Project
avrg: 2D miR_HR1_12
Navigation
Downloads
Results for E2f2_E2f5
Z-value: 10.53
Transcription factors associated with E2f2_E2f5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
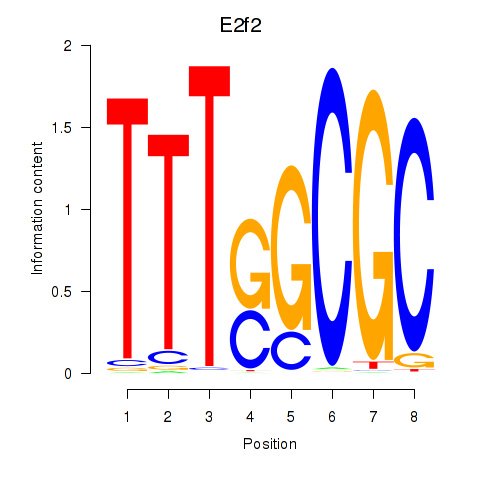
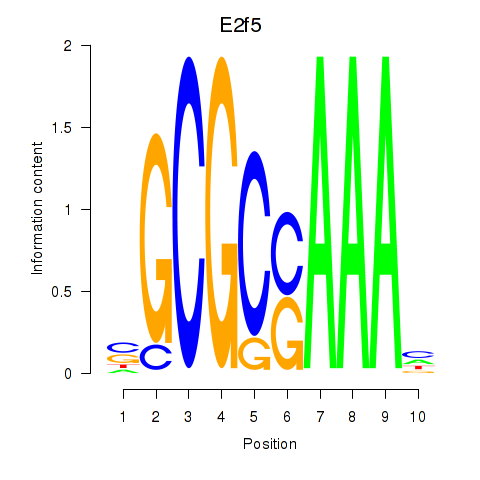
E2f2
|
ENSMUSG00000018983.9 | E2F transcription factor 2 |
|
E2f5
|
ENSMUSG00000027552.8 | E2F transcription factor 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f2 | mm10_v2_chr4_+_136172367_136172395 | 0.93 | 1.5e-05 | Click! |
| E2f5 | mm10_v2_chr3_+_14578609_14578687 | 0.84 | 7.1e-04 | Click! |
Activity profile of E2f2_E2f5 motif
Sorted Z-values of E2f2_E2f5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_55329723 | 38.42 |
ENSMUST00000021941.7
|
Mxd3
|
Max dimerization protein 3 |
| chr10_-_69352886 | 37.46 |
ENSMUST00000119827.1
ENSMUST00000020099.5 |
Cdk1
|
cyclin-dependent kinase 1 |
| chr8_+_75109528 | 28.49 |
ENSMUST00000164309.1
|
Mcm5
|
minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) |
| chr4_+_126556935 | 28.02 |
ENSMUST00000048391.8
|
Clspn
|
claspin |
| chr17_+_56304313 | 25.38 |
ENSMUST00000113035.1
ENSMUST00000113039.2 ENSMUST00000142387.1 |
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr10_+_110745433 | 22.75 |
ENSMUST00000174857.1
ENSMUST00000073781.5 ENSMUST00000173471.1 ENSMUST00000173634.1 |
E2f7
|
E2F transcription factor 7 |
| chr9_+_122951051 | 22.59 |
ENSMUST00000040717.5
|
Kif15
|
kinesin family member 15 |
| chr5_-_138171813 | 22.28 |
ENSMUST00000155902.1
ENSMUST00000148879.1 |
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr1_-_128359610 | 21.79 |
ENSMUST00000027601.4
|
Mcm6
|
minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S. cerevisiae) |
| chr2_-_113848655 | 21.12 |
ENSMUST00000102545.1
ENSMUST00000110948.1 |
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr2_-_113848601 | 20.42 |
ENSMUST00000110949.2
|
Arhgap11a
|
Rho GTPase activating protein 11A |
| chr15_-_58135047 | 20.41 |
ENSMUST00000038194.3
|
Atad2
|
ATPase family, AAA domain containing 2 |
| chr2_+_72476159 | 19.67 |
ENSMUST00000102691.4
|
Cdca7
|
cell division cycle associated 7 |
| chr12_+_24708241 | 18.91 |
ENSMUST00000020980.5
|
Rrm2
|
ribonucleotide reductase M2 |
| chr2_+_72476225 | 18.57 |
ENSMUST00000157019.1
|
Cdca7
|
cell division cycle associated 7 |
| chr4_+_136172367 | 18.37 |
ENSMUST00000061721.5
|
E2f2
|
E2F transcription factor 2 |
| chr12_-_11265768 | 18.22 |
ENSMUST00000166117.1
|
Gen1
|
Gen homolog 1, endonuclease (Drosophila) |
| chr1_-_20820213 | 18.19 |
ENSMUST00000053266.9
|
Mcm3
|
minichromosome maintenance deficient 3 (S. cerevisiae) |
| chr4_+_134468320 | 18.19 |
ENSMUST00000030636.4
ENSMUST00000127279.1 ENSMUST00000105867.1 |
Stmn1
|
stathmin 1 |
| chr15_-_55090422 | 17.96 |
ENSMUST00000110231.1
ENSMUST00000023059.6 |
Dscc1
|
defective in sister chromatid cohesion 1 homolog (S. cerevisiae) |
| chr4_+_126556994 | 17.49 |
ENSMUST00000147675.1
|
Clspn
|
claspin |
| chr2_+_150909565 | 16.04 |
ENSMUST00000028948.4
|
Gins1
|
GINS complex subunit 1 (Psf1 homolog) |
| chr1_-_86359455 | 15.63 |
ENSMUST00000027438.6
|
Ncl
|
nucleolin |
| chrX_-_93632113 | 15.58 |
ENSMUST00000006856.2
|
Pola1
|
polymerase (DNA directed), alpha 1 |
| chr13_-_24761440 | 14.37 |
ENSMUST00000176890.1
ENSMUST00000175689.1 |
Gmnn
|
geminin |
| chr4_+_115000156 | 14.12 |
ENSMUST00000030490.6
|
Stil
|
Scl/Tal1 interrupting locus |
| chr1_-_191575534 | 13.94 |
ENSMUST00000027933.5
|
Dtl
|
denticleless homolog (Drosophila) |
| chr13_-_24761861 | 13.76 |
ENSMUST00000006898.3
ENSMUST00000110382.2 |
Gmnn
|
geminin |
| chr11_+_80089385 | 13.69 |
ENSMUST00000108239.1
ENSMUST00000017694.5 |
Atad5
|
ATPase family, AAA domain containing 5 |
| chr6_-_88898664 | 13.52 |
ENSMUST00000058011.6
|
Mcm2
|
minichromosome maintenance deficient 2 mitotin (S. cerevisiae) |
| chr10_-_5805412 | 12.88 |
ENSMUST00000019907.7
|
Fbxo5
|
F-box protein 5 |
| chr2_+_119112793 | 12.87 |
ENSMUST00000140939.1
ENSMUST00000028795.3 |
Rad51
|
RAD51 homolog |
| chr7_-_48881596 | 12.53 |
ENSMUST00000119223.1
|
E2f8
|
E2F transcription factor 8 |
| chr7_-_48881032 | 12.10 |
ENSMUST00000058745.8
|
E2f8
|
E2F transcription factor 8 |
| chr12_-_69228167 | 12.06 |
ENSMUST00000021359.5
|
Pole2
|
polymerase (DNA directed), epsilon 2 (p59 subunit) |
| chr5_-_138172383 | 11.99 |
ENSMUST00000000505.9
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr3_+_98013503 | 11.75 |
ENSMUST00000079812.6
|
Notch2
|
notch 2 |
| chr12_+_116405397 | 11.16 |
ENSMUST00000084828.3
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr17_+_56303396 | 10.97 |
ENSMUST00000113038.1
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr9_-_20952838 | 10.86 |
ENSMUST00000004202.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr8_+_105348163 | 10.36 |
ENSMUST00000073149.5
|
Slc9a5
|
solute carrier family 9 (sodium/hydrogen exchanger), member 5 |
| chr5_+_110286306 | 10.27 |
ENSMUST00000007296.5
ENSMUST00000112482.1 |
Pole
|
polymerase (DNA directed), epsilon |
| chr7_-_44548733 | 10.11 |
ENSMUST00000145956.1
ENSMUST00000049343.8 |
Pold1
|
polymerase (DNA directed), delta 1, catalytic subunit |
| chr2_+_132816141 | 9.88 |
ENSMUST00000028831.8
ENSMUST00000066559.5 |
Mcm8
|
minichromosome maintenance deficient 8 (S. cerevisiae) |
| chr9_+_65890237 | 9.71 |
ENSMUST00000045802.6
|
2810417H13Rik
|
RIKEN cDNA 2810417H13 gene |
| chr4_+_115000174 | 9.68 |
ENSMUST00000129957.1
|
Stil
|
Scl/Tal1 interrupting locus |
| chr3_+_104638658 | 9.56 |
ENSMUST00000046212.1
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chr15_+_102296256 | 9.31 |
ENSMUST00000064924.4
|
Espl1
|
extra spindle poles-like 1 (S. cerevisiae) |
| chr13_+_92354783 | 9.25 |
ENSMUST00000022218.4
|
Dhfr
|
dihydrofolate reductase |
| chr12_+_24708984 | 8.64 |
ENSMUST00000154588.1
|
Rrm2
|
ribonucleotide reductase M2 |
| chr15_-_9140374 | 8.46 |
ENSMUST00000096482.3
ENSMUST00000110585.2 |
Skp2
|
S-phase kinase-associated protein 2 (p45) |
| chr14_-_31019055 | 8.05 |
ENSMUST00000037739.6
|
Gnl3
|
guanine nucleotide binding protein-like 3 (nucleolar) |
| chr5_+_110839973 | 7.95 |
ENSMUST00000066160.1
|
Chek2
|
checkpoint kinase 2 |
| chr8_-_53638945 | 7.88 |
ENSMUST00000047768.4
|
Neil3
|
nei like 3 (E. coli) |
| chr9_+_64281575 | 7.69 |
ENSMUST00000034964.6
|
Tipin
|
timeless interacting protein |
| chr18_+_10617768 | 7.50 |
ENSMUST00000002551.3
|
Snrpd1
|
small nuclear ribonucleoprotein D1 |
| chr16_-_18811615 | 6.90 |
ENSMUST00000096990.3
|
Cdc45
|
cell division cycle 45 |
| chr6_+_113531675 | 6.89 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr17_+_56303321 | 6.65 |
ENSMUST00000001258.8
|
Uhrf1
|
ubiquitin-like, containing PHD and RING finger domains, 1 |
| chr3_+_88532314 | 6.57 |
ENSMUST00000172699.1
|
Mex3a
|
mex3 homolog A (C. elegans) |
| chr6_+_51470633 | 6.55 |
ENSMUST00000114445.1
ENSMUST00000114446.1 ENSMUST00000141711.1 |
Cbx3
|
chromobox 3 |
| chr6_-_47594967 | 6.53 |
ENSMUST00000081721.6
ENSMUST00000114618.1 ENSMUST00000114616.1 |
Ezh2
|
enhancer of zeste homolog 2 (Drosophila) |
| chr19_+_8723478 | 6.48 |
ENSMUST00000180819.1
ENSMUST00000181422.1 |
Snhg1
|
small nucleolar RNA host gene (non-protein coding) 1 |
| chr16_-_15637277 | 6.33 |
ENSMUST00000023353.3
|
Mcm4
|
minichromosome maintenance deficient 4 homolog (S. cerevisiae) |
| chr9_+_103305156 | 6.11 |
ENSMUST00000035164.3
|
Topbp1
|
topoisomerase (DNA) II binding protein 1 |
| chrX_-_8074720 | 6.10 |
ENSMUST00000115636.3
ENSMUST00000115638.3 |
Suv39h1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr5_-_110286159 | 5.94 |
ENSMUST00000031472.5
|
Pxmp2
|
peroxisomal membrane protein 2 |
| chr1_+_175880775 | 5.79 |
ENSMUST00000039725.6
|
Exo1
|
exonuclease 1 |
| chr10_+_128232065 | 5.68 |
ENSMUST00000055539.4
ENSMUST00000105244.1 ENSMUST00000105243.2 ENSMUST00000125289.1 ENSMUST00000105242.1 |
Timeless
|
timeless circadian clock 1 |
| chr6_+_51470510 | 5.59 |
ENSMUST00000031862.7
|
Cbx3
|
chromobox 3 |
| chr4_+_132768325 | 5.46 |
ENSMUST00000102561.4
|
Rpa2
|
replication protein A2 |
| chr6_-_148946146 | 5.45 |
ENSMUST00000132696.1
|
Fam60a
|
family with sequence similarity 60, member A |
| chr9_-_97018823 | 5.41 |
ENSMUST00000055433.4
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr5_+_88764983 | 5.34 |
ENSMUST00000031311.9
|
Dck
|
deoxycytidine kinase |
| chr2_-_5012716 | 5.32 |
ENSMUST00000027980.7
|
Mcm10
|
minichromosome maintenance deficient 10 (S. cerevisiae) |
| chr2_+_24949747 | 5.01 |
ENSMUST00000028350.3
|
Zmynd19
|
zinc finger, MYND domain containing 19 |
| chr7_-_19359477 | 4.97 |
ENSMUST00000047036.8
|
Cd3eap
|
CD3E antigen, epsilon polypeptide associated protein |
| chr7_+_102441685 | 4.90 |
ENSMUST00000033283.9
|
Rrm1
|
ribonucleotide reductase M1 |
| chrX_+_42151002 | 4.84 |
ENSMUST00000123245.1
|
Stag2
|
stromal antigen 2 |
| chr17_+_23726336 | 4.69 |
ENSMUST00000024701.7
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr7_+_13278778 | 4.67 |
ENSMUST00000098814.4
ENSMUST00000146998.1 ENSMUST00000185145.1 |
Lig1
|
ligase I, DNA, ATP-dependent |
| chr12_+_11265867 | 4.56 |
ENSMUST00000020931.5
|
Smc6
|
structural maintenance of chromosomes 6 |
| chr13_-_64153194 | 4.54 |
ENSMUST00000059817.4
ENSMUST00000117241.1 |
Zfp367
|
zinc finger protein 367 |
| chr6_-_8259098 | 4.48 |
ENSMUST00000012627.4
|
Rpa3
|
replication protein A3 |
| chr7_-_38107490 | 4.40 |
ENSMUST00000108023.3
|
Ccne1
|
cyclin E1 |
| chr2_-_154569845 | 4.33 |
ENSMUST00000103145.4
|
E2f1
|
E2F transcription factor 1 |
| chr4_+_11191354 | 4.29 |
ENSMUST00000170901.1
|
Ccne2
|
cyclin E2 |
| chr3_+_116594959 | 4.28 |
ENSMUST00000029571.8
|
Sass6
|
spindle assembly 6 homolog (C. elegans) |
| chr7_-_4789541 | 4.07 |
ENSMUST00000168578.1
|
Tmem238
|
transmembrane protein 238 |
| chr1_+_86526688 | 4.05 |
ENSMUST00000045897.8
|
Ptma
|
prothymosin alpha |
| chr11_+_98907801 | 3.98 |
ENSMUST00000092706.6
|
Cdc6
|
cell division cycle 6 |
| chr19_-_53371766 | 3.87 |
ENSMUST00000086887.1
|
Gm10197
|
predicted gene 10197 |
| chr4_-_133967953 | 3.85 |
ENSMUST00000102553.4
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr19_-_10203880 | 3.81 |
ENSMUST00000142241.1
ENSMUST00000116542.2 ENSMUST00000025651.5 ENSMUST00000156291.1 |
Fen1
|
flap structure specific endonuclease 1 |
| chr18_+_56707725 | 3.78 |
ENSMUST00000025486.8
|
Lmnb1
|
lamin B1 |
| chr9_+_106477269 | 3.76 |
ENSMUST00000047721.8
|
Rrp9
|
RRP9, small subunit (SSU) processome component, homolog (yeast) |
| chr9_+_44334685 | 3.76 |
ENSMUST00000052686.2
|
H2afx
|
H2A histone family, member X |
| chr19_+_53600377 | 3.58 |
ENSMUST00000025930.9
|
Smc3
|
structural maintenance of chromosomes 3 |
| chr4_-_43010226 | 3.53 |
ENSMUST00000030165.4
|
Fancg
|
Fanconi anemia, complementation group G |
| chr15_+_55557399 | 3.51 |
ENSMUST00000022998.7
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chr2_-_132253227 | 3.43 |
ENSMUST00000028817.6
|
Pcna
|
proliferating cell nuclear antigen |
| chr9_+_109875541 | 3.39 |
ENSMUST00000094324.3
|
Cdc25a
|
cell division cycle 25A |
| chr6_+_51470339 | 3.35 |
ENSMUST00000094623.3
|
Cbx3
|
chromobox 3 |
| chr3_-_30509462 | 3.34 |
ENSMUST00000173899.1
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr10_-_80577285 | 3.33 |
ENSMUST00000038558.8
|
Klf16
|
Kruppel-like factor 16 |
| chr8_-_105707933 | 3.20 |
ENSMUST00000013299.9
|
Enkd1
|
enkurin domain containing 1 |
| chr19_-_5964132 | 3.20 |
ENSMUST00000025752.7
ENSMUST00000165143.1 |
Pola2
|
polymerase (DNA directed), alpha 2 |
| chr12_+_112644828 | 3.18 |
ENSMUST00000021728.4
ENSMUST00000109755.3 |
Siva1
|
SIVA1, apoptosis-inducing factor |
| chr10_+_7667503 | 3.17 |
ENSMUST00000040135.8
|
Nup43
|
nucleoporin 43 |
| chr16_-_11203259 | 3.13 |
ENSMUST00000119953.1
|
Rsl1d1
|
ribosomal L1 domain containing 1 |
| chr15_+_55557575 | 3.13 |
ENSMUST00000170046.1
|
Mtbp
|
Mdm2, transformed 3T3 cell double minute p53 binding protein |
| chr1_-_9700209 | 3.09 |
ENSMUST00000088658.4
|
Mybl1
|
myeloblastosis oncogene-like 1 |
| chr9_-_36726374 | 3.09 |
ENSMUST00000172702.2
ENSMUST00000172742.1 ENSMUST00000034625.5 |
Chek1
|
checkpoint kinase 1 |
| chr4_-_132843111 | 3.07 |
ENSMUST00000105919.1
ENSMUST00000030702.7 |
Ppp1r8
|
protein phosphatase 1, regulatory (inhibitor) subunit 8 |
| chr8_+_95633500 | 2.96 |
ENSMUST00000034094.9
|
Gins3
|
GINS complex subunit 3 (Psf3 homolog) |
| chr14_-_20388822 | 2.95 |
ENSMUST00000022345.6
|
Dnajc9
|
DnaJ (Hsp40) homolog, subfamily C, member 9 |
| chr7_-_127260677 | 2.92 |
ENSMUST00000035276.4
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr1_-_156474249 | 2.90 |
ENSMUST00000051396.6
|
Soat1
|
sterol O-acyltransferase 1 |
| chr13_-_35906324 | 2.90 |
ENSMUST00000174230.1
ENSMUST00000171686.2 |
Rpp40
|
ribonuclease P 40 subunit |
| chr17_-_71475285 | 2.87 |
ENSMUST00000127430.1
|
Smchd1
|
SMC hinge domain containing 1 |
| chr2_-_157204483 | 2.87 |
ENSMUST00000029170.7
|
Rbl1
|
retinoblastoma-like 1 (p107) |
| chr15_+_8109313 | 2.76 |
ENSMUST00000163765.1
|
Nup155
|
nucleoporin 155 |
| chr1_+_157412352 | 2.73 |
ENSMUST00000061537.5
|
2810025M15Rik
|
RIKEN cDNA 2810025M15 gene |
| chr11_-_35980473 | 2.68 |
ENSMUST00000018993.6
|
Wwc1
|
WW, C2 and coiled-coil domain containing 1 |
| chr15_+_9140527 | 2.66 |
ENSMUST00000090380.4
|
Lmbrd2
|
LMBR1 domain containing 2 |
| chr17_+_87975044 | 2.62 |
ENSMUST00000005503.3
|
Msh6
|
mutS homolog 6 (E. coli) |
| chr5_+_114130386 | 2.59 |
ENSMUST00000031587.6
|
Ung
|
uracil DNA glycosylase |
| chr7_+_79392305 | 2.58 |
ENSMUST00000117227.1
ENSMUST00000118959.1 ENSMUST00000036865.6 |
Fanci
|
Fanconi anemia, complementation group I |
| chr7_+_29309429 | 2.58 |
ENSMUST00000137848.1
|
Dpf1
|
D4, zinc and double PHD fingers family 1 |
| chr15_-_98093245 | 2.57 |
ENSMUST00000180657.1
|
Senp1
|
SUMO1/sentrin specific peptidase 1 |
| chr2_-_34799618 | 2.56 |
ENSMUST00000113086.2
ENSMUST00000118108.1 |
Rabepk
|
Rab9 effector protein with kelch motifs |
| chr8_+_70594466 | 2.56 |
ENSMUST00000019283.9
|
Isyna1
|
myo-inositol 1-phosphate synthase A1 |
| chrX_+_94234594 | 2.53 |
ENSMUST00000153900.1
|
Klhl15
|
kelch-like 15 |
| chr10_-_7956223 | 2.52 |
ENSMUST00000146444.1
|
Tab2
|
TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| chr10_+_13090788 | 2.52 |
ENSMUST00000121646.1
ENSMUST00000121325.1 ENSMUST00000121766.1 |
Plagl1
|
pleiomorphic adenoma gene-like 1 |
| chr13_-_106936907 | 2.52 |
ENSMUST00000080856.7
|
Ipo11
|
importin 11 |
| chr11_-_100712429 | 2.48 |
ENSMUST00000006973.5
ENSMUST00000103118.3 |
Kat2a
|
K(lysine) acetyltransferase 2A |
| chrX_-_157492280 | 2.45 |
ENSMUST00000112529.1
|
Sms
|
spermine synthase |
| chr1_+_131910458 | 2.41 |
ENSMUST00000062264.6
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr4_+_32615473 | 2.40 |
ENSMUST00000178925.1
ENSMUST00000029950.3 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr16_-_5255923 | 2.39 |
ENSMUST00000139584.1
ENSMUST00000064635.5 |
Fam86
|
family with sequence similarity 86 |
| chr6_+_14901344 | 2.37 |
ENSMUST00000115477.1
|
Foxp2
|
forkhead box P2 |
| chr17_-_23740301 | 2.36 |
ENSMUST00000024702.3
|
Paqr4
|
progestin and adipoQ receptor family member IV |
| chr7_+_110122299 | 2.35 |
ENSMUST00000033326.8
|
Wee1
|
WEE 1 homolog 1 (S. pombe) |
| chr9_-_21291124 | 2.32 |
ENSMUST00000086374.6
|
Cdkn2d
|
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr10_+_80356459 | 2.31 |
ENSMUST00000039836.8
ENSMUST00000105351.1 |
Plk5
|
polo-like kinase 5 |
| chr7_-_122101735 | 2.30 |
ENSMUST00000139456.1
ENSMUST00000106471.2 ENSMUST00000123296.1 ENSMUST00000033157.3 |
Ndufab1
|
NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1 |
| chr2_+_85037212 | 2.28 |
ENSMUST00000077798.6
|
Ssrp1
|
structure specific recognition protein 1 |
| chr2_-_152398046 | 2.26 |
ENSMUST00000063332.8
ENSMUST00000182625.1 |
Sox12
|
SRY-box containing gene 12 |
| chr16_-_18876655 | 2.22 |
ENSMUST00000023391.8
|
Mrpl40
|
mitochondrial ribosomal protein L40 |
| chr7_+_110018301 | 2.15 |
ENSMUST00000084731.3
|
Ipo7
|
importin 7 |
| chr12_+_111166485 | 2.13 |
ENSMUST00000139162.1
|
Traf3
|
TNF receptor-associated factor 3 |
| chr3_+_10088173 | 2.10 |
ENSMUST00000061419.7
|
Gm9833
|
predicted gene 9833 |
| chr15_-_55557748 | 2.09 |
ENSMUST00000172387.1
|
Mrpl13
|
mitochondrial ribosomal protein L13 |
| chr1_-_38129618 | 2.08 |
ENSMUST00000027251.6
|
Rev1
|
REV1 homolog (S. cerevisiae) |
| chr9_-_13827029 | 2.08 |
ENSMUST00000148086.1
ENSMUST00000034398.5 |
Cep57
|
centrosomal protein 57 |
| chr11_+_88047302 | 2.04 |
ENSMUST00000139129.2
|
Srsf1
|
serine/arginine-rich splicing factor 1 |
| chr7_+_66109474 | 2.01 |
ENSMUST00000036372.6
|
Chsy1
|
chondroitin sulfate synthase 1 |
| chr19_+_46075842 | 2.00 |
ENSMUST00000165017.1
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr11_-_72207413 | 2.00 |
ENSMUST00000108505.1
|
4933427D14Rik
|
RIKEN cDNA 4933427D14 gene |
| chr4_+_109280365 | 1.97 |
ENSMUST00000177089.1
ENSMUST00000175776.1 ENSMUST00000132165.2 |
Eps15
|
epidermal growth factor receptor pathway substrate 15 |
| chr3_+_127553462 | 1.96 |
ENSMUST00000043108.4
|
4930422G04Rik
|
RIKEN cDNA 4930422G04 gene |
| chr2_+_55437100 | 1.95 |
ENSMUST00000112633.2
ENSMUST00000112632.1 |
Kcnj3
|
potassium inwardly-rectifying channel, subfamily J, member 3 |
| chr13_-_96132568 | 1.89 |
ENSMUST00000161263.1
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr5_-_149051604 | 1.89 |
ENSMUST00000093196.4
|
Hmgb1
|
high mobility group box 1 |
| chrX_+_99821021 | 1.82 |
ENSMUST00000096363.2
|
Tmem28
|
transmembrane protein 28 |
| chr10_-_60219260 | 1.80 |
ENSMUST00000135158.2
|
Chst3
|
carbohydrate (chondroitin 6/keratan) sulfotransferase 3 |
| chr13_-_47106176 | 1.76 |
ENSMUST00000021807.6
ENSMUST00000135278.1 |
Dek
|
DEK oncogene (DNA binding) |
| chr16_-_45742888 | 1.76 |
ENSMUST00000128348.1
ENSMUST00000066983.6 |
Abhd10
|
abhydrolase domain containing 10 |
| chr2_+_25054355 | 1.75 |
ENSMUST00000100334.4
ENSMUST00000152122.1 ENSMUST00000116574.3 ENSMUST00000006646.8 |
Nsmf
|
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr12_-_80643799 | 1.75 |
ENSMUST00000166931.1
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr2_-_154569720 | 1.74 |
ENSMUST00000000894.5
|
E2f1
|
E2F transcription factor 1 |
| chrX_+_68678541 | 1.73 |
ENSMUST00000088546.5
|
Fmr1
|
fragile X mental retardation syndrome 1 |
| chr16_-_18248697 | 1.73 |
ENSMUST00000115645.3
|
Ranbp1
|
RAN binding protein 1 |
| chr17_+_88440711 | 1.69 |
ENSMUST00000112238.2
ENSMUST00000155640.1 |
Foxn2
|
forkhead box N2 |
| chr5_-_136135989 | 1.68 |
ENSMUST00000150406.1
ENSMUST00000006301.4 |
Lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr5_+_143909964 | 1.68 |
ENSMUST00000148011.1
ENSMUST00000110709.3 |
Pms2
|
postmeiotic segregation increased 2 (S. cerevisiae) |
| chr11_+_101119938 | 1.68 |
ENSMUST00000043680.8
|
Tubg1
|
tubulin, gamma 1 |
| chr4_-_133967893 | 1.67 |
ENSMUST00000100472.3
ENSMUST00000136327.1 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr7_+_46397648 | 1.66 |
ENSMUST00000160433.1
|
Kcnc1
|
potassium voltage gated channel, Shaw-related subfamily, member 1 |
| chr17_+_87672523 | 1.64 |
ENSMUST00000172855.1
|
Msh2
|
mutS homolog 2 (E. coli) |
| chr7_-_100121512 | 1.64 |
ENSMUST00000032969.7
|
Pold3
|
polymerase (DNA-directed), delta 3, accessory subunit |
| chr2_-_34913976 | 1.64 |
ENSMUST00000028232.3
|
Phf19
|
PHD finger protein 19 |
| chr10_+_107271827 | 1.62 |
ENSMUST00000020057.8
ENSMUST00000105280.3 |
Lin7a
|
lin-7 homolog A (C. elegans) |
| chr18_+_31789120 | 1.62 |
ENSMUST00000025106.3
|
Polr2d
|
polymerase (RNA) II (DNA directed) polypeptide D |
| chr17_-_94749874 | 1.60 |
ENSMUST00000171284.1
|
Mettl4
|
methyltransferase like 4 |
| chr11_-_73177002 | 1.59 |
ENSMUST00000108480.1
ENSMUST00000054952.3 |
Emc6
|
ER membrane protein complex subunit 6 |
| chr19_-_5457397 | 1.59 |
ENSMUST00000179549.1
|
Ccdc85b
|
coiled-coil domain containing 85B |
| chr8_-_78508876 | 1.50 |
ENSMUST00000049245.7
|
Rbmxl1
|
RNA binding motif protein, X linked-like-1 |
| chr7_-_126396715 | 1.50 |
ENSMUST00000075671.4
|
Nfatc2ip
|
nuclear factor of activated T cells, cytoplasmic, calcineurin dependent 2 interacting protein |
| chr11_+_79993062 | 1.50 |
ENSMUST00000017692.8
ENSMUST00000163272.1 |
Suz12
|
suppressor of zeste 12 homolog (Drosophila) |
| chr19_-_7217549 | 1.48 |
ENSMUST00000039758.4
|
Cox8a
|
cytochrome c oxidase subunit VIIIa |
| chr12_+_57230406 | 1.48 |
ENSMUST00000139049.1
ENSMUST00000145003.1 ENSMUST00000123498.1 |
Prps1l3
Mipol1
|
phosphoribosyl pyrophosphate synthetase 1-like 3 mirror-image polydactyly gene 1 homolog (human) |
| chr2_+_18677002 | 1.47 |
ENSMUST00000028071.6
|
Bmi1
|
Bmi1 polycomb ring finger oncogene |
| chr15_-_81871883 | 1.47 |
ENSMUST00000023117.8
|
Phf5a
|
PHD finger protein 5A |
| chr1_+_181150926 | 1.46 |
ENSMUST00000134115.1
ENSMUST00000111059.1 |
Cnih4
|
cornichon homolog 4 (Drosophila) |
| chr9_-_115310421 | 1.46 |
ENSMUST00000035010.8
|
Stt3b
|
STT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) |
| chr7_-_133122615 | 1.44 |
ENSMUST00000167218.1
|
Ctbp2
|
C-terminal binding protein 2 |
| chr1_+_155558112 | 1.44 |
ENSMUST00000080138.6
ENSMUST00000097529.3 ENSMUST00000035560.3 |
Acbd6
|
acyl-Coenzyme A binding domain containing 6 |
| chr3_-_88950271 | 1.42 |
ENSMUST00000174402.1
ENSMUST00000174077.1 |
Dap3
|
death associated protein 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f2_E2f5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 15.8 | 47.4 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 10.1 | 10.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 8.9 | 88.8 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 7.3 | 29.3 | GO:0006272 | leading strand elongation(GO:0006272) |
| 4.7 | 51.6 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 4.0 | 35.9 | GO:0090166 | Golgi disassembly(GO:0090166) |
| 3.9 | 11.7 | GO:1990705 | intrahepatic bile duct development(GO:0035622) cholangiocyte proliferation(GO:1990705) |
| 3.6 | 18.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 3.6 | 53.9 | GO:0090308 | regulation of methylation-dependent chromatin silencing(GO:0090308) |
| 3.5 | 24.4 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 3.4 | 20.4 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 3.2 | 12.9 | GO:0007056 | spindle assembly involved in female meiosis(GO:0007056) |
| 3.2 | 32.0 | GO:1902969 | mitotic DNA replication(GO:1902969) |
| 3.1 | 77.8 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 2.9 | 2.9 | GO:0009211 | pyrimidine deoxyribonucleoside triphosphate metabolic process(GO:0009211) |
| 2.6 | 18.2 | GO:0070495 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 2.4 | 14.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 2.3 | 13.9 | GO:0019985 | translesion synthesis(GO:0019985) |
| 2.2 | 23.8 | GO:0033504 | floor plate development(GO:0033504) |
| 2.1 | 6.4 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 2.0 | 18.2 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 2.0 | 15.6 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 1.9 | 7.7 | GO:0000076 | DNA replication checkpoint(GO:0000076) |
| 1.9 | 5.7 | GO:1904976 | response to bleomycin(GO:1904975) cellular response to bleomycin(GO:1904976) |
| 1.8 | 9.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 1.8 | 32.4 | GO:0009263 | deoxyribonucleotide biosynthetic process(GO:0009263) |
| 1.6 | 1.6 | GO:0031990 | mRNA export from nucleus in response to heat stress(GO:0031990) |
| 1.3 | 5.3 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 1.3 | 14.6 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 1.3 | 6.6 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
| 1.3 | 3.8 | GO:0031662 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 1.2 | 13.7 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 1.1 | 11.2 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 1.0 | 3.1 | GO:0099578 | regulation of translation at synapse, modulating synaptic transmission(GO:0099547) regulation of translation at postsynapse, modulating synaptic transmission(GO:0099578) positive regulation of intracellular transport of viral material(GO:1901254) |
| 1.0 | 11.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 1.0 | 9.7 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 1.0 | 2.9 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.9 | 2.8 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.9 | 14.3 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.8 | 2.5 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.8 | 2.4 | GO:0060382 | release from viral latency(GO:0019046) regulation of DNA strand elongation(GO:0060382) |
| 0.8 | 3.8 | GO:0043137 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.7 | 2.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.7 | 4.3 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.7 | 8.5 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.7 | 10.4 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.7 | 6.9 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.7 | 2.0 | GO:0060365 | orbitofrontal cortex development(GO:0021769) fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development(GO:0035607) coronal suture morphogenesis(GO:0060365) |
| 0.7 | 5.4 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.6 | 2.6 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.6 | 1.9 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) |
| 0.6 | 6.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.6 | 1.8 | GO:0019389 | glucuronoside metabolic process(GO:0019389) |
| 0.6 | 8.1 | GO:1904816 | positive regulation of protein localization to chromosome, telomeric region(GO:1904816) |
| 0.5 | 29.3 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.5 | 4.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.5 | 1.5 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.5 | 16.6 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.4 | 1.8 | GO:0099527 | postsynapse to nucleus signaling pathway(GO:0099527) |
| 0.4 | 1.7 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.4 | 1.6 | GO:0006297 | nucleotide-excision repair, DNA gap filling(GO:0006297) |
| 0.4 | 1.2 | GO:0032954 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.4 | 1.9 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.4 | 2.7 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.4 | 3.8 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.3 | 2.4 | GO:0036337 | Fas signaling pathway(GO:0036337) |
| 0.3 | 2.3 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.3 | 1.6 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.3 | 1.0 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) endothelial cell-cell adhesion(GO:0071603) |
| 0.3 | 2.4 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.3 | 1.5 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.3 | 1.5 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.3 | 1.2 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.3 | 2.9 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.3 | 2.3 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.3 | 2.8 | GO:0075509 | receptor-mediated endocytosis of virus by host cell(GO:0019065) endocytosis involved in viral entry into host cell(GO:0075509) |
| 0.3 | 4.1 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.3 | 3.0 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.3 | 3.2 | GO:1901030 | positive regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901030) |
| 0.3 | 0.8 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.3 | 1.8 | GO:0044838 | cell quiescence(GO:0044838) regulation of double-strand break repair via nonhomologous end joining(GO:2001032) |
| 0.2 | 8.6 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.2 | 1.0 | GO:0032466 | negative regulation of cytokinesis(GO:0032466) |
| 0.2 | 0.2 | GO:0072554 | blood vessel lumenization(GO:0072554) |
| 0.2 | 0.9 | GO:0034086 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.2 | 2.5 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.2 | 2.5 | GO:0001682 | tRNA 5'-leader removal(GO:0001682) |
| 0.2 | 0.7 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.2 | 5.0 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.2 | 2.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.2 | 2.6 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.2 | 6.3 | GO:0007099 | centriole replication(GO:0007099) |
| 0.2 | 0.6 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.2 | 1.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.2 | 0.7 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.2 | 1.1 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.2 | 5.5 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.2 | 1.6 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.2 | 4.1 | GO:0043486 | histone exchange(GO:0043486) |
| 0.2 | 0.5 | GO:0048377 | neural plate mediolateral regionalization(GO:0021998) mesoderm structural organization(GO:0048338) paraxial mesoderm structural organization(GO:0048352) lateral mesodermal cell fate commitment(GO:0048372) lateral mesodermal cell fate specification(GO:0048377) regulation of lateral mesodermal cell fate specification(GO:0048378) positive regulation of cardiac ventricle development(GO:1904414) |
| 0.2 | 1.4 | GO:0048386 | positive regulation of retinoic acid receptor signaling pathway(GO:0048386) |
| 0.2 | 2.6 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 1.6 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.1 | 1.4 | GO:1990035 | calcium ion import across plasma membrane(GO:0098703) calcium ion import into cell(GO:1990035) |
| 0.1 | 1.1 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.1 | 0.3 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.1 | 3.4 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.1 | 1.7 | GO:1903818 | positive regulation of voltage-gated potassium channel activity(GO:1903818) |
| 0.1 | 0.9 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.1 | 0.8 | GO:0072697 | protein localization to cell cortex(GO:0072697) |
| 0.1 | 1.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.1 | 3.1 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.1 | 0.6 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 2.6 | GO:0048026 | positive regulation of mRNA splicing, via spliceosome(GO:0048026) |
| 0.1 | 1.2 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.1 | 0.8 | GO:0035562 | negative regulation of chromatin binding(GO:0035562) |
| 0.1 | 0.2 | GO:0023035 | CD40 signaling pathway(GO:0023035) |
| 0.1 | 2.3 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.1 | 1.5 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.1 | 0.7 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.1 | 0.5 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.1 | 0.7 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.1 | 0.5 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.1 | 0.9 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) phosphate ion transmembrane transport(GO:0035435) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.1 | 2.1 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.1 | 0.5 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 1.1 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.2 | GO:0070535 | histone H2A K63-linked ubiquitination(GO:0070535) |
| 0.1 | 2.8 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.1 | 15.8 | GO:0007018 | microtubule-based movement(GO:0007018) |
| 0.1 | 0.2 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.1 | 0.5 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 1.3 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.4 | GO:1900364 | negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.0 | 0.4 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.0 | 0.3 | GO:0043415 | positive regulation of skeletal muscle tissue regeneration(GO:0043415) |
| 0.0 | 0.8 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 1.4 | GO:0061003 | positive regulation of dendritic spine morphogenesis(GO:0061003) |
| 0.0 | 0.6 | GO:0045899 | positive regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045899) |
| 0.0 | 0.8 | GO:0021511 | spinal cord patterning(GO:0021511) |
| 0.0 | 2.3 | GO:0021510 | spinal cord development(GO:0021510) |
| 0.0 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.5 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 3.8 | GO:0006364 | rRNA processing(GO:0006364) |
| 0.0 | 0.6 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.8 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.5 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0015855 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.5 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 2.3 | GO:0008637 | apoptotic mitochondrial changes(GO:0008637) |
| 0.0 | 1.4 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.6 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.2 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.3 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 1.4 | GO:0035308 | negative regulation of protein dephosphorylation(GO:0035308) |
| 0.0 | 0.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.0 | 0.5 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.4 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.6 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.5 | GO:0006261 | DNA-dependent DNA replication(GO:0006261) |
| 0.0 | 0.3 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 8.9 | 133.0 | GO:0042555 | MCM complex(GO:0042555) |
| 6.5 | 32.4 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 4.5 | 22.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 4.2 | 12.5 | GO:0000811 | GINS complex(GO:0000811) |
| 4.0 | 19.9 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 3.8 | 18.8 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 3.4 | 10.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 3.1 | 15.6 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 2.0 | 18.0 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 1.9 | 15.5 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 1.6 | 4.7 | GO:0035061 | interchromatin granule(GO:0035061) |
| 1.5 | 7.5 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 1.4 | 4.3 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 1.4 | 74.6 | GO:0005657 | replication fork(GO:0005657) |
| 1.3 | 8.9 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 1.2 | 6.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 1.2 | 6.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 1.1 | 11.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.8 | 6.2 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.8 | 3.1 | GO:1902737 | viral replication complex(GO:0019034) dendritic filopodium(GO:1902737) |
| 0.8 | 42.1 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.7 | 2.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.7 | 4.3 | GO:0098536 | deuterosome(GO:0098536) |
| 0.7 | 4.9 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.7 | 6.1 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.7 | 7.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.6 | 3.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.6 | 2.9 | GO:0001740 | Barr body(GO:0001740) |
| 0.6 | 12.9 | GO:0000800 | lateral element(GO:0000800) |
| 0.6 | 2.8 | GO:0034709 | methylosome(GO:0034709) |
| 0.6 | 2.8 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.5 | 3.8 | GO:0005638 | lamin filament(GO:0005638) |
| 0.4 | 9.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.4 | 2.9 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.4 | 1.5 | GO:1903349 | omegasome membrane(GO:1903349) |
| 0.3 | 60.0 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.3 | 23.3 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.3 | 5.5 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.3 | 5.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 3.2 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.3 | 1.5 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.2 | 1.7 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.2 | 1.7 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.2 | 1.6 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.2 | 2.8 | GO:0030122 | AP-2 adaptor complex(GO:0030122) |
| 0.2 | 2.6 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.2 | 5.9 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.2 | 3.0 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.2 | 1.4 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.2 | 0.7 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 1.7 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.1 | 9.3 | GO:0072686 | mitotic spindle(GO:0072686) |
| 0.1 | 1.7 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.1 | 2.0 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.1 | 4.2 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 1.1 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 3.6 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.1 | 0.7 | GO:0070187 | telosome(GO:0070187) |
| 0.1 | 6.2 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.1 | 1.2 | GO:0070938 | contractile ring(GO:0070938) |
| 0.1 | 10.5 | GO:0005814 | centriole(GO:0005814) |
| 0.1 | 2.6 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.1 | 9.4 | GO:0016605 | PML body(GO:0016605) |
| 0.1 | 12.5 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.1 | 1.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 2.0 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.1 | 6.8 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 13.0 | GO:0005819 | spindle(GO:0005819) |
| 0.1 | 25.6 | GO:0005667 | transcription factor complex(GO:0005667) |
| 0.1 | 1.4 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.1 | 3.2 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.1 | 1.8 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.1 | 1.5 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.1 | 5.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.1 | 0.5 | GO:0000801 | central element(GO:0000801) |
| 0.1 | 2.3 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.1 | 3.2 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.1 | 0.8 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 3.8 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.1 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.1 | 2.0 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 0.6 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.8 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 16.8 | GO:0005813 | centrosome(GO:0005813) |
| 0.0 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.4 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 2.1 | GO:0005747 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.0 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 6.8 | GO:0005874 | microtubule(GO:0005874) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 42.5 | GO:0005654 | nucleoplasm(GO:0005654) |
| 0.0 | 1.9 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.5 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.3 | GO:0030665 | clathrin-coated vesicle membrane(GO:0030665) |
| 0.0 | 0.0 | GO:0031251 | PAN complex(GO:0031251) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 7.6 | 45.5 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 7.2 | 43.0 | GO:0031493 | nucleosomal histone binding(GO:0031493) |
| 6.5 | 32.4 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 3.9 | 7.9 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 3.9 | 54.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 3.3 | 19.8 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 3.1 | 12.5 | GO:0043138 | 3'-5' DNA helicase activity(GO:0043138) |
| 2.4 | 9.6 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 2.4 | 19.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 2.3 | 40.5 | GO:0035173 | histone kinase activity(GO:0035173) |
| 2.0 | 51.2 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 1.9 | 30.8 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 1.8 | 9.0 | GO:0032135 | DNA insertion or deletion binding(GO:0032135) |
| 1.8 | 62.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 1.6 | 9.6 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 1.5 | 5.9 | GO:0036033 | mediator complex binding(GO:0036033) |
| 1.4 | 12.9 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 1.4 | 12.5 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 1.3 | 5.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 1.2 | 6.1 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 1.1 | 8.0 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 1.0 | 8.7 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.9 | 14.4 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.9 | 11.7 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.9 | 2.6 | GO:0070139 | ubiquitin-like protein-specific endopeptidase activity(GO:0070137) SUMO-specific endopeptidase activity(GO:0070139) |
| 0.8 | 10.4 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.7 | 2.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.7 | 0.7 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.6 | 2.6 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.6 | 1.9 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.6 | 2.5 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.6 | 1.8 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.5 | 11.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.5 | 4.7 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.5 | 9.5 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.5 | 6.1 | GO:0098505 | single-stranded telomeric DNA binding(GO:0043047) G-rich strand telomeric DNA binding(GO:0098505) |
| 0.5 | 3.8 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.4 | 4.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 3.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.4 | 2.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.4 | 1.8 | GO:0034481 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) chondroitin sulfotransferase activity(GO:0034481) |
| 0.3 | 2.4 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.3 | 11.4 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.3 | 76.6 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.3 | 1.9 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.3 | 2.9 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.3 | 4.4 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.3 | 11.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.3 | 1.6 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.3 | 2.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.5 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.2 | 0.9 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.2 | 23.3 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.2 | 2.5 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.2 | 10.7 | GO:0035064 | methylated histone binding(GO:0035064) |
| 0.2 | 1.6 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.2 | 27.9 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 1.2 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.2 | 0.9 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.2 | 1.5 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.2 | 23.4 | GO:0042393 | histone binding(GO:0042393) |
| 0.2 | 0.5 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.2 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.2 | 0.6 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.2 | 3.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.1 | 1.5 | GO:0031730 | CCR5 chemokine receptor binding(GO:0031730) |
| 0.1 | 2.8 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.3 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.1 | 2.8 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.1 | 1.7 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.1 | 2.9 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.5 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 9.3 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.1 | 4.3 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 3.4 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 31.9 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 1.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 0.8 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 35.8 | GO:0005096 | GTPase activator activity(GO:0005096) |
| 0.1 | 2.8 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 15.0 | GO:0001047 | core promoter binding(GO:0001047) |
| 0.1 | 2.1 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.1 | 4.2 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.1 | 0.7 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.1 | 0.6 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.0 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 1.3 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.5 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.5 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.5 | GO:0098821 | BMP receptor activity(GO:0098821) |
| 0.0 | 0.5 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.8 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.6 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 1.3 | GO:0008536 | Ran GTPase binding(GO:0008536) |
| 0.0 | 0.8 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 2.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.7 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 19.5 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 0.1 | GO:0030519 | snoRNP binding(GO:0030519) |
| 0.0 | 5.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 1.7 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 2.4 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 1.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 2.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 2.3 | GO:0072341 | modified amino acid binding(GO:0072341) |
| 0.0 | 0.9 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.4 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.0 | 2.0 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 1.5 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.2 | GO:0016594 | glycine binding(GO:0016594) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 5.1 | 46.3 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 3.0 | 142.0 | PID ATR PATHWAY | ATR signaling pathway |
| 2.4 | 33.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 1.3 | 105.6 | PID E2F PATHWAY | E2F transcription factor network |
| 0.8 | 13.6 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.7 | 23.8 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.6 | 16.4 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.5 | 53.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.4 | 20.8 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.4 | 8.2 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.3 | 8.7 | PID ATM PATHWAY | ATM pathway |
| 0.3 | 17.0 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.2 | 2.9 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.2 | 14.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.2 | 2.4 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 3.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 3.1 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.1 | 4.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.1 | 3.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.1 | 2.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.1 | 3.8 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.1 | 3.5 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 1.2 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 0.5 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 2.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.9 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.3 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.0 | 0.3 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 9.5 | 151.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 6.9 | 124.0 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 4.6 | 46.5 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 3.1 | 44.0 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 1.9 | 11.7 | REACTOME DNA STRAND ELONGATION | Genes involved in DNA strand elongation |
| 1.5 | 13.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.4 | 16.6 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 1.1 | 6.9 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 1.1 | 12.0 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.9 | 5.3 | REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | Genes involved in Activation of the pre-replicative complex |
| 0.9 | 9.6 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.8 | 22.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.4 | 4.7 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.4 | 6.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.4 | 4.8 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.3 | 4.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.3 | 16.7 | REACTOME MRNA SPLICING MINOR PATHWAY | Genes involved in mRNA Splicing - Minor Pathway |
| 0.3 | 10.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.3 | 5.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.2 | 7.1 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |
| 0.2 | 1.7 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.2 | 4.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.2 | 3.0 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.2 | 37.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.2 | 3.8 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.2 | 2.5 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.2 | 0.9 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 2.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.1 | 2.0 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 0.7 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.1 | 1.9 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.1 | 1.0 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 3.4 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.7 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 2.8 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.1 | 4.3 | REACTOME RNA POL I TRANSCRIPTION | Genes involved in RNA Polymerase I Transcription |
| 0.1 | 4.0 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 1.9 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.1 | 1.1 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 1.9 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 2.7 | REACTOME FACTORS INVOLVED IN MEGAKARYOCYTE DEVELOPMENT AND PLATELET PRODUCTION | Genes involved in Factors involved in megakaryocyte development and platelet production |
| 0.0 | 0.5 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.4 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.1 | REACTOME CYCLIN E ASSOCIATED EVENTS DURING G1 S TRANSITION | Genes involved in Cyclin E associated events during G1/S transition |
| 0.0 | 1.1 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 5.0 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 1.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 0.4 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |