Project
GFI1 WT vs 36n/n vs KD
Navigation
Downloads
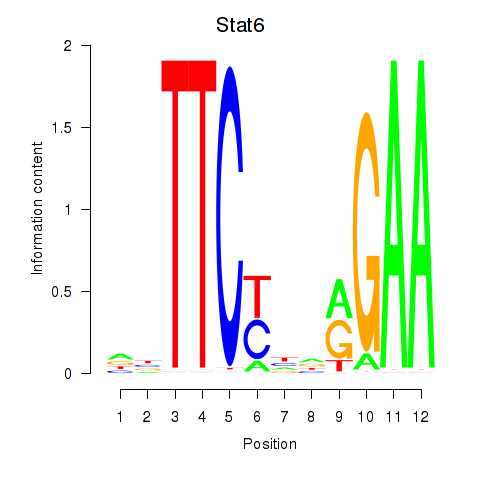
Results for Stat6
Z-value: 0.47
Transcription factors associated with Stat6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Stat6
|
ENSMUSG00000002147.19 | signal transducer and activator of transcription 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Stat6 | mm39_v1_chr10_+_127478844_127478914 | 0.07 | 9.1e-01 | Click! |
Activity profile of Stat6 motif
Sorted Z-values of Stat6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_87684548 | 0.59 |
ENSMUST00000143021.9
|
Mpo
|
myeloperoxidase |
| chr11_+_87684299 | 0.46 |
ENSMUST00000020779.11
|
Mpo
|
myeloperoxidase |
| chr14_-_56499690 | 0.46 |
ENSMUST00000015581.6
|
Gzmb
|
granzyme B |
| chr7_-_126399208 | 0.38 |
ENSMUST00000133514.8
ENSMUST00000151137.8 |
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_126399778 | 0.37 |
ENSMUST00000141355.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_-_126399574 | 0.34 |
ENSMUST00000106348.8
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr17_-_34406193 | 0.33 |
ENSMUST00000173831.3
|
Psmb9
|
proteasome (prosome, macropain) subunit, beta type 9 (large multifunctional peptidase 2) |
| chr5_-_140368482 | 0.32 |
ENSMUST00000196566.5
|
Snx8
|
sorting nexin 8 |
| chr18_+_50186349 | 0.31 |
ENSMUST00000148159.3
|
Tnfaip8
|
tumor necrosis factor, alpha-induced protein 8 |
| chr5_+_117501557 | 0.29 |
ENSMUST00000111959.2
|
Wsb2
|
WD repeat and SOCS box-containing 2 |
| chrX_+_133657312 | 0.28 |
ENSMUST00000081834.10
ENSMUST00000086880.11 ENSMUST00000086884.5 |
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr5_+_21577640 | 0.28 |
ENSMUST00000035799.6
|
Fgl2
|
fibrinogen-like protein 2 |
| chr17_+_29309942 | 0.28 |
ENSMUST00000119901.9
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A (P21) |
| chr17_-_90217868 | 0.26 |
ENSMUST00000086423.6
|
Gm10184
|
predicted pseudogene 10184 |
| chr8_-_48443525 | 0.26 |
ENSMUST00000057561.9
|
Wwc2
|
WW, C2 and coiled-coil domain containing 2 |
| chr10_+_80905869 | 0.24 |
ENSMUST00000005057.7
|
Thop1
|
thimet oligopeptidase 1 |
| chr18_+_66591604 | 0.23 |
ENSMUST00000025399.9
ENSMUST00000237161.2 ENSMUST00000236933.2 |
Pmaip1
|
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr7_+_140462343 | 0.22 |
ENSMUST00000163610.9
ENSMUST00000164681.8 |
Psmd13
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 |
| chr17_+_34406762 | 0.22 |
ENSMUST00000041633.15
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr14_+_26722319 | 0.22 |
ENSMUST00000035433.10
|
Hesx1
|
homeobox gene expressed in ES cells |
| chr1_-_44157916 | 0.21 |
ENSMUST00000027213.14
ENSMUST00000065767.9 |
Poglut2
|
protein O-glucosyltransferase 2 |
| chr10_-_53952686 | 0.21 |
ENSMUST00000220088.2
|
Man1a
|
mannosidase 1, alpha |
| chr5_+_31212165 | 0.21 |
ENSMUST00000202795.4
ENSMUST00000201182.4 ENSMUST00000200953.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr17_+_34406523 | 0.20 |
ENSMUST00000170086.8
|
Tap1
|
transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) |
| chr6_+_57679455 | 0.20 |
ENSMUST00000072954.8
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr6_-_4086914 | 0.19 |
ENSMUST00000049166.5
|
Bet1
|
Bet1 golgi vesicular membrane trafficking protein |
| chr6_+_57679621 | 0.19 |
ENSMUST00000050077.15
|
Lancl2
|
LanC (bacterial lantibiotic synthetase component C)-like 2 |
| chr5_+_31212110 | 0.19 |
ENSMUST00000013773.12
ENSMUST00000201838.4 |
Cad
|
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr16_-_18232202 | 0.19 |
ENSMUST00000165430.8
ENSMUST00000147720.3 |
Comt
|
catechol-O-methyltransferase |
| chr9_+_74959259 | 0.19 |
ENSMUST00000170310.2
ENSMUST00000166549.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr16_-_30152708 | 0.19 |
ENSMUST00000229503.2
|
Atp13a3
|
ATPase type 13A3 |
| chr5_+_114924106 | 0.18 |
ENSMUST00000137519.2
|
Ankrd13a
|
ankyrin repeat domain 13a |
| chr1_+_179788037 | 0.18 |
ENSMUST00000097453.9
ENSMUST00000111117.8 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr3_+_14706781 | 0.17 |
ENSMUST00000029071.9
|
Car13
|
carbonic anhydrase 13 |
| chr2_-_125701059 | 0.17 |
ENSMUST00000110463.8
ENSMUST00000028635.6 |
Cops2
|
COP9 signalosome subunit 2 |
| chr18_+_32970363 | 0.17 |
ENSMUST00000166214.9
|
Wdr36
|
WD repeat domain 36 |
| chr2_-_113678945 | 0.17 |
ENSMUST00000110949.9
|
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr3_+_90383425 | 0.17 |
ENSMUST00000001042.10
|
Ilf2
|
interleukin enhancer binding factor 2 |
| chr2_-_14060840 | 0.17 |
ENSMUST00000074854.9
|
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr18_+_32970278 | 0.17 |
ENSMUST00000053663.11
|
Wdr36
|
WD repeat domain 36 |
| chr11_-_55310724 | 0.16 |
ENSMUST00000108858.8
ENSMUST00000141530.2 |
Sparc
|
secreted acidic cysteine rich glycoprotein |
| chr4_-_116982804 | 0.16 |
ENSMUST00000183310.2
|
Btbd19
|
BTB (POZ) domain containing 19 |
| chr7_+_80863314 | 0.16 |
ENSMUST00000026672.8
|
Pde8a
|
phosphodiesterase 8A |
| chr9_+_74945101 | 0.15 |
ENSMUST00000167885.8
ENSMUST00000169188.2 |
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr1_-_175453117 | 0.15 |
ENSMUST00000027810.14
|
Fh1
|
fumarate hydratase 1 |
| chr15_-_81810349 | 0.15 |
ENSMUST00000023113.7
|
Polr3h
|
polymerase (RNA) III (DNA directed) polypeptide H |
| chr5_-_148329615 | 0.15 |
ENSMUST00000138257.8
|
Slc7a1
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
| chr19_+_46611826 | 0.15 |
ENSMUST00000111855.5
|
Wbp1l
|
WW domain binding protein 1 like |
| chr11_+_94219046 | 0.15 |
ENSMUST00000021227.6
|
Ankrd40
|
ankyrin repeat domain 40 |
| chr10_+_126914755 | 0.15 |
ENSMUST00000039259.7
ENSMUST00000217941.2 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr16_-_56533179 | 0.14 |
ENSMUST00000136394.8
|
Tfg
|
Trk-fused gene |
| chr1_+_134110142 | 0.14 |
ENSMUST00000082060.10
ENSMUST00000153856.8 ENSMUST00000133701.8 ENSMUST00000132873.8 |
Chil1
|
chitinase-like 1 |
| chr8_-_111808488 | 0.14 |
ENSMUST00000188466.7
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr2_-_113678999 | 0.14 |
ENSMUST00000102545.8
ENSMUST00000110948.8 |
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr10_-_100425067 | 0.14 |
ENSMUST00000218821.2
ENSMUST00000054471.10 |
4930430F08Rik
|
RIKEN cDNA 4930430F08 gene |
| chr8_-_111808674 | 0.14 |
ENSMUST00000039597.14
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr14_+_20753074 | 0.14 |
ENSMUST00000071215.5
ENSMUST00000224633.2 |
Chchd1
|
coiled-coil-helix-coiled-coil-helix domain containing 1 |
| chr8_-_85414220 | 0.13 |
ENSMUST00000238449.2
ENSMUST00000238687.2 |
Nacc1
|
nucleus accumbens associated 1, BEN and BTB (POZ) domain containing |
| chr14_-_54648057 | 0.13 |
ENSMUST00000200545.2
ENSMUST00000227967.2 |
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr7_-_44465998 | 0.12 |
ENSMUST00000209072.2
ENSMUST00000047356.11 |
Atf5
|
activating transcription factor 5 |
| chr8_+_106428256 | 0.12 |
ENSMUST00000093195.7
ENSMUST00000211888.2 ENSMUST00000212430.2 |
Pard6a
|
par-6 family cell polarity regulator alpha |
| chrX_-_139857424 | 0.12 |
ENSMUST00000033805.15
ENSMUST00000112978.2 |
Psmd10
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 10 |
| chr3_-_96201248 | 0.12 |
ENSMUST00000029748.8
|
Fcgr1
|
Fc receptor, IgG, high affinity I |
| chr9_+_120758282 | 0.12 |
ENSMUST00000130466.8
|
Ctnnb1
|
catenin (cadherin associated protein), beta 1 |
| chr6_+_48695535 | 0.12 |
ENSMUST00000127537.6
ENSMUST00000052503.8 |
Gm28053
Gimap7
|
predicted gene, 28053 GTPase, IMAP family member 7 |
| chr1_+_179788675 | 0.11 |
ENSMUST00000076687.12
ENSMUST00000097450.10 ENSMUST00000212756.2 |
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr14_+_21126204 | 0.11 |
ENSMUST00000224069.2
|
Adk
|
adenosine kinase |
| chr11_-_3864664 | 0.11 |
ENSMUST00000109995.2
ENSMUST00000051207.2 |
Slc35e4
|
solute carrier family 35, member E4 |
| chr1_-_46927230 | 0.11 |
ENSMUST00000185520.2
|
Slc39a10
|
solute carrier family 39 (zinc transporter), member 10 |
| chr4_+_116565898 | 0.11 |
ENSMUST00000135499.8
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr14_-_104760051 | 0.10 |
ENSMUST00000022716.4
ENSMUST00000228448.2 ENSMUST00000227640.2 |
Obi1
|
ORC ubiquitin ligase 1 |
| chr19_-_8690330 | 0.10 |
ENSMUST00000206598.2
|
Slc3a2
|
solute carrier family 3 (activators of dibasic and neutral amino acid transport), member 2 |
| chr13_-_77283534 | 0.10 |
ENSMUST00000159462.3
ENSMUST00000151524.9 |
Slf1
|
SMC5-SMC6 complex localization factor 1 |
| chr7_+_127160751 | 0.10 |
ENSMUST00000190278.3
|
Tmem265
|
transmembrane protein 265 |
| chr6_+_134807097 | 0.10 |
ENSMUST00000046303.12
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr4_+_43406435 | 0.09 |
ENSMUST00000098106.9
ENSMUST00000139198.2 |
Rusc2
|
RUN and SH3 domain containing 2 |
| chr3_+_67799510 | 0.09 |
ENSMUST00000063263.5
ENSMUST00000182006.4 |
Iqcj
Iqschfp
|
IQ motif containing J Iqcj and Schip1 fusion protein |
| chr15_+_6451721 | 0.09 |
ENSMUST00000163082.2
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr5_-_88823049 | 0.09 |
ENSMUST00000133532.8
ENSMUST00000150438.2 |
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr18_+_42669322 | 0.09 |
ENSMUST00000236418.2
|
Tcerg1
|
transcription elongation regulator 1 (CA150) |
| chr11_-_60111391 | 0.09 |
ENSMUST00000020846.8
|
Srebf1
|
sterol regulatory element binding transcription factor 1 |
| chr15_-_82128538 | 0.09 |
ENSMUST00000229747.2
ENSMUST00000230408.2 |
Cenpm
|
centromere protein M |
| chr1_+_134109888 | 0.09 |
ENSMUST00000156873.8
|
Chil1
|
chitinase-like 1 |
| chr5_+_8710059 | 0.09 |
ENSMUST00000047753.5
|
Abcb1a
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1A |
| chr9_+_74945021 | 0.09 |
ENSMUST00000168301.2
|
Arpp19
|
cAMP-regulated phosphoprotein 19 |
| chr15_+_25774070 | 0.09 |
ENSMUST00000125667.3
|
Myo10
|
myosin X |
| chr11_+_76297969 | 0.08 |
ENSMUST00000021203.7
ENSMUST00000152183.2 |
Timm22
|
translocase of inner mitochondrial membrane 22 |
| chr2_+_118730823 | 0.08 |
ENSMUST00000151162.2
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr5_+_57875309 | 0.08 |
ENSMUST00000191837.6
ENSMUST00000068110.10 |
Pcdh7
|
protocadherin 7 |
| chr15_-_82128888 | 0.08 |
ENSMUST00000089155.6
ENSMUST00000089157.11 |
Cenpm
|
centromere protein M |
| chr15_+_51741138 | 0.08 |
ENSMUST00000136129.2
|
Utp23
|
UTP23 small subunit processome component |
| chr2_-_132657891 | 0.08 |
ENSMUST00000039554.7
|
Trmt6
|
tRNA methyltransferase 6 |
| chr4_+_116565819 | 0.08 |
ENSMUST00000106463.8
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr11_+_81926394 | 0.07 |
ENSMUST00000000193.6
|
Ccl2
|
chemokine (C-C motif) ligand 2 |
| chr18_+_24338729 | 0.07 |
ENSMUST00000170243.8
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr5_-_24650164 | 0.07 |
ENSMUST00000115043.8
ENSMUST00000115041.2 ENSMUST00000030800.13 |
Fastk
|
Fas-activated serine/threonine kinase |
| chr3_-_8988854 | 0.07 |
ENSMUST00000042148.6
|
Mrps28
|
mitochondrial ribosomal protein S28 |
| chr11_-_83421333 | 0.07 |
ENSMUST00000035938.3
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr7_+_28050077 | 0.07 |
ENSMUST00000082134.6
|
Rps16
|
ribosomal protein S16 |
| chr4_-_149747644 | 0.07 |
ENSMUST00000105689.8
|
Pik3cd
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| chr6_+_34840057 | 0.07 |
ENSMUST00000074949.4
|
Tmem140
|
transmembrane protein 140 |
| chr1_-_23948764 | 0.06 |
ENSMUST00000129254.8
|
Smap1
|
small ArfGAP 1 |
| chr15_+_32920869 | 0.06 |
ENSMUST00000022871.7
|
Sdc2
|
syndecan 2 |
| chr1_+_88154727 | 0.06 |
ENSMUST00000061013.13
ENSMUST00000113130.8 |
Mroh2a
|
maestro heat-like repeat family member 2A |
| chr2_-_14060774 | 0.06 |
ENSMUST00000114753.8
ENSMUST00000091429.12 |
Hacd1
|
3-hydroxyacyl-CoA dehydratase 1 |
| chr10_-_44334711 | 0.05 |
ENSMUST00000039174.11
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr11_-_106670309 | 0.05 |
ENSMUST00000021060.6
|
Polg2
|
polymerase (DNA directed), gamma 2, accessory subunit |
| chr4_+_150939521 | 0.05 |
ENSMUST00000030811.2
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr6_+_127430668 | 0.05 |
ENSMUST00000039680.7
|
Parp11
|
poly (ADP-ribose) polymerase family, member 11 |
| chr11_+_78079243 | 0.05 |
ENSMUST00000002128.14
ENSMUST00000150941.8 |
Rab34
|
RAB34, member RAS oncogene family |
| chr4_-_43454561 | 0.05 |
ENSMUST00000107926.8
ENSMUST00000107925.8 |
Cd72
|
CD72 antigen |
| chr6_+_71299758 | 0.05 |
ENSMUST00000065248.9
|
Cd8b1
|
CD8 antigen, beta chain 1 |
| chr5_+_24633206 | 0.05 |
ENSMUST00000115049.9
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr9_+_43978290 | 0.05 |
ENSMUST00000034508.14
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr11_-_69576363 | 0.04 |
ENSMUST00000018896.14
|
Tnfsf13
|
tumor necrosis factor (ligand) superfamily, member 13 |
| chr12_-_59266511 | 0.04 |
ENSMUST00000043204.8
|
Fbxo33
|
F-box protein 33 |
| chr10_-_78187545 | 0.04 |
ENSMUST00000105390.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr1_+_74811045 | 0.04 |
ENSMUST00000006716.8
|
Wnt6
|
wingless-type MMTV integration site family, member 6 |
| chr18_+_37827413 | 0.04 |
ENSMUST00000193414.2
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chrX_+_149981074 | 0.04 |
ENSMUST00000184730.8
ENSMUST00000184392.8 ENSMUST00000096285.5 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr12_-_15866763 | 0.04 |
ENSMUST00000020922.8
ENSMUST00000221215.2 ENSMUST00000221518.2 |
Trib2
|
tribbles pseudokinase 2 |
| chr18_-_36859732 | 0.04 |
ENSMUST00000061829.8
|
Cd14
|
CD14 antigen |
| chr6_-_125290809 | 0.04 |
ENSMUST00000032489.8
|
Ltbr
|
lymphotoxin B receptor |
| chr7_-_97228589 | 0.04 |
ENSMUST00000151840.2
ENSMUST00000135998.8 ENSMUST00000144858.8 ENSMUST00000146605.8 ENSMUST00000072725.12 ENSMUST00000138060.3 ENSMUST00000154853.8 ENSMUST00000136757.8 ENSMUST00000124552.3 |
Aamdc
|
adipogenesis associated Mth938 domain containing |
| chr2_-_101459274 | 0.04 |
ENSMUST00000099682.9
|
Iftap
|
intraflagellar transport associated protein |
| chr4_+_120523758 | 0.04 |
ENSMUST00000094814.6
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr1_+_171668173 | 0.04 |
ENSMUST00000136479.8
|
Cd84
|
CD84 antigen |
| chr1_+_59555423 | 0.03 |
ENSMUST00000114243.8
|
Gm973
|
predicted gene 973 |
| chr5_+_122239030 | 0.03 |
ENSMUST00000139213.8
ENSMUST00000111751.8 ENSMUST00000155612.8 |
Myl2
|
myosin, light polypeptide 2, regulatory, cardiac, slow |
| chr6_+_8520006 | 0.03 |
ENSMUST00000162567.8
ENSMUST00000161217.8 |
Glcci1
|
glucocorticoid induced transcript 1 |
| chr4_+_116565706 | 0.03 |
ENSMUST00000030452.13
ENSMUST00000106462.9 |
Ccdc163
|
coiled-coil domain containing 163 |
| chr16_-_22084700 | 0.03 |
ENSMUST00000161286.8
|
Tra2b
|
transformer 2 beta |
| chr3_-_116601451 | 0.03 |
ENSMUST00000159670.3
|
Agl
|
amylo-1,6-glucosidase, 4-alpha-glucanotransferase |
| chr10_-_44334683 | 0.03 |
ENSMUST00000105490.3
|
Prdm1
|
PR domain containing 1, with ZNF domain |
| chr13_+_43938251 | 0.03 |
ENSMUST00000015540.4
|
Cd83
|
CD83 antigen |
| chr5_+_90708962 | 0.03 |
ENSMUST00000094615.8
ENSMUST00000200765.2 |
Albfm1
|
albumin superfamily member 1 |
| chr19_+_12647803 | 0.03 |
ENSMUST00000207341.3
ENSMUST00000208494.3 ENSMUST00000208657.3 |
Olfr1442
|
olfactory receptor 1442 |
| chr1_-_157072060 | 0.03 |
ENSMUST00000129880.8
|
Rasal2
|
RAS protein activator like 2 |
| chr1_+_131890679 | 0.02 |
ENSMUST00000191034.2
ENSMUST00000177943.8 |
Gm29103
Slc45a3
|
predicted gene 29103 solute carrier family 45, member 3 |
| chr1_+_171594690 | 0.02 |
ENSMUST00000015460.5
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr6_+_48695571 | 0.02 |
ENSMUST00000204785.2
|
Gimap7
|
GTPase, IMAP family member 7 |
| chr3_-_87170903 | 0.02 |
ENSMUST00000090986.11
|
Fcrls
|
Fc receptor-like S, scavenger receptor |
| chr1_+_44158111 | 0.02 |
ENSMUST00000155917.8
|
Bivm
|
basic, immunoglobulin-like variable motif containing |
| chr18_+_37488174 | 0.02 |
ENSMUST00000192867.2
ENSMUST00000051163.3 |
Pcdhb8
|
protocadherin beta 8 |
| chr10_+_43777777 | 0.02 |
ENSMUST00000054418.12
|
Rtn4ip1
|
reticulon 4 interacting protein 1 |
| chr9_+_43978369 | 0.02 |
ENSMUST00000177054.8
|
Usp2
|
ubiquitin specific peptidase 2 |
| chr11_+_108477903 | 0.02 |
ENSMUST00000146912.9
|
Cep112
|
centrosomal protein 112 |
| chr14_+_79825131 | 0.02 |
ENSMUST00000054908.10
|
Sugt1
|
SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) |
| chr16_-_58850641 | 0.02 |
ENSMUST00000216415.2
ENSMUST00000206463.3 |
Olfr186
|
olfactory receptor 186 |
| chr15_+_25933632 | 0.02 |
ENSMUST00000228327.2
|
Retreg1
|
reticulophagy regulator 1 |
| chrX_+_162694397 | 0.02 |
ENSMUST00000140845.2
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr3_+_103739877 | 0.02 |
ENSMUST00000062945.12
|
Bcl2l15
|
BCLl2-like 15 |
| chr14_-_73286535 | 0.02 |
ENSMUST00000169168.3
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr15_+_59520199 | 0.02 |
ENSMUST00000067543.8
|
Trib1
|
tribbles pseudokinase 1 |
| chr11_+_108811626 | 0.01 |
ENSMUST00000140821.2
|
Axin2
|
axin 2 |
| chr7_-_109330915 | 0.01 |
ENSMUST00000035372.3
|
Ascl3
|
achaete-scute family bHLH transcription factor 3 |
| chr1_+_146373352 | 0.01 |
ENSMUST00000132847.8
ENSMUST00000166814.8 |
Brinp3
|
bone morphogenetic protein/retinoic acid inducible neural specific 3 |
| chr14_-_73286504 | 0.01 |
ENSMUST00000044664.12
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr13_-_21798192 | 0.01 |
ENSMUST00000051874.6
|
Olfr1362
|
olfactory receptor 1362 |
| chr9_+_39580355 | 0.01 |
ENSMUST00000073433.2
|
Olfr963
|
olfactory receptor 963 |
| chr5_+_14075281 | 0.01 |
ENSMUST00000073957.8
|
Sema3e
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
| chr9_-_38971960 | 0.01 |
ENSMUST00000055567.3
|
Olfr937
|
olfactory receptor 937 |
| chr10_-_79940168 | 0.01 |
ENSMUST00000219260.2
|
Sbno2
|
strawberry notch 2 |
| chr9_+_39372729 | 0.01 |
ENSMUST00000080329.2
|
Olfr954
|
olfactory receptor 954 |
| chr5_-_6926523 | 0.01 |
ENSMUST00000164784.2
|
Zfp804b
|
zinc finger protein 804B |
| chr5_-_72893941 | 0.01 |
ENSMUST00000169534.6
|
Txk
|
TXK tyrosine kinase |
| chr7_-_46902594 | 0.01 |
ENSMUST00000098436.4
|
Mrgpra9
|
MAS-related GPR, member A9 |
| chr7_+_102576837 | 0.01 |
ENSMUST00000098214.2
|
Olfr572
|
olfactory receptor 572 |
| chrX_-_104918911 | 0.01 |
ENSMUST00000200471.2
|
Atrx
|
ATRX, chromatin remodeler |
| chr6_-_141719536 | 0.01 |
ENSMUST00000148411.2
|
Gm5724
|
predicted gene 5724 |
| chr6_+_34840151 | 0.01 |
ENSMUST00000202010.2
|
Tmem140
|
transmembrane protein 140 |
| chr2_-_147028309 | 0.01 |
ENSMUST00000067075.7
|
Nkx2-2
|
NK2 homeobox 2 |
| chr7_+_88079534 | 0.01 |
ENSMUST00000208478.2
|
Rab38
|
RAB38, member RAS oncogene family |
| chr1_-_130867810 | 0.01 |
ENSMUST00000112465.2
ENSMUST00000187410.7 ENSMUST00000187916.7 |
Il19
|
interleukin 19 |
| chr7_-_46902575 | 0.01 |
ENSMUST00000179005.3
|
Mrgpra9
|
MAS-related GPR, member A9 |
| chr10_+_97442727 | 0.01 |
ENSMUST00000105286.4
|
Kera
|
keratocan |
| chr15_-_93417380 | 0.01 |
ENSMUST00000109255.3
|
Prickle1
|
prickle planar cell polarity protein 1 |
| chrX_+_55939732 | 0.01 |
ENSMUST00000136396.8
|
Adgrg4
|
adhesion G protein-coupled receptor G4 |
| chr2_+_87082184 | 0.01 |
ENSMUST00000081986.5
|
Olfr1115
|
olfactory receptor 1115 |
| chr7_-_106547519 | 0.01 |
ENSMUST00000055923.8
|
Olfr710
|
olfactory receptor 710 |
| chr11_+_108811168 | 0.01 |
ENSMUST00000052915.14
|
Axin2
|
axin 2 |
| chr4_+_135870808 | 0.01 |
ENSMUST00000008016.3
|
Id3
|
inhibitor of DNA binding 3 |
| chr19_+_12245527 | 0.01 |
ENSMUST00000073507.3
|
Olfr235
|
olfactory receptor 235 |
| chr14_-_100522101 | 0.01 |
ENSMUST00000228216.2
|
Klf12
|
Kruppel-like factor 12 |
| chr3_-_144738526 | 0.01 |
ENSMUST00000029919.7
|
Clca1
|
chloride channel accessory 1 |
| chr19_+_12824046 | 0.01 |
ENSMUST00000189517.2
|
Gm5244
|
predicted pseudogene 5244 |
| chr9_-_39423014 | 0.01 |
ENSMUST00000051653.2
|
Olfr957
|
olfactory receptor 957 |
| chr4_-_94817025 | 0.01 |
ENSMUST00000030309.6
|
Eqtn
|
equatorin, sperm acrosome associated |
| chr2_+_163067283 | 0.01 |
ENSMUST00000099110.10
ENSMUST00000165937.8 |
Tox2
|
TOX high mobility group box family member 2 |
| chr11_-_99907030 | 0.01 |
ENSMUST00000018399.3
|
Krt33a
|
keratin 33A |
| chr19_-_10582672 | 0.01 |
ENSMUST00000236478.2
ENSMUST00000236950.2 |
Tkfc
|
triokinase, FMN cyclase |
| chr17_+_37777099 | 0.01 |
ENSMUST00000031086.5
|
Olfr109
|
olfactory receptor 109 |
| chr19_+_12186932 | 0.01 |
ENSMUST00000072316.5
|
Olfr1431
|
olfactory receptor 1431 |
| chr11_+_96242422 | 0.01 |
ENSMUST00000100523.7
|
Hoxb2
|
homeobox B2 |
| chr10_-_78187524 | 0.01 |
ENSMUST00000166360.8
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr2_-_87466089 | 0.01 |
ENSMUST00000090711.3
|
Olfr1132
|
olfactory receptor 1132 |
| chr8_+_45960931 | 0.01 |
ENSMUST00000171337.10
ENSMUST00000067107.15 |
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_+_70393195 | 0.01 |
ENSMUST00000130998.8
|
Gad1
|
glutamate decarboxylase 1 |
| chr16_-_35691914 | 0.01 |
ENSMUST00000042665.9
|
Parp14
|
poly (ADP-ribose) polymerase family, member 14 |
| chr4_-_94817056 | 0.01 |
ENSMUST00000107097.9
|
Eqtn
|
equatorin, sperm acrosome associated |
Network of associatons between targets according to the STRING database.
First level regulatory network of Stat6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0002148 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.1 | 0.4 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.1 | 0.4 | GO:1901421 | positive regulation of response to alcohol(GO:1901421) |
| 0.1 | 0.5 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.1 | 1.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.4 | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process(GO:0006207) pyrimidine nucleobase biosynthetic process(GO:0019856) |
| 0.1 | 0.2 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.1 | GO:1904499 | regulation of chromatin-mediated maintenance of transcription(GO:1904499) positive regulation of chromatin-mediated maintenance of transcription(GO:1904501) regulation of euchromatin binding(GO:1904793) |
| 0.0 | 0.2 | GO:0045914 | regulation of sulfur amino acid metabolic process(GO:0031335) negative regulation of catecholamine metabolic process(GO:0045914) negative regulation of dopamine metabolic process(GO:0045963) |
| 0.0 | 0.1 | GO:0006169 | adenosine salvage(GO:0006169) dATP biosynthetic process(GO:0006175) |
| 0.0 | 0.2 | GO:1900740 | regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900739) positive regulation of protein insertion into mitochondrial membrane involved in apoptotic signaling pathway(GO:1900740) |
| 0.0 | 0.1 | GO:0033082 | regulation of extrathymic T cell differentiation(GO:0033082) |
| 0.0 | 0.3 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0090264 | immune complex clearance by monocytes and macrophages(GO:0002436) monocyte homeostasis(GO:0035702) regulation of immune complex clearance by monocytes and macrophages(GO:0090264) positive regulation of immune complex clearance by monocytes and macrophages(GO:0090265) |
| 0.0 | 0.1 | GO:2001025 | positive regulation of response to drug(GO:2001025) |
| 0.0 | 0.1 | GO:0035697 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.1 | GO:0060356 | leucine import(GO:0060356) |
| 0.0 | 0.3 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.3 | GO:0071493 | cellular response to UV-B(GO:0071493) |
| 0.0 | 0.2 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0034184 | positive regulation of maintenance of mitotic sister chromatid cohesion(GO:0034184) protein localization to site of double-strand break(GO:1990166) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.0 | GO:0002426 | immunoglobulin production in mucosal tissue(GO:0002426) |
| 0.0 | 0.0 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0010616 | negative regulation of cardiac muscle adaptation(GO:0010616) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.4 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:1905171 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.1 | GO:0003062 | regulation of heart rate by chemical signal(GO:0003062) |
| 0.0 | 0.1 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.1 | 0.5 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.1 | 1.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.4 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 1.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.1 | 0.4 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.1 | 1.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.3 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.2 | GO:0080023 | 3R-hydroxyacyl-CoA dehydratase activity(GO:0080023) |
| 0.1 | 0.2 | GO:0016206 | catechol O-methyltransferase activity(GO:0016206) |
| 0.0 | 0.1 | GO:0004001 | adenosine kinase activity(GO:0004001) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0030943 | mitochondrion targeting sequence binding(GO:0030943) |
| 0.0 | 0.1 | GO:0030298 | receptor signaling protein tyrosine kinase activator activity(GO:0030298) CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.2 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 1.1 | GO:0004601 | peroxidase activity(GO:0004601) |
| 0.0 | 0.3 | GO:0004298 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 1.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.4 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 1.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |