ENSMUST00000226059.2
ENSMUST00000077453.13
|
Dapk1
|
death associated protein kinase 1
|
|
chr5_-_31684036
Show fit
|
0.32 |
ENSMUST00000202421.2
ENSMUST00000201769.4
ENSMUST00000065388.11
|
Supt7l
|
SPT7-like, STAGA complex gamma subunit
|
|
chr7_-_140087224
Show fit
|
0.32 |
ENSMUST00000209873.2
ENSMUST00000064392.8
ENSMUST00000215768.2
ENSMUST00000215340.2
|
Olfr536
|
olfactory receptor 536
|
|
chr12_+_59178258
Show fit
|
0.32 |
ENSMUST00000177162.8
|
Mia2
|
MIA SH3 domain ER export factor 2
|
|
chr2_-_51039112
Show fit
|
0.32 |
ENSMUST00000154545.2
ENSMUST00000017288.9
|
Rnd3
|
Rho family GTPase 3
|
|
chr7_+_102965522
Show fit
|
0.31 |
ENSMUST00000216456.2
|
Olfr597
|
olfactory receptor 597
|
|
chr17_-_24199523
Show fit
|
0.29 |
ENSMUST00000234765.2
|
Prss30
|
protease, serine 30
|
|
chr15_+_54975814
Show fit
|
0.29 |
ENSMUST00000100660.11
|
Deptor
|
DEP domain containing MTOR-interacting protein
|
|
chr7_-_30234481
Show fit
|
0.29 |
ENSMUST00000207858.2
|
Arhgap33
|
Rho GTPase activating protein 33
|
|
chr13_+_49697919
Show fit
|
0.28 |
ENSMUST00000177948.2
ENSMUST00000021820.14
|
Aspn
|
asporin
|
|
chr15_-_50746202
Show fit
|
0.28 |
ENSMUST00000184885.8
|
Trps1
|
transcriptional repressor GATA binding 1
|
|
chr8_-_85500010
Show fit
|
0.28 |
ENSMUST00000109764.8
|
Nfix
|
nuclear factor I/X
|
|
chr3_+_94745009
Show fit
|
0.28 |
ENSMUST00000107266.8
ENSMUST00000042402.12
ENSMUST00000107269.2
|
Pogz
|
pogo transposable element with ZNF domain
|
|
chr13_+_60749995
Show fit
|
0.28 |
ENSMUST00000044083.9
|
Dapk1
|
death associated protein kinase 1
|
|
chr2_-_111456842
Show fit
|
0.27 |
ENSMUST00000213398.2
|
Olfr1297
|
olfactory receptor 1297
|
|
chr6_+_127430630
Show fit
|
0.26 |
ENSMUST00000112193.8
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11
|
|
chr12_+_4819022
Show fit
|
0.25 |
ENSMUST00000219503.2
|
Pfn4
|
profilin family, member 4
|
|
chr4_+_146033882
Show fit
|
0.24 |
ENSMUST00000105730.2
ENSMUST00000091878.6
|
Zfp987
|
zinc finger protein 987
|
|
chr3_+_66892979
Show fit
|
0.24 |
ENSMUST00000162362.8
ENSMUST00000065074.14
ENSMUST00000065047.13
|
Rsrc1
|
arginine/serine-rich coiled-coil 1
|
|
chr18_-_9282754
Show fit
|
0.23 |
ENSMUST00000041007.4
|
Gjd4
|
gap junction protein, delta 4
|
|
chr3_-_15491482
Show fit
|
0.23 |
ENSMUST00000099201.9
ENSMUST00000194144.3
ENSMUST00000192700.3
|
Sirpb1a
|
signal-regulatory protein beta 1A
|
|
chr13_+_112604037
Show fit
|
0.23 |
ENSMUST00000183868.8
|
Il6st
|
interleukin 6 signal transducer
|
|
chr9_-_50650663
Show fit
|
0.22 |
ENSMUST00000117093.2
ENSMUST00000121634.8
|
Dixdc1
|
DIX domain containing 1
|
|
chr6_-_127746390
Show fit
|
0.22 |
ENSMUST00000032500.9
|
Prmt8
|
protein arginine N-methyltransferase 8
|
|
chr7_-_119461027
Show fit
|
0.22 |
ENSMUST00000137888.2
ENSMUST00000142120.2
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae)
|
|
chr11_-_87783073
Show fit
|
0.22 |
ENSMUST00000213672.2
ENSMUST00000213928.2
|
Olfr462
|
olfactory receptor 462
|
|
chr13_-_14787602
Show fit
|
0.21 |
ENSMUST00000220621.2
|
Mrpl32
|
mitochondrial ribosomal protein L32
|
|
chr11_-_60618022
Show fit
|
0.21 |
ENSMUST00000002889.5
|
Flii
|
flightless I actin binding protein
|
|
chr9_+_109865810
Show fit
|
0.20 |
ENSMUST00000163190.8
|
Map4
|
microtubule-associated protein 4
|
|
chr9_+_38259893
Show fit
|
0.20 |
ENSMUST00000216304.2
|
Olfr898
|
olfactory receptor 898
|
|
chr5_+_110751362
Show fit
|
0.20 |
ENSMUST00000165856.3
|
Galnt9
|
polypeptide N-acetylgalactosaminyltransferase 9
|
|
chr7_-_85610837
Show fit
|
0.20 |
ENSMUST00000232886.2
ENSMUST00000232758.2
ENSMUST00000166355.3
|
Vmn2r74
|
vomeronasal 2, receptor 74
|
|
chr17_+_34783242
Show fit
|
0.19 |
ENSMUST00000015612.14
|
Notch4
|
notch 4
|
|
chr17_-_43187280
Show fit
|
0.19 |
ENSMUST00000024709.9
ENSMUST00000233476.2
|
Cd2ap
|
CD2-associated protein
|
|
chr3_+_138058139
Show fit
|
0.19 |
ENSMUST00000090166.5
|
Adh6b
|
alcohol dehydrogenase 6B (class V)
|
|
chr3_+_94744844
Show fit
|
0.18 |
ENSMUST00000107270.9
|
Pogz
|
pogo transposable element with ZNF domain
|
|
chr5_-_147831610
Show fit
|
0.18 |
ENSMUST00000118527.8
ENSMUST00000031655.4
ENSMUST00000138244.2
|
Slc46a3
|
solute carrier family 46, member 3
|
|
chr4_-_147787010
Show fit
|
0.18 |
ENSMUST00000117638.2
|
Zfp534
|
zinc finger protein 534
|
|
chr16_-_56891711
Show fit
|
0.18 |
ENSMUST00000067173.8
ENSMUST00000227043.2
|
Tmem45a2
|
transmembrane protein 45A2
|
|
chr8_-_105350898
Show fit
|
0.18 |
ENSMUST00000212882.2
ENSMUST00000163783.4
|
Cdh16
|
cadherin 16
|
|
chr4_-_147726953
Show fit
|
0.17 |
ENSMUST00000133006.2
ENSMUST00000037565.14
ENSMUST00000105720.8
|
Zfp979
|
zinc finger protein 979
|
|
chr1_-_131127825
Show fit
|
0.16 |
ENSMUST00000068564.15
|
Rassf5
|
Ras association (RalGDS/AF-6) domain family member 5
|
|
chr6_-_41751648
Show fit
|
0.16 |
ENSMUST00000214752.2
|
Olfr459
|
olfactory receptor 459
|
|
chr19_+_55730696
Show fit
|
0.15 |
ENSMUST00000153888.9
ENSMUST00000127233.9
ENSMUST00000061496.17
ENSMUST00000111656.8
ENSMUST00000111657.11
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box
|
|
chr4_+_147056433
Show fit
|
0.15 |
ENSMUST00000146688.3
|
Zfp989
|
zinc finger protein 989
|
|
chr6_-_82916655
Show fit
|
0.14 |
ENSMUST00000000641.15
|
Sema4f
|
sema domain, immunoglobulin domain (Ig), TM domain, and short cytoplasmic domain
|
|
chr10_+_128089965
Show fit
|
0.14 |
ENSMUST00000060782.5
ENSMUST00000218722.2
|
Apon
|
apolipoprotein N
|
|
chr3_-_79439181
Show fit
|
0.14 |
ENSMUST00000239298.2
|
Fnip2
|
folliculin interacting protein 2
|
|
chr2_-_111942878
Show fit
|
0.14 |
ENSMUST00000217078.2
|
Olfr1315-ps1
|
olfactory receptor 1315, pseudogene 1
|
|
chr1_-_63153675
Show fit
|
0.14 |
ENSMUST00000097718.9
|
Ino80d
|
INO80 complex subunit D
|
|
chr15_-_79212400
Show fit
|
0.14 |
ENSMUST00000173163.8
ENSMUST00000047816.15
ENSMUST00000172403.9
ENSMUST00000173632.8
|
Pla2g6
|
phospholipase A2, group VI
|
|
chr3_-_10273628
Show fit
|
0.14 |
ENSMUST00000029041.6
|
Fabp4
|
fatty acid binding protein 4, adipocyte
|
|
chr7_-_23907518
Show fit
|
0.13 |
ENSMUST00000086006.12
|
Zfp111
|
zinc finger protein 111
|
|
chr9_-_40015750
Show fit
|
0.13 |
ENSMUST00000213858.2
|
Olfr984
|
olfactory receptor 984
|
|
chr4_+_147445744
Show fit
|
0.13 |
ENSMUST00000133078.8
ENSMUST00000154154.2
|
Zfp978
|
zinc finger protein 978
|
|
chr2_-_88562710
Show fit
|
0.13 |
ENSMUST00000213118.2
|
Olfr1197
|
olfactory receptor 1197
|
|
chr10_+_129184719
Show fit
|
0.13 |
ENSMUST00000213970.2
|
Olfr782
|
olfactory receptor 782
|
|
chr6_+_29694181
Show fit
|
0.13 |
ENSMUST00000046750.14
ENSMUST00000115250.4
|
Tspan33
|
tetraspanin 33
|
|
chr12_-_32000534
Show fit
|
0.13 |
ENSMUST00000172314.9
|
Hbp1
|
high mobility group box transcription factor 1
|
|
chr16_-_43484494
Show fit
|
0.13 |
ENSMUST00000096065.6
|
Tigit
|
T cell immunoreceptor with Ig and ITIM domains
|
|
chr11_-_99447672
Show fit
|
0.12 |
ENSMUST00000092699.3
|
Krtap3-2
|
keratin associated protein 3-2
|
|
chr1_+_138201460
Show fit
|
0.12 |
ENSMUST00000027643.6
|
Atp6v1g3
|
ATPase, H+ transporting, lysosomal V1 subunit G3
|
|
chr12_-_32000209
Show fit
|
0.12 |
ENSMUST00000176084.2
ENSMUST00000176103.8
ENSMUST00000167458.9
|
Hbp1
|
high mobility group box transcription factor 1
|
|
chr9_-_48876290
Show fit
|
0.12 |
ENSMUST00000008734.5
|
Htr3b
|
5-hydroxytryptamine (serotonin) receptor 3B
|
|
chr10_-_63926044
Show fit
|
0.11 |
ENSMUST00000105439.2
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3
|
|
chr7_+_127503251
Show fit
|
0.11 |
ENSMUST00000071056.14
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase
|
|
chr10_-_125225298
Show fit
|
0.11 |
ENSMUST00000210780.2
|
Slc16a7
|
solute carrier family 16 (monocarboxylic acid transporters), member 7
|
|
chr4_+_43383449
Show fit
|
0.11 |
ENSMUST00000135216.2
ENSMUST00000152322.8
|
Rusc2
|
RUN and SH3 domain containing 2
|
|
chr10_-_45346297
Show fit
|
0.10 |
ENSMUST00000079390.7
|
Lin28b
|
lin-28 homolog B (C. elegans)
|
|
chr13_+_35059285
Show fit
|
0.10 |
ENSMUST00000077853.5
|
Prpf4b
|
pre-mRNA processing factor 4B
|
|
chr18_+_36431732
Show fit
|
0.10 |
ENSMUST00000210490.3
|
Igip
|
IgA inducing protein
|
|
chr2_-_86722536
Show fit
|
0.10 |
ENSMUST00000111576.3
ENSMUST00000217403.2
|
Olfr1097
|
olfactory receptor 1097
|
|
chr17_+_45866618
Show fit
|
0.10 |
ENSMUST00000024742.9
ENSMUST00000233929.2
|
Nfkbie
|
nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, epsilon
|
|
chr5_-_65492907
Show fit
|
0.10 |
ENSMUST00000203581.3
|
Rfc1
|
replication factor C (activator 1) 1
|
|
chr7_-_73191484
Show fit
|
0.09 |
ENSMUST00000197642.2
ENSMUST00000026895.14
ENSMUST00000169922.9
|
Chd2
|
chromodomain helicase DNA binding protein 2
|
|
chr8_-_33374282
Show fit
|
0.09 |
ENSMUST00000209107.4
ENSMUST00000209022.3
|
Nrg1
|
neuregulin 1
|
|
chr6_-_85879510
Show fit
|
0.08 |
ENSMUST00000159755.8
|
Nat8f4
|
N-acetyltransferase 8 (GCN5-related) family member 4
|
|
chr4_+_135639117
Show fit
|
0.08 |
ENSMUST00000068830.4
|
Cnr2
|
cannabinoid receptor 2 (macrophage)
|
|
chr2_-_63014622
Show fit
|
0.08 |
ENSMUST00000075052.10
ENSMUST00000112454.8
|
Kcnh7
|
potassium voltage-gated channel, subfamily H (eag-related), member 7
|
|
chr8_-_33374825
Show fit
|
0.08 |
ENSMUST00000238791.2
|
Nrg1
|
neuregulin 1
|
|
chr4_-_115797118
Show fit
|
0.08 |
ENSMUST00000124071.9
ENSMUST00000084338.7
|
Dmbx1
|
diencephalon/mesencephalon homeobox 1
|
|
chr1_+_167425953
Show fit
|
0.08 |
ENSMUST00000015987.10
|
Rxrg
|
retinoid X receptor gamma
|
|
chr9_-_19968858
Show fit
|
0.08 |
ENSMUST00000216538.2
ENSMUST00000212098.3
|
Olfr867
|
olfactory receptor 867
|
|
chr11_+_72580823
Show fit
|
0.08 |
ENSMUST00000155998.2
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1
|
|
chr19_+_55730242
Show fit
|
0.07 |
ENSMUST00000111662.11
ENSMUST00000041717.14
|
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box
|
|
chr7_-_126408280
Show fit
|
0.07 |
ENSMUST00000207534.3
|
Aldoa
|
aldolase A, fructose-bisphosphate
|
|
chr12_+_56742413
Show fit
|
0.07 |
ENSMUST00000001538.10
|
Pax9
|
paired box 9
|
|
chr14_-_75368321
Show fit
|
0.07 |
ENSMUST00000022574.5
|
Lrrc63
|
leucine rich repeat containing 63
|
|
chr3_-_95763160
Show fit
|
0.07 |
ENSMUST00000015892.14
|
Prpf3
|
pre-mRNA processing factor 3
|
|
chr15_+_102927366
Show fit
|
0.07 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4
|
|
chr11_-_24025054
Show fit
|
0.07 |
ENSMUST00000068360.2
|
A830031A19Rik
|
RIKEN cDNA A830031A19 gene
|
|
chr4_+_147576874
Show fit
|
0.06 |
ENSMUST00000105721.9
|
Zfp982
|
zinc finger protein 982
|
|
chr6_+_97968737
Show fit
|
0.06 |
ENSMUST00000043628.13
ENSMUST00000203938.2
|
Mitf
|
melanogenesis associated transcription factor
|
|
chr13_+_119625345
Show fit
|
0.06 |
ENSMUST00000099147.5
|
Tmem267
|
transmembrane protein 267
|
|
chr4_+_52964547
Show fit
|
0.06 |
ENSMUST00000215010.2
ENSMUST00000215127.2
|
Olfr270
|
olfactory receptor 270
|
|
chr6_+_127049865
Show fit
|
0.06 |
ENSMUST00000000186.9
|
Fgf23
|
fibroblast growth factor 23
|
|
chr12_+_108572015
Show fit
|
0.06 |
ENSMUST00000109854.9
|
Evl
|
Ena-vasodilator stimulated phosphoprotein
|
|
chr11_+_78389913
Show fit
|
0.05 |
ENSMUST00000017488.5
|
Vtn
|
vitronectin
|
|
chr16_-_35951553
Show fit
|
0.05 |
ENSMUST00000161638.2
ENSMUST00000096090.3
|
Csta1
|
cystatin A1
|
|
chr11_+_75239259
Show fit
|
0.05 |
ENSMUST00000044530.3
|
Smyd4
|
SET and MYND domain containing 4
|
|
chr2_+_38229270
Show fit
|
0.05 |
ENSMUST00000143783.9
|
Lhx2
|
LIM homeobox protein 2
|
|
chr7_-_3669882
Show fit
|
0.05 |
ENSMUST00000145034.3
|
Tmc4
|
transmembrane channel-like gene family 4
|
|
chr7_-_89590404
Show fit
|
0.05 |
ENSMUST00000153470.9
|
Hikeshi
|
heat shock protein nuclear import factor
|
|
chr7_-_89590334
Show fit
|
0.05 |
ENSMUST00000207309.2
ENSMUST00000130609.3
|
Hikeshi
|
heat shock protein nuclear import factor
|
|
chr17_-_90395771
Show fit
|
0.05 |
ENSMUST00000197268.5
ENSMUST00000173917.8
|
Nrxn1
|
neurexin I
|
|
chr9_-_86577940
Show fit
|
0.05 |
ENSMUST00000034989.15
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic
|
|
chr10_-_129509659
Show fit
|
0.05 |
ENSMUST00000213294.2
ENSMUST00000216067.3
ENSMUST00000203424.2
|
Olfr801
|
olfactory receptor 801
|
|
chr14_+_67982630
Show fit
|
0.04 |
ENSMUST00000223929.2
ENSMUST00000111095.4
|
Gnrh1
|
gonadotropin releasing hormone 1
|
|
chr17_+_43671314
Show fit
|
0.04 |
ENSMUST00000226087.2
|
Adgrf5
|
adhesion G protein-coupled receptor F5
|
|
chr3_-_16060491
Show fit
|
0.04 |
ENSMUST00000108347.3
|
Gm5150
|
predicted gene 5150
|
|
chr14_+_26616514
Show fit
|
0.04 |
ENSMUST00000238987.2
ENSMUST00000239004.2
ENSMUST00000165929.4
ENSMUST00000090337.12
|
Asb14
|
ankyrin repeat and SOCS box-containing 14
|
|
chr15_+_6673167
Show fit
|
0.04 |
ENSMUST00000163073.2
|
Fyb
|
FYN binding protein
|
|
chr6_+_116483477
Show fit
|
0.04 |
ENSMUST00000075756.3
|
Olfr212
|
olfactory receptor 212
|