Project
GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for Thra
Z-value: 0.39
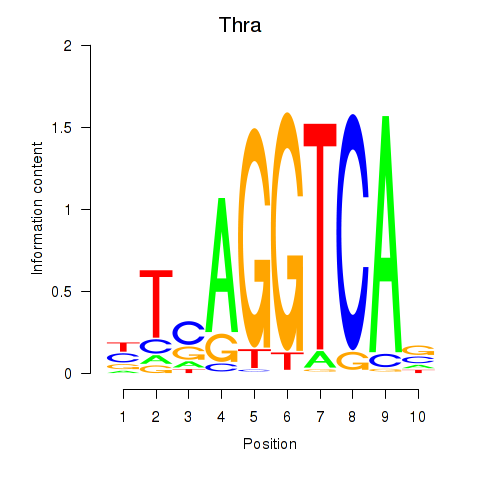
Transcription factors associated with Thra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Thra
|
ENSMUSG00000058756.14 | thyroid hormone receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Thra | mm39_v1_chr11_+_98632953_98632984 | -0.48 | 4.2e-01 | Click! |
Activity profile of Thra motif
Sorted Z-values of Thra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_113223839 | 0.34 |
ENSMUST00000194738.6
ENSMUST00000178282.3 |
Igha
|
immunoglobulin heavy constant alpha |
| chr7_-_142050663 | 0.31 |
ENSMUST00000238347.2
|
Prr33
|
proline rich 33 |
| chr14_+_58308004 | 0.29 |
ENSMUST00000165526.9
|
Fgf9
|
fibroblast growth factor 9 |
| chr13_+_44883270 | 0.27 |
ENSMUST00000172830.8
|
Jarid2
|
jumonji, AT rich interactive domain 2 |
| chr7_+_3339077 | 0.26 |
ENSMUST00000203566.3
|
Myadm
|
myeloid-associated differentiation marker |
| chr7_+_3339059 | 0.26 |
ENSMUST00000096744.8
|
Myadm
|
myeloid-associated differentiation marker |
| chr11_-_70146156 | 0.25 |
ENSMUST00000108574.3
ENSMUST00000000329.9 |
Alox12
|
arachidonate 12-lipoxygenase |
| chr16_-_23339548 | 0.24 |
ENSMUST00000089883.7
|
Masp1
|
mannan-binding lectin serine peptidase 1 |
| chr11_+_49447705 | 0.22 |
ENSMUST00000215360.2
|
Olfr1380
|
olfactory receptor 1380 |
| chr10_+_79984097 | 0.22 |
ENSMUST00000099492.10
ENSMUST00000042057.12 |
Midn
|
midnolin |
| chr1_-_170133901 | 0.22 |
ENSMUST00000179801.3
|
Gm7694
|
predicted gene 7694 |
| chr4_+_85123654 | 0.22 |
ENSMUST00000030212.15
ENSMUST00000107189.8 ENSMUST00000107184.8 |
Sh3gl2
|
SH3-domain GRB2-like 2 |
| chr17_-_36220924 | 0.21 |
ENSMUST00000141662.8
ENSMUST00000056034.13 ENSMUST00000077494.13 ENSMUST00000149277.8 ENSMUST00000061052.12 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr13_+_95833359 | 0.21 |
ENSMUST00000022182.5
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr4_-_62278761 | 0.20 |
ENSMUST00000107461.2
ENSMUST00000084528.10 |
Fkbp15
|
FK506 binding protein 15 |
| chr9_-_119812829 | 0.20 |
ENSMUST00000216929.2
|
Csrnp1
|
cysteine-serine-rich nuclear protein 1 |
| chr2_+_71884943 | 0.19 |
ENSMUST00000028525.6
|
Rapgef4
|
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr7_-_73187369 | 0.18 |
ENSMUST00000172704.6
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr14_+_25459690 | 0.18 |
ENSMUST00000007961.15
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr7_+_79674562 | 0.18 |
ENSMUST00000164056.9
ENSMUST00000166250.8 |
Zfp710
|
zinc finger protein 710 |
| chr5_+_33021042 | 0.17 |
ENSMUST00000149350.8
ENSMUST00000118698.8 ENSMUST00000150130.8 ENSMUST00000049780.13 ENSMUST00000087897.11 ENSMUST00000119705.8 ENSMUST00000125574.8 |
Depdc5
|
DEP domain containing 5 |
| chr5_+_34527230 | 0.17 |
ENSMUST00000180376.8
|
Fam193a
|
family with sequence homology 193, member A |
| chr16_+_24540599 | 0.15 |
ENSMUST00000115314.4
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr8_+_72860884 | 0.15 |
ENSMUST00000210435.4
|
Olfr374
|
olfactory receptor 374 |
| chr7_+_30159137 | 0.15 |
ENSMUST00000006825.9
|
Nphs1
|
nephrosis 1, nephrin |
| chr4_+_62581857 | 0.14 |
ENSMUST00000126338.2
|
Rgs3
|
regulator of G-protein signaling 3 |
| chr8_+_121264161 | 0.14 |
ENSMUST00000118136.2
|
Gse1
|
genetic suppressor element 1, coiled-coil protein |
| chr3_+_122523219 | 0.14 |
ENSMUST00000200389.2
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr14_-_50586329 | 0.14 |
ENSMUST00000216634.2
|
Olfr735
|
olfactory receptor 735 |
| chr3_+_122522592 | 0.14 |
ENSMUST00000066728.10
|
Pde5a
|
phosphodiesterase 5A, cGMP-specific |
| chr14_+_25459630 | 0.14 |
ENSMUST00000162645.8
|
Zmiz1
|
zinc finger, MIZ-type containing 1 |
| chr4_-_41048124 | 0.13 |
ENSMUST00000030136.13
|
Aqp7
|
aquaporin 7 |
| chr6_+_136509922 | 0.13 |
ENSMUST00000187429.4
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr4_-_144921567 | 0.12 |
ENSMUST00000036579.14
|
Vps13d
|
vacuolar protein sorting 13D |
| chr16_-_18904240 | 0.12 |
ENSMUST00000103746.3
|
Iglv1
|
immunoglobulin lambda variable 1 |
| chr4_-_139695337 | 0.12 |
ENSMUST00000105031.4
|
Klhdc7a
|
kelch domain containing 7A |
| chr11_+_97206542 | 0.12 |
ENSMUST00000019026.10
ENSMUST00000132168.2 |
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chrX_-_48823936 | 0.12 |
ENSMUST00000215373.3
|
Olfr1321
|
olfactory receptor 1321 |
| chr9_+_110848339 | 0.12 |
ENSMUST00000198884.5
ENSMUST00000196777.5 ENSMUST00000196209.5 ENSMUST00000035077.8 ENSMUST00000196122.3 |
Ltf
|
lactotransferrin |
| chr6_+_11907808 | 0.11 |
ENSMUST00000155037.4
|
Phf14
|
PHD finger protein 14 |
| chr9_+_108685555 | 0.11 |
ENSMUST00000035218.9
ENSMUST00000195323.2 ENSMUST00000194819.2 |
Nckipsd
|
NCK interacting protein with SH3 domain |
| chrX_-_165368675 | 0.11 |
ENSMUST00000000412.3
|
Egfl6
|
EGF-like-domain, multiple 6 |
| chr18_+_82932747 | 0.11 |
ENSMUST00000071233.7
|
Zfp516
|
zinc finger protein 516 |
| chr6_+_22288220 | 0.11 |
ENSMUST00000128245.8
ENSMUST00000031681.10 ENSMUST00000148639.2 |
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr4_-_20778847 | 0.11 |
ENSMUST00000102998.4
|
Nkain3
|
Na+/K+ transporting ATPase interacting 3 |
| chr11_+_67477347 | 0.11 |
ENSMUST00000108682.9
|
Gas7
|
growth arrest specific 7 |
| chr7_+_107194446 | 0.11 |
ENSMUST00000040056.15
ENSMUST00000208956.2 |
Ppfibp2
|
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr13_+_43276323 | 0.10 |
ENSMUST00000136576.8
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr4_+_124774692 | 0.10 |
ENSMUST00000059343.7
|
Epha10
|
Eph receptor A10 |
| chr9_+_64142483 | 0.10 |
ENSMUST00000039011.4
|
Uchl4
|
ubiquitin carboxyl-terminal esterase L4 |
| chr5_-_137623331 | 0.10 |
ENSMUST00000031732.14
|
Fbxo24
|
F-box protein 24 |
| chr7_-_100164007 | 0.10 |
ENSMUST00000207405.2
|
Dnajb13
|
DnaJ heat shock protein family (Hsp40) member B13 |
| chr17_-_36220518 | 0.10 |
ENSMUST00000141132.2
|
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr15_-_102419115 | 0.10 |
ENSMUST00000171565.8
|
Map3k12
|
mitogen-activated protein kinase kinase kinase 12 |
| chr7_-_140676623 | 0.10 |
ENSMUST00000209352.2
|
Sigirr
|
single immunoglobulin and toll-interleukin 1 receptor (TIR) domain |
| chr4_-_41275091 | 0.09 |
ENSMUST00000030143.13
ENSMUST00000108068.8 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr17_+_15163466 | 0.09 |
ENSMUST00000164837.3
ENSMUST00000174004.2 |
1600012H06Rik
|
RIKEN cDNA 1600012H06 gene |
| chr5_-_25200745 | 0.09 |
ENSMUST00000076306.12
|
Prkag2
|
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
| chr17_+_15163446 | 0.09 |
ENSMUST00000052691.9
|
1600012H06Rik
|
RIKEN cDNA 1600012H06 gene |
| chr11_-_101875575 | 0.09 |
ENSMUST00000176261.2
ENSMUST00000143177.2 ENSMUST00000003612.13 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chrX_+_98086187 | 0.09 |
ENSMUST00000036606.14
|
Stard8
|
START domain containing 8 |
| chr17_+_35235552 | 0.08 |
ENSMUST00000007245.8
ENSMUST00000172499.2 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr2_+_112096154 | 0.08 |
ENSMUST00000110991.9
|
Slc12a6
|
solute carrier family 12, member 6 |
| chr1_-_84817976 | 0.08 |
ENSMUST00000190067.7
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr1_+_74582044 | 0.08 |
ENSMUST00000113749.8
ENSMUST00000067916.13 ENSMUST00000113747.8 ENSMUST00000113750.8 |
Plcd4
|
phospholipase C, delta 4 |
| chr4_-_118266416 | 0.08 |
ENSMUST00000075406.12
|
Szt2
|
SZT2 subunit of KICSTOR complex |
| chr2_+_145627900 | 0.08 |
ENSMUST00000110005.8
ENSMUST00000094480.11 |
Rin2
|
Ras and Rab interactor 2 |
| chr9_+_107454114 | 0.08 |
ENSMUST00000112387.9
ENSMUST00000123005.8 ENSMUST00000010195.14 ENSMUST00000144392.2 |
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr9_+_121780054 | 0.08 |
ENSMUST00000043011.9
ENSMUST00000214536.3 ENSMUST00000215990.3 |
Gask1a
|
golgi associated kinase 1A |
| chr7_+_24990596 | 0.08 |
ENSMUST00000164820.2
|
Cic
|
capicua transcriptional repressor |
| chr7_-_140793990 | 0.08 |
ENSMUST00000026573.7
ENSMUST00000170841.9 |
Lmntd2
|
lamin tail domain containing 2 |
| chr11_-_104332528 | 0.08 |
ENSMUST00000106971.2
|
Kansl1
|
KAT8 regulatory NSL complex subunit 1 |
| chr14_-_20133246 | 0.08 |
ENSMUST00000059666.6
|
Saysd1
|
SAYSVFN motif domain containing 1 |
| chr15_-_100449668 | 0.08 |
ENSMUST00000229581.2
ENSMUST00000231138.2 |
Tfcp2
|
transcription factor CP2 |
| chr3_+_89970088 | 0.08 |
ENSMUST00000238911.2
ENSMUST00000029551.3 |
1700094D03Rik
|
RIKEN cDNA 1700094D03 gene |
| chr14_-_73563212 | 0.08 |
ENSMUST00000022701.7
|
Rb1
|
RB transcriptional corepressor 1 |
| chr4_-_141327146 | 0.07 |
ENSMUST00000141518.8
ENSMUST00000127455.8 ENSMUST00000105784.8 |
Fblim1
|
filamin binding LIM protein 1 |
| chr14_-_47426863 | 0.07 |
ENSMUST00000089959.7
|
Gch1
|
GTP cyclohydrolase 1 |
| chr3_-_89300936 | 0.07 |
ENSMUST00000124783.8
ENSMUST00000126027.8 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr16_+_91282121 | 0.07 |
ENSMUST00000023689.11
ENSMUST00000117748.8 |
Ifnar1
|
interferon (alpha and beta) receptor 1 |
| chr15_-_74624811 | 0.07 |
ENSMUST00000189128.2
ENSMUST00000023259.15 |
Lynx1
|
Ly6/neurotoxin 1 |
| chr9_+_44151962 | 0.07 |
ENSMUST00000092426.5
ENSMUST00000217221.2 ENSMUST00000213891.2 |
Ccdc153
|
coiled-coil domain containing 153 |
| chr7_-_126574959 | 0.07 |
ENSMUST00000206296.2
ENSMUST00000205437.3 |
Cdiptos
|
CDIP transferase, opposite strand |
| chr19_-_46315543 | 0.07 |
ENSMUST00000223917.2
ENSMUST00000224447.2 ENSMUST00000041391.5 ENSMUST00000096029.12 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr3_+_86131970 | 0.07 |
ENSMUST00000192145.6
ENSMUST00000194759.6 ENSMUST00000107635.7 |
Lrba
|
LPS-responsive beige-like anchor |
| chr5_+_53747556 | 0.07 |
ENSMUST00000037618.13
ENSMUST00000201912.4 |
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr6_-_116050081 | 0.07 |
ENSMUST00000173548.3
|
Tmcc1
|
transmembrane and coiled coil domains 1 |
| chr14_-_55995912 | 0.06 |
ENSMUST00000001497.9
|
Cideb
|
cell death-inducing DNA fragmentation factor, alpha subunit-like effector B |
| chr15_-_5273645 | 0.06 |
ENSMUST00000120563.2
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr12_+_111679689 | 0.06 |
ENSMUST00000040519.12
ENSMUST00000163220.10 ENSMUST00000162316.2 |
Coa8
|
cytochrome c oxidase assembly factor 8 |
| chr15_-_5273659 | 0.06 |
ENSMUST00000047379.15
|
Ptger4
|
prostaglandin E receptor 4 (subtype EP4) |
| chr11_+_31822211 | 0.06 |
ENSMUST00000020543.13
ENSMUST00000109412.9 |
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr7_-_30755007 | 0.06 |
ENSMUST00000206474.2
ENSMUST00000205807.2 ENSMUST00000039909.13 ENSMUST00000206305.2 ENSMUST00000205439.2 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr11_-_85030761 | 0.06 |
ENSMUST00000108075.9
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr7_-_119461027 | 0.06 |
ENSMUST00000137888.2
ENSMUST00000142120.2 |
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chrX_+_72760183 | 0.06 |
ENSMUST00000002084.14
|
Abcd1
|
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr10_-_81436671 | 0.06 |
ENSMUST00000151858.8
ENSMUST00000142948.8 ENSMUST00000072020.9 |
Tle6
|
transducin-like enhancer of split 6 |
| chr10_+_7556948 | 0.06 |
ENSMUST00000165952.9
|
Lats1
|
large tumor suppressor |
| chrX_+_143902497 | 0.06 |
ENSMUST00000096301.5
|
Rtl4
|
retrotransposon Gag like 4 |
| chr10_-_80895949 | 0.06 |
ENSMUST00000219401.2
ENSMUST00000220317.2 ENSMUST00000005067.6 ENSMUST00000218208.2 |
Sgta
|
small glutamine-rich tetratricopeptide repeat (TPR)-containing, alpha |
| chr6_-_71800788 | 0.06 |
ENSMUST00000065103.4
|
Mrpl35
|
mitochondrial ribosomal protein L35 |
| chr10_+_126814542 | 0.05 |
ENSMUST00000105256.10
|
Ctdsp2
|
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr18_+_42408418 | 0.05 |
ENSMUST00000091920.6
ENSMUST00000046972.14 ENSMUST00000236240.2 |
Rbm27
|
RNA binding motif protein 27 |
| chr15_-_82796308 | 0.05 |
ENSMUST00000109510.10
ENSMUST00000048966.7 |
Tcf20
|
transcription factor 20 |
| chr11_-_40624200 | 0.05 |
ENSMUST00000020579.9
|
Hmmr
|
hyaluronan mediated motility receptor (RHAMM) |
| chr10_-_78300802 | 0.05 |
ENSMUST00000041616.15
|
Pdxk
|
pyridoxal (pyridoxine, vitamin B6) kinase |
| chr11_+_67477501 | 0.05 |
ENSMUST00000108680.2
|
Gas7
|
growth arrest specific 7 |
| chr17_+_85928459 | 0.05 |
ENSMUST00000162695.3
|
Six3
|
sine oculis-related homeobox 3 |
| chr10_+_84412490 | 0.05 |
ENSMUST00000020223.8
|
Tcp11l2
|
t-complex 11 (mouse) like 2 |
| chr7_-_80052491 | 0.05 |
ENSMUST00000122232.8
|
Furin
|
furin (paired basic amino acid cleaving enzyme) |
| chr11_+_51541728 | 0.04 |
ENSMUST00000117859.8
ENSMUST00000064493.6 |
D930048N14Rik
|
RIKEN cDNA D930048N14 gene |
| chr11_+_44409775 | 0.04 |
ENSMUST00000019333.10
|
Rnf145
|
ring finger protein 145 |
| chr17_+_48571298 | 0.04 |
ENSMUST00000059873.14
ENSMUST00000154335.8 ENSMUST00000136272.8 ENSMUST00000125426.8 ENSMUST00000153420.2 |
Treml4
|
triggering receptor expressed on myeloid cells-like 4 |
| chr13_-_81781238 | 0.04 |
ENSMUST00000126444.8
ENSMUST00000128585.9 ENSMUST00000146749.2 ENSMUST00000095585.11 |
Adgrv1
|
adhesion G protein-coupled receptor V1 |
| chr7_+_18962301 | 0.04 |
ENSMUST00000161711.2
|
Opa3
|
optic atrophy 3 |
| chrX_+_100492684 | 0.04 |
ENSMUST00000033674.6
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr17_-_24863907 | 0.04 |
ENSMUST00000234505.2
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr7_-_102566717 | 0.04 |
ENSMUST00000214160.2
ENSMUST00000215773.2 |
Olfr571
|
olfactory receptor 571 |
| chr3_-_95214443 | 0.04 |
ENSMUST00000015846.9
|
Anxa9
|
annexin A9 |
| chr15_-_100449839 | 0.04 |
ENSMUST00000009877.8
ENSMUST00000229696.2 |
Tfcp2
|
transcription factor CP2 |
| chr16_+_20513658 | 0.04 |
ENSMUST00000056518.13
|
Fam131a
|
family with sequence similarity 131, member A |
| chr11_-_95037089 | 0.04 |
ENSMUST00000021241.8
|
Dlx4
|
distal-less homeobox 4 |
| chr4_+_131600918 | 0.04 |
ENSMUST00000053819.6
|
Srsf4
|
serine and arginine-rich splicing factor 4 |
| chr1_-_134006847 | 0.04 |
ENSMUST00000020692.7
|
Btg2
|
BTG anti-proliferation factor 2 |
| chr17_-_36207965 | 0.04 |
ENSMUST00000150056.2
ENSMUST00000156817.2 ENSMUST00000146451.8 ENSMUST00000148482.8 |
2310061I04Rik
|
RIKEN cDNA 2310061I04 gene |
| chr2_-_79959178 | 0.04 |
ENSMUST00000102654.11
ENSMUST00000102655.10 |
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent |
| chr10_-_79940168 | 0.04 |
ENSMUST00000219260.2
|
Sbno2
|
strawberry notch 2 |
| chr2_-_119308094 | 0.03 |
ENSMUST00000110808.2
ENSMUST00000049920.14 |
Ino80
|
INO80 complex subunit |
| chr4_-_126096376 | 0.03 |
ENSMUST00000106142.8
ENSMUST00000169403.8 ENSMUST00000130334.2 |
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr7_+_18946448 | 0.03 |
ENSMUST00000060225.6
|
Gpr4
|
G protein-coupled receptor 4 |
| chr2_-_89779008 | 0.03 |
ENSMUST00000214846.2
|
Olfr1259
|
olfactory receptor 1259 |
| chr11_+_120348919 | 0.03 |
ENSMUST00000058370.14
ENSMUST00000175970.8 ENSMUST00000176120.2 |
Ccdc137
|
coiled-coil domain containing 137 |
| chr11_+_115366470 | 0.03 |
ENSMUST00000035240.7
|
Armc7
|
armadillo repeat containing 7 |
| chr2_-_34716199 | 0.03 |
ENSMUST00000113075.8
ENSMUST00000113080.9 ENSMUST00000091020.10 |
Fbxw2
|
F-box and WD-40 domain protein 2 |
| chr4_-_141327253 | 0.03 |
ENSMUST00000147785.8
|
Fblim1
|
filamin binding LIM protein 1 |
| chr2_+_130248398 | 0.03 |
ENSMUST00000055421.6
|
Tmem239
|
transmembrane 239 |
| chr12_+_84498196 | 0.03 |
ENSMUST00000137170.3
|
Lin52
|
lin-52 homolog (C. elegans) |
| chr15_+_99600475 | 0.03 |
ENSMUST00000228984.2
ENSMUST00000229845.2 |
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr17_+_85928244 | 0.03 |
ENSMUST00000176081.3
|
Six3
|
sine oculis-related homeobox 3 |
| chr15_-_76606361 | 0.03 |
ENSMUST00000049956.5
|
Lrrc24
|
leucine rich repeat containing 24 |
| chr8_+_73197718 | 0.03 |
ENSMUST00000064853.13
ENSMUST00000121902.2 |
1700030K09Rik
|
RIKEN cDNA 1700030K09 gene |
| chr12_+_102435383 | 0.03 |
ENSMUST00000179218.9
|
Golga5
|
golgi autoantigen, golgin subfamily a, 5 |
| chr15_+_73594965 | 0.03 |
ENSMUST00000165541.8
ENSMUST00000167582.8 |
Ptp4a3
|
protein tyrosine phosphatase 4a3 |
| chr5_-_143255713 | 0.03 |
ENSMUST00000161448.8
|
Zfp316
|
zinc finger protein 316 |
| chr13_+_55357585 | 0.03 |
ENSMUST00000224973.2
ENSMUST00000099490.3 |
Nsd1
|
nuclear receptor-binding SET-domain protein 1 |
| chr12_-_81379903 | 0.03 |
ENSMUST00000085238.13
ENSMUST00000182208.8 |
Slc8a3
|
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr1_+_131838220 | 0.02 |
ENSMUST00000189946.7
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr14_+_47069667 | 0.02 |
ENSMUST00000140114.3
ENSMUST00000133989.8 |
Cgrrf1
|
cell growth regulator with ring finger domain 1 |
| chr17_+_28794615 | 0.02 |
ENSMUST00000232862.2
ENSMUST00000080780.8 |
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr19_-_20931566 | 0.02 |
ENSMUST00000039500.4
|
Tmc1
|
transmembrane channel-like gene family 1 |
| chr16_+_17095416 | 0.02 |
ENSMUST00000232364.2
|
Tmem191c
|
transmembrane protein 191C |
| chr19_-_6036171 | 0.02 |
ENSMUST00000041827.8
|
Slc22a20
|
solute carrier family 22 (organic anion transporter), member 20 |
| chr19_-_10460238 | 0.02 |
ENSMUST00000235392.2
ENSMUST00000237522.2 ENSMUST00000038842.5 |
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr18_+_65158873 | 0.02 |
ENSMUST00000226058.2
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated gene 4-like |
| chr16_+_91282183 | 0.02 |
ENSMUST00000129878.2
|
Ifnar1
|
interferon (alpha and beta) receptor 1 |
| chrX_-_7834057 | 0.02 |
ENSMUST00000033502.14
|
Gata1
|
GATA binding protein 1 |
| chr15_+_102927366 | 0.02 |
ENSMUST00000165375.3
|
Hoxc4
|
homeobox C4 |
| chr13_+_95012107 | 0.02 |
ENSMUST00000022195.13
|
Otp
|
orthopedia homeobox |
| chr3_+_31150982 | 0.02 |
ENSMUST00000118204.2
|
Skil
|
SKI-like |
| chr14_-_55344004 | 0.02 |
ENSMUST00000036041.15
|
Ap1g2
|
adaptor protein complex AP-1, gamma 2 subunit |
| chr6_+_120813199 | 0.02 |
ENSMUST00000009256.4
|
Bcl2l13
|
BCL2-like 13 (apoptosis facilitator) |
| chr2_+_32340459 | 0.02 |
ENSMUST00000048431.3
|
Naif1
|
nuclear apoptosis inducing factor 1 |
| chr17_+_28794727 | 0.02 |
ENSMUST00000232974.2
|
Lhfpl5
|
lipoma HMGIC fusion partner-like 5 |
| chr16_-_10608766 | 0.02 |
ENSMUST00000050864.7
|
Prm3
|
protamine 3 |
| chr7_+_100122192 | 0.01 |
ENSMUST00000032958.14
ENSMUST00000107059.2 |
Ucp3
|
uncoupling protein 3 (mitochondrial, proton carrier) |
| chr18_-_64794338 | 0.01 |
ENSMUST00000025482.10
|
Atp8b1
|
ATPase, class I, type 8B, member 1 |
| chr5_-_123804745 | 0.01 |
ENSMUST00000149410.2
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr5_-_74226394 | 0.01 |
ENSMUST00000145016.3
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr15_+_99600149 | 0.01 |
ENSMUST00000229236.2
|
Smarcd1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
| chr17_+_34863779 | 0.01 |
ENSMUST00000174796.2
|
Fkbpl
|
FK506 binding protein-like |
| chr17_-_24863956 | 0.01 |
ENSMUST00000019684.13
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr10_-_61946724 | 0.01 |
ENSMUST00000142821.8
ENSMUST00000124615.8 ENSMUST00000064050.5 ENSMUST00000125704.8 ENSMUST00000142796.8 |
Fam241b
|
family with sequence similarity 241, member B |
| chr19_-_5821373 | 0.01 |
ENSMUST00000236297.2
ENSMUST00000236773.2 ENSMUST00000025890.10 |
Scyl1
|
SCY1-like 1 (S. cerevisiae) |
| chr10_-_17823736 | 0.01 |
ENSMUST00000037879.8
|
Heca
|
hdc homolog, cell cycle regulator |
| chr5_-_73025775 | 0.01 |
ENSMUST00000071944.13
ENSMUST00000073843.13 ENSMUST00000113594.8 |
Tec
|
tec protein tyrosine kinase |
| chr4_+_110254858 | 0.01 |
ENSMUST00000106589.9
ENSMUST00000106587.9 ENSMUST00000106591.8 ENSMUST00000106592.8 |
Agbl4
|
ATP/GTP binding protein-like 4 |
| chr4_-_126096551 | 0.01 |
ENSMUST00000080919.12
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr15_-_73702827 | 0.01 |
ENSMUST00000238402.2
|
Mroh5
|
maestro heat-like repeat family member 5 |
| chr11_+_72332167 | 0.01 |
ENSMUST00000045633.6
|
Mybbp1a
|
MYB binding protein (P160) 1a |
| chr7_+_83234118 | 0.01 |
ENSMUST00000039317.14
ENSMUST00000164944.2 |
Tmc3
|
transmembrane channel-like gene family 3 |
| chr16_-_31094095 | 0.01 |
ENSMUST00000060188.14
|
Ppp1r2
|
protein phosphatase 1, regulatory inhibitor subunit 2 |
| chr10_-_93871567 | 0.01 |
ENSMUST00000118077.8
ENSMUST00000118205.8 ENSMUST00000047711.13 ENSMUST00000123201.8 ENSMUST00000119818.8 |
Vezt
|
vezatin, adherens junctions transmembrane protein |
| chr11_+_101875095 | 0.01 |
ENSMUST00000176722.8
ENSMUST00000175972.2 |
Cfap97d1
|
CFAP97 domain containing 1 |
| chr7_+_139673300 | 0.01 |
ENSMUST00000026540.9
|
Prap1
|
proline-rich acidic protein 1 |
| chr4_-_49597425 | 0.01 |
ENSMUST00000150664.2
|
Pgap4
|
post-GPI attachment to proteins GalNAc transferase 4 |
| chr11_+_97340962 | 0.01 |
ENSMUST00000107601.8
|
Arhgap23
|
Rho GTPase activating protein 23 |
| chr6_+_29396574 | 0.01 |
ENSMUST00000115275.8
ENSMUST00000145310.8 ENSMUST00000096084.12 |
Ccdc136
|
coiled-coil domain containing 136 |
| chr1_+_171975625 | 0.01 |
ENSMUST00000191689.6
ENSMUST00000192704.6 ENSMUST00000074144.11 |
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr3_+_63202687 | 0.01 |
ENSMUST00000194836.6
ENSMUST00000191633.6 |
Mme
|
membrane metallo endopeptidase |
| chr12_-_98703664 | 0.01 |
ENSMUST00000170188.8
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr19_+_53781721 | 0.01 |
ENSMUST00000162910.2
|
Rbm20
|
RNA binding motif protein 20 |
| chr16_-_19060440 | 0.01 |
ENSMUST00000103751.3
|
Iglv3
|
immunoglobulin lambda variable 3 |
| chr6_+_113508636 | 0.01 |
ENSMUST00000036340.12
ENSMUST00000204827.3 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr11_+_70655035 | 0.00 |
ENSMUST00000060444.6
|
Zfp3
|
zinc finger protein 3 |
| chr7_-_104835219 | 0.00 |
ENSMUST00000214712.2
ENSMUST00000217432.2 ENSMUST00000209409.3 |
Olfr685
|
olfactory receptor 685 |
| chr10_-_62322551 | 0.00 |
ENSMUST00000105447.11
|
Vps26a
|
VPS26 retromer complex component A |
| chr5_-_21629661 | 0.00 |
ENSMUST00000115245.8
ENSMUST00000030552.7 |
Ccdc146
|
coiled-coil domain containing 146 |
| chr5_-_113216632 | 0.00 |
ENSMUST00000031295.8
|
Crybb2
|
crystallin, beta B2 |
| chr19_-_46314945 | 0.00 |
ENSMUST00000225781.2
ENSMUST00000223903.2 |
Psd
|
pleckstrin and Sec7 domain containing |
| chr11_+_75541324 | 0.00 |
ENSMUST00000102505.10
|
Myo1c
|
myosin IC |
Network of associatons between targets according to the STRING database.
First level regulatory network of Thra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.5 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.1 | 0.3 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.3 | GO:0060282 | positive regulation of oocyte development(GO:0060282) |
| 0.0 | 0.1 | GO:0033624 | negative regulation of integrin activation(GO:0033624) regulation of matrix metallopeptidase secretion(GO:1904464) matrix metallopeptidase secretion(GO:1990773) positive regulation of ovarian follicle development(GO:2000386) regulation of antral ovarian follicle growth(GO:2000387) positive regulation of antral ovarian follicle growth(GO:2000388) negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.2 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.2 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:1900228 | biofilm formation(GO:0042710) single-species biofilm formation(GO:0044010) single-species biofilm formation in or on host organism(GO:0044407) membrane disruption in other organism(GO:0051673) regulation of single-species biofilm formation(GO:1900190) negative regulation of single-species biofilm formation(GO:1900191) regulation of single-species biofilm formation in or on host organism(GO:1900228) negative regulation of single-species biofilm formation in or on host organism(GO:1900229) |
| 0.0 | 0.1 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0097402 | neuroblast migration(GO:0097402) |
| 0.0 | 0.1 | GO:0070295 | glycerol transport(GO:0015793) renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:2000583 | regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0090403 | oxidative stress-induced premature senescence(GO:0090403) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 0.2 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:1903944 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.0 | 0.3 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.3 | GO:0051574 | positive regulation of histone H3-K9 methylation(GO:0051574) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.3 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0001827 | inner cell mass cell fate commitment(GO:0001827) |
| 0.0 | 0.3 | GO:0002385 | mucosal immune response(GO:0002385) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:1905068 | positive regulation of canonical Wnt signaling pathway involved in cardiac muscle cell fate commitment(GO:1901297) positive regulation of canonical Wnt signaling pathway involved in heart development(GO:1905068) |
| 0.0 | 0.2 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.0 | GO:0090472 | dibasic protein processing(GO:0090472) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.1 | GO:0044279 | other organism cell membrane(GO:0044218) other organism membrane(GO:0044279) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0002142 | stereocilia ankle link complex(GO:0002142) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.2 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0004637 | phosphoribosylamine-glycine ligase activity(GO:0004637) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0070279 | vitamin B6 binding(GO:0070279) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.1 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |