Project
avrg: 2D miR_HR1_12
Navigation
Downloads
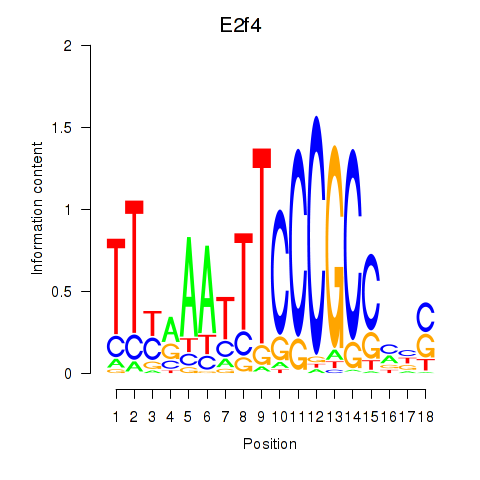
Results for E2f4
Z-value: 5.65
Transcription factors associated with E2f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
E2f4
|
ENSMUSG00000014859.8 | E2F transcription factor 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| E2f4 | mm10_v2_chr8_+_105297663_105297742 | 0.92 | 2.9e-05 | Click! |
Activity profile of E2f4 motif
Sorted Z-values of E2f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_134510999 | 14.68 |
ENSMUST00000105866.2
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr18_-_34751502 | 13.12 |
ENSMUST00000060710.7
|
Cdc25c
|
cell division cycle 25C |
| chr1_-_169531343 | 13.08 |
ENSMUST00000028000.7
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr19_+_6084983 | 12.74 |
ENSMUST00000025704.2
|
Cdca5
|
cell division cycle associated 5 |
| chr1_-_169531447 | 12.28 |
ENSMUST00000111368.1
|
Nuf2
|
NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) |
| chr1_-_189688074 | 12.02 |
ENSMUST00000171929.1
ENSMUST00000165962.1 |
Cenpf
|
centromere protein F |
| chr14_-_47418407 | 11.86 |
ENSMUST00000043296.3
|
Dlgap5
|
discs, large (Drosophila) homolog-associated protein 5 |
| chr6_+_124830217 | 11.81 |
ENSMUST00000131847.1
ENSMUST00000151674.1 |
Cdca3
|
cell division cycle associated 3 |
| chr8_-_53638945 | 10.83 |
ENSMUST00000047768.4
|
Neil3
|
nei like 3 (E. coli) |
| chrX_+_164980592 | 10.42 |
ENSMUST00000101082.4
ENSMUST00000167446.1 ENSMUST00000057150.6 |
Fancb
|
Fanconi anemia, complementation group B |
| chr4_-_132345715 | 10.39 |
ENSMUST00000084250.4
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr4_-_132345686 | 10.34 |
ENSMUST00000030726.6
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr13_-_73937761 | 9.83 |
ENSMUST00000022053.8
|
Trip13
|
thyroid hormone receptor interactor 13 |
| chr11_-_101551837 | 9.49 |
ENSMUST00000017290.4
|
Brca1
|
breast cancer 1 |
| chr10_-_88146867 | 9.29 |
ENSMUST00000164121.1
ENSMUST00000164803.1 ENSMUST00000168163.1 ENSMUST00000048518.9 |
Parpbp
|
PARP1 binding protein |
| chr17_-_25727364 | 8.89 |
ENSMUST00000170070.1
ENSMUST00000048054.7 |
Chtf18
|
CTF18, chromosome transmission fidelity factor 18 |
| chr17_-_24251382 | 8.48 |
ENSMUST00000115390.3
|
Ccnf
|
cyclin F |
| chr4_-_116123618 | 7.96 |
ENSMUST00000102704.3
ENSMUST00000102705.3 |
Rad54l
|
RAD54 like (S. cerevisiae) |
| chr7_-_92874196 | 7.49 |
ENSMUST00000032877.9
|
4632434I11Rik
|
RIKEN cDNA 4632434I11 gene |
| chr14_-_87141206 | 7.02 |
ENSMUST00000022599.7
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr14_-_87141114 | 6.94 |
ENSMUST00000168889.1
|
Diap3
|
diaphanous homolog 3 (Drosophila) |
| chr18_+_34751803 | 6.74 |
ENSMUST00000181453.1
ENSMUST00000181641.1 |
2010110K18Rik
|
RIKEN cDNA 2010110K18 gene |
| chr12_+_117843873 | 6.68 |
ENSMUST00000176735.1
ENSMUST00000177339.1 |
Cdca7l
|
cell division cycle associated 7 like |
| chr16_-_90727329 | 6.45 |
ENSMUST00000099554.4
|
Mis18a
|
MIS18 kinetochore protein homolog A (S. pombe) |
| chr9_+_107950952 | 5.95 |
ENSMUST00000049348.3
|
Traip
|
TRAF-interacting protein |
| chr9_+_106477269 | 5.84 |
ENSMUST00000047721.8
|
Rrp9
|
RRP9, small subunit (SSU) processome component, homolog (yeast) |
| chr1_-_57377476 | 5.57 |
ENSMUST00000181949.1
|
4930558J18Rik
|
RIKEN cDNA 4930558J18 gene |
| chr1_-_33669745 | 5.47 |
ENSMUST00000027312.9
|
Prim2
|
DNA primase, p58 subunit |
| chr2_+_163054682 | 5.05 |
ENSMUST00000018005.3
|
Mybl2
|
myeloblastosis oncogene-like 2 |
| chr4_+_11558914 | 5.05 |
ENSMUST00000178703.1
ENSMUST00000095145.5 ENSMUST00000108306.2 ENSMUST00000070755.6 |
Rad54b
|
RAD54 homolog B (S. cerevisiae) |
| chr7_-_38107490 | 4.68 |
ENSMUST00000108023.3
|
Ccne1
|
cyclin E1 |
| chr6_+_113531675 | 4.66 |
ENSMUST00000036340.5
ENSMUST00000101051.2 |
Fancd2
|
Fanconi anemia, complementation group D2 |
| chr11_+_16951371 | 4.59 |
ENSMUST00000109635.1
ENSMUST00000061327.1 |
Fbxo48
|
F-box protein 48 |
| chr11_+_98907801 | 4.51 |
ENSMUST00000092706.6
|
Cdc6
|
cell division cycle 6 |
| chr7_-_127260677 | 4.35 |
ENSMUST00000035276.4
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr4_-_133967235 | 4.20 |
ENSMUST00000123234.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr4_-_133967296 | 3.86 |
ENSMUST00000105893.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr10_+_88147061 | 3.79 |
ENSMUST00000169309.1
|
Nup37
|
nucleoporin 37 |
| chr13_+_23535411 | 3.72 |
ENSMUST00000080859.5
|
Hist1h3g
|
histone cluster 1, H3g |
| chr8_+_18595526 | 3.69 |
ENSMUST00000146819.1
|
Mcph1
|
microcephaly, primary autosomal recessive 1 |
| chr10_+_88146992 | 3.67 |
ENSMUST00000052355.7
|
Nup37
|
nucleoporin 37 |
| chr2_+_30286406 | 3.51 |
ENSMUST00000138666.1
ENSMUST00000113634.2 |
Nup188
|
nucleoporin 188 |
| chr18_+_56707725 | 3.50 |
ENSMUST00000025486.8
|
Lmnb1
|
lamin B1 |
| chr15_-_76639840 | 3.49 |
ENSMUST00000166974.1
ENSMUST00000168185.1 |
Tonsl
|
tonsoku-like, DNA repair protein |
| chr4_-_59783800 | 3.33 |
ENSMUST00000107526.1
ENSMUST00000095063.4 |
Inip
|
INTS3 and NABP interacting protein |
| chr1_+_157412352 | 3.11 |
ENSMUST00000061537.5
|
2810025M15Rik
|
RIKEN cDNA 2810025M15 gene |
| chr6_-_56704673 | 3.04 |
ENSMUST00000170382.2
|
Lsm5
|
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr8_+_124023394 | 3.00 |
ENSMUST00000034457.8
|
Urb2
|
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
| chr17_+_87975044 | 2.96 |
ENSMUST00000005503.3
|
Msh6
|
mutS homolog 6 (E. coli) |
| chr5_-_138171813 | 2.94 |
ENSMUST00000155902.1
ENSMUST00000148879.1 |
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr5_-_138172383 | 2.93 |
ENSMUST00000000505.9
|
Mcm7
|
minichromosome maintenance deficient 7 (S. cerevisiae) |
| chr4_-_133967893 | 2.90 |
ENSMUST00000100472.3
ENSMUST00000136327.1 |
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr10_-_117792663 | 2.78 |
ENSMUST00000167943.1
ENSMUST00000064848.5 |
Nup107
|
nucleoporin 107 |
| chr5_+_88764983 | 2.71 |
ENSMUST00000031311.9
|
Dck
|
deoxycytidine kinase |
| chrX_-_8074720 | 2.69 |
ENSMUST00000115636.3
ENSMUST00000115638.3 |
Suv39h1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr8_+_18595131 | 2.68 |
ENSMUST00000039412.8
|
Mcph1
|
microcephaly, primary autosomal recessive 1 |
| chr1_+_179803376 | 2.52 |
ENSMUST00000097454.2
|
Gm10518
|
predicted gene 10518 |
| chr3_+_88553716 | 2.42 |
ENSMUST00000008748.6
|
Ubqln4
|
ubiquilin 4 |
| chr19_+_46075842 | 2.41 |
ENSMUST00000165017.1
|
Nolc1
|
nucleolar and coiled-body phosphoprotein 1 |
| chr7_+_97371604 | 2.34 |
ENSMUST00000098300.4
|
Alg8
|
asparagine-linked glycosylation 8 (alpha-1,3-glucosyltransferase) |
| chr17_+_88440711 | 2.31 |
ENSMUST00000112238.2
ENSMUST00000155640.1 |
Foxn2
|
forkhead box N2 |
| chr5_+_130257029 | 2.20 |
ENSMUST00000100662.3
ENSMUST00000040213.6 |
Tyw1
|
tRNA-yW synthesizing protein 1 homolog (S. cerevisiae) |
| chr2_+_181319714 | 2.19 |
ENSMUST00000098971.4
ENSMUST00000054622.8 ENSMUST00000108814.1 ENSMUST00000048608.9 ENSMUST00000108815.1 |
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr14_-_54517353 | 2.19 |
ENSMUST00000023873.5
|
Prmt5
|
protein arginine N-methyltransferase 5 |
| chr7_-_4789541 | 2.16 |
ENSMUST00000168578.1
|
Tmem238
|
transmembrane protein 238 |
| chr5_-_25705791 | 2.13 |
ENSMUST00000030773.7
|
Xrcc2
|
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr19_+_8735808 | 2.12 |
ENSMUST00000049424.9
|
Wdr74
|
WD repeat domain 74 |
| chr1_-_179803625 | 2.12 |
ENSMUST00000027768.7
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr4_-_3835595 | 2.03 |
ENSMUST00000138502.1
|
Rps20
|
ribosomal protein S20 |
| chr9_+_72438519 | 2.01 |
ENSMUST00000184604.1
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr13_-_23745511 | 1.97 |
ENSMUST00000091752.2
|
Hist1h3c
|
histone cluster 1, H3c |
| chr12_+_99884498 | 1.92 |
ENSMUST00000153627.1
|
Tdp1
|
tyrosyl-DNA phosphodiesterase 1 |
| chr4_-_133967953 | 1.90 |
ENSMUST00000102553.4
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr10_+_127677064 | 1.90 |
ENSMUST00000118612.1
ENSMUST00000048099.4 |
Tmem194
|
transmembrane protein 194 |
| chr2_+_181319806 | 1.86 |
ENSMUST00000153112.1
|
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr2_-_3512746 | 1.86 |
ENSMUST00000056700.7
ENSMUST00000027961.5 |
Hspa14
Hspa14
|
heat shock protein 14 heat shock protein 14 |
| chr19_-_5366626 | 1.84 |
ENSMUST00000025762.8
|
Banf1
|
barrier to autointegration factor 1 |
| chrX_+_36112110 | 1.80 |
ENSMUST00000033418.7
|
Il13ra1
|
interleukin 13 receptor, alpha 1 |
| chr15_+_57912199 | 1.79 |
ENSMUST00000022992.6
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr5_-_33652339 | 1.71 |
ENSMUST00000075670.6
|
Slbp
|
stem-loop binding protein |
| chr7_-_16286010 | 1.69 |
ENSMUST00000145519.2
|
Ccdc9
|
coiled-coil domain containing 9 |
| chr18_-_60848911 | 1.57 |
ENSMUST00000177172.1
ENSMUST00000175934.1 ENSMUST00000176630.1 |
Tcof1
|
Treacher Collins Franceschetti syndrome 1, homolog |
| chr17_-_66101466 | 1.55 |
ENSMUST00000024909.8
ENSMUST00000147484.1 ENSMUST00000143987.1 |
Ndufv2
|
NADH dehydrogenase (ubiquinone) flavoprotein 2 |
| chr2_+_126152141 | 1.54 |
ENSMUST00000170908.1
|
Dtwd1
|
DTW domain containing 1 |
| chr9_-_123678782 | 1.43 |
ENSMUST00000170591.1
ENSMUST00000171647.1 |
Slc6a20a
|
solute carrier family 6 (neurotransmitter transporter), member 20A |
| chr6_-_148831395 | 1.42 |
ENSMUST00000145960.1
|
Ipo8
|
importin 8 |
| chr17_+_50698525 | 1.39 |
ENSMUST00000061681.7
|
Gm7334
|
predicted gene 7334 |
| chr17_+_34982099 | 1.39 |
ENSMUST00000007266.7
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr2_+_140395446 | 1.36 |
ENSMUST00000110061.1
|
Macrod2
|
MACRO domain containing 2 |
| chr14_-_33447142 | 1.34 |
ENSMUST00000111944.3
ENSMUST00000022504.5 ENSMUST00000111945.2 |
Mapk8
|
mitogen-activated protein kinase 8 |
| chrX_+_105079735 | 1.34 |
ENSMUST00000033577.4
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr13_-_69533839 | 1.32 |
ENSMUST00000044081.7
|
Papd7
|
PAP associated domain containing 7 |
| chr17_+_34982154 | 1.29 |
ENSMUST00000173004.1
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr9_+_72438534 | 1.26 |
ENSMUST00000034746.8
|
Mns1
|
meiosis-specific nuclear structural protein 1 |
| chr4_+_108834601 | 1.24 |
ENSMUST00000030296.8
|
Txndc12
|
thioredoxin domain containing 12 (endoplasmic reticulum) |
| chr12_+_106010263 | 1.18 |
ENSMUST00000021539.8
ENSMUST00000085026.4 ENSMUST00000072040.5 |
Vrk1
|
vaccinia related kinase 1 |
| chr1_+_57377593 | 1.17 |
ENSMUST00000042734.2
|
1700066M21Rik
|
RIKEN cDNA 1700066M21 gene |
| chr5_+_30232581 | 1.17 |
ENSMUST00000145167.1
|
Ept1
|
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr17_-_87025353 | 1.14 |
ENSMUST00000024957.6
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chrY_+_90784738 | 1.09 |
ENSMUST00000179483.1
|
Erdr1
|
erythroid differentiation regulator 1 |
| chr2_+_174076296 | 1.07 |
ENSMUST00000155000.1
ENSMUST00000134876.1 ENSMUST00000147038.1 |
Stx16
|
syntaxin 16 |
| chr14_+_55578123 | 1.06 |
ENSMUST00000174484.1
|
Psme1
|
proteasome (prosome, macropain) activator subunit 1 (PA28 alpha) |
| chr11_+_87405049 | 1.03 |
ENSMUST00000060835.5
|
Tex14
|
testis expressed gene 14 |
| chr7_+_24884651 | 1.03 |
ENSMUST00000153451.2
ENSMUST00000108429.1 |
Rps19
|
ribosomal protein S19 |
| chr11_-_106999369 | 1.03 |
ENSMUST00000106768.1
ENSMUST00000144834.1 |
Kpna2
|
karyopherin (importin) alpha 2 |
| chrX_+_105079761 | 1.01 |
ENSMUST00000119477.1
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr1_+_132007606 | 1.01 |
ENSMUST00000086556.5
|
Elk4
|
ELK4, member of ETS oncogene family |
| chr4_-_132463873 | 1.00 |
ENSMUST00000102567.3
|
Med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr17_+_34981847 | 0.93 |
ENSMUST00000114011.4
|
Lsm2
|
LSM2 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chrX_+_38600626 | 0.92 |
ENSMUST00000000365.2
|
Mcts1
|
malignant T cell amplified sequence 1 |
| chr7_+_24884611 | 0.91 |
ENSMUST00000108428.1
|
Rps19
|
ribosomal protein S19 |
| chr14_+_8002890 | 0.91 |
ENSMUST00000166497.1
|
Abhd6
|
abhydrolase domain containing 6 |
| chr10_-_81427114 | 0.89 |
ENSMUST00000078185.7
ENSMUST00000020461.8 ENSMUST00000105321.3 |
Nfic
|
nuclear factor I/C |
| chr9_+_44334685 | 0.87 |
ENSMUST00000052686.2
|
H2afx
|
H2A histone family, member X |
| chr15_+_102406143 | 0.86 |
ENSMUST00000170884.1
ENSMUST00000165924.1 ENSMUST00000163709.1 ENSMUST00000001326.6 |
Sp1
|
trans-acting transcription factor 1 |
| chr18_-_39490649 | 0.85 |
ENSMUST00000115567.1
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr9_-_103365769 | 0.83 |
ENSMUST00000035484.4
ENSMUST00000072249.6 |
Cdv3
|
carnitine deficiency-associated gene expressed in ventricle 3 |
| chr6_+_70726430 | 0.83 |
ENSMUST00000103410.1
|
Igkc
|
immunoglobulin kappa constant |
| chr14_+_8002949 | 0.81 |
ENSMUST00000026313.3
|
Abhd6
|
abhydrolase domain containing 6 |
| chr12_-_118198917 | 0.79 |
ENSMUST00000084806.6
|
Dnah11
|
dynein, axonemal, heavy chain 11 |
| chrX_-_12762069 | 0.79 |
ENSMUST00000096495.4
ENSMUST00000076016.5 |
Med14
|
mediator complex subunit 14 |
| chr11_-_106999482 | 0.78 |
ENSMUST00000018506.6
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr11_+_101552188 | 0.77 |
ENSMUST00000147239.1
|
Nbr1
|
neighbor of Brca1 gene 1 |
| chr5_-_33652296 | 0.77 |
ENSMUST00000151081.1
ENSMUST00000101354.3 |
Slbp
|
stem-loop binding protein |
| chr3_-_107969162 | 0.74 |
ENSMUST00000004136.8
ENSMUST00000106678.1 |
Gstm3
|
glutathione S-transferase, mu 3 |
| chr8_-_123158268 | 0.71 |
ENSMUST00000000755.7
|
Sult5a1
|
sulfotransferase family 5A, member 1 |
| chr19_+_4214238 | 0.70 |
ENSMUST00000046506.6
|
Clcf1
|
cardiotrophin-like cytokine factor 1 |
| chr2_+_101678403 | 0.68 |
ENSMUST00000004949.7
|
Traf6
|
TNF receptor-associated factor 6 |
| chr9_+_99456243 | 0.67 |
ENSMUST00000163199.2
|
1600029I14Rik
|
RIKEN cDNA 1600029I14 gene |
| chr9_+_44134562 | 0.67 |
ENSMUST00000034650.8
ENSMUST00000098852.2 |
Mcam
|
melanoma cell adhesion molecule |
| chr7_-_16285454 | 0.66 |
ENSMUST00000174270.1
|
Ccdc9
|
coiled-coil domain containing 9 |
| chr11_-_88864534 | 0.64 |
ENSMUST00000018572.4
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr3_-_142881942 | 0.63 |
ENSMUST00000043812.8
|
Pkn2
|
protein kinase N2 |
| chr15_-_38078842 | 0.63 |
ENSMUST00000110336.2
|
Ubr5
|
ubiquitin protein ligase E3 component n-recognin 5 |
| chr9_+_57708534 | 0.58 |
ENSMUST00000043990.7
ENSMUST00000142807.1 |
Edc3
|
enhancer of mRNA decapping 3 homolog (S. cerevisiae) |
| chr3_+_121953213 | 0.56 |
ENSMUST00000037958.7
ENSMUST00000128366.1 |
Arhgap29
|
Rho GTPase activating protein 29 |
| chr2_-_26503814 | 0.53 |
ENSMUST00000028288.4
|
Notch1
|
notch 1 |
| chr1_+_9798123 | 0.50 |
ENSMUST00000168907.1
ENSMUST00000166384.1 |
Sgk3
|
serum/glucocorticoid regulated kinase 3 |
| chr11_-_6606053 | 0.43 |
ENSMUST00000045713.3
|
Nacad
|
NAC alpha domain containing |
| chrX_+_162901226 | 0.36 |
ENSMUST00000101095.2
|
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr2_+_153649617 | 0.32 |
ENSMUST00000109771.1
|
Dnmt3b
|
DNA methyltransferase 3B |
| chr12_-_4233958 | 0.32 |
ENSMUST00000111169.3
ENSMUST00000020981.5 |
Cenpo
|
centromere protein O |
| chr14_+_31019159 | 0.30 |
ENSMUST00000112094.1
ENSMUST00000144009.1 |
Pbrm1
|
polybromo 1 |
| chr10_-_91123955 | 0.29 |
ENSMUST00000164505.1
ENSMUST00000170810.1 ENSMUST00000076694.6 |
Slc25a3
|
solute carrier family 25 (mitochondrial carrier, phosphate carrier), member 3 |
| chr2_-_145935014 | 0.29 |
ENSMUST00000001818.4
|
Crnkl1
|
Crn, crooked neck-like 1 (Drosophila) |
| chrX_+_162901567 | 0.29 |
ENSMUST00000112303.1
ENSMUST00000033727.7 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr9_-_82975475 | 0.28 |
ENSMUST00000034787.5
|
Phip
|
pleckstrin homology domain interacting protein |
| chr2_-_166713758 | 0.25 |
ENSMUST00000036719.5
|
Prex1
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
| chrX_+_162901762 | 0.24 |
ENSMUST00000112302.1
ENSMUST00000112301.1 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr8_-_64205970 | 0.23 |
ENSMUST00000066166.4
|
Tll1
|
tolloid-like |
| chr4_+_44004438 | 0.22 |
ENSMUST00000107846.3
|
Clta
|
clathrin, light polypeptide (Lca) |
| chr1_-_9748376 | 0.22 |
ENSMUST00000057438.6
|
Vcpip1
|
valosin containing protein (p97)/p47 complex interacting protein 1 |
| chr15_+_12824841 | 0.21 |
ENSMUST00000090292.5
|
Drosha
|
drosha, ribonuclease type III |
| chr17_-_17624458 | 0.20 |
ENSMUST00000041047.2
|
Lnpep
|
leucyl/cystinyl aminopeptidase |
| chr1_+_52630692 | 0.15 |
ENSMUST00000165859.1
|
Tmem194b
|
transmembrane protein 194B |
| chr5_-_66080971 | 0.14 |
ENSMUST00000127275.1
ENSMUST00000113724.1 |
Rbm47
|
RNA binding motif protein 47 |
| chr9_-_44134481 | 0.11 |
ENSMUST00000180670.1
|
Gm10687
|
predicted gene 10687 |
| chr11_+_119602981 | 0.11 |
ENSMUST00000026671.6
|
Rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr15_+_12824815 | 0.09 |
ENSMUST00000169061.1
|
Drosha
|
drosha, ribonuclease type III |
| chr5_+_144100387 | 0.08 |
ENSMUST00000041804.7
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chr8_-_69974367 | 0.07 |
ENSMUST00000116463.2
|
Gatad2a
|
GATA zinc finger domain containing 2A |
| chr7_+_97579868 | 0.05 |
ENSMUST00000042399.7
ENSMUST00000107153.1 |
Rsf1
|
remodeling and spacing factor 1 |
| chr8_-_64849818 | 0.05 |
ENSMUST00000034017.7
|
Klhl2
|
kelch-like 2, Mayven |
| chr9_-_65580040 | 0.04 |
ENSMUST00000068944.7
|
Plekho2
|
pleckstrin homology domain containing, family O member 2 |
| chr15_-_39943963 | 0.03 |
ENSMUST00000110305.2
|
Lrp12
|
low density lipoprotein-related protein 12 |
| chr1_+_136052750 | 0.01 |
ENSMUST00000160641.1
|
Cacna1s
|
calcium channel, voltage-dependent, L type, alpha 1S subunit |
| chr17_+_25727726 | 0.00 |
ENSMUST00000047273.1
|
Rpusd1
|
RNA pseudouridylate synthase domain containing 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of E2f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 12.7 | GO:0071921 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 3.2 | 9.5 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 2.6 | 10.4 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 1.4 | 5.5 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 1.2 | 9.8 | GO:0007144 | female meiosis I(GO:0007144) |
| 1.2 | 8.5 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 1.2 | 3.5 | GO:0031660 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031660) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G2/M transition of mitotic cell cycle(GO:0031662) |
| 1.1 | 4.3 | GO:0009223 | pyrimidine nucleotide catabolic process(GO:0006244) pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 1.1 | 6.5 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 1.1 | 6.4 | GO:0060623 | regulation of chromosome condensation(GO:0060623) |
| 1.0 | 4.1 | GO:1902990 | mitotic telomere maintenance via semi-conservative replication(GO:1902990) negative regulation of t-circle formation(GO:1904430) |
| 1.0 | 8.9 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.9 | 2.8 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.8 | 9.3 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.7 | 26.4 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.7 | 3.5 | GO:0090521 | glomerular visceral epithelial cell migration(GO:0090521) |
| 0.7 | 2.7 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.7 | 5.9 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.6 | 16.0 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.5 | 2.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.5 | 2.4 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.5 | 1.9 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.5 | 4.7 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.4 | 1.8 | GO:0035962 | response to interleukin-13(GO:0035962) cellular response to interleukin-13(GO:0035963) |
| 0.4 | 2.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.4 | 13.1 | GO:1902751 | positive regulation of cell cycle G2/M phase transition(GO:1902751) |
| 0.4 | 13.4 | GO:0040034 | regulation of development, heterochronic(GO:0040034) |
| 0.4 | 24.4 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.4 | 2.4 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.4 | 6.0 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.4 | 1.4 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.3 | 9.2 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.3 | 1.3 | GO:2000017 | positive regulation of determination of dorsal identity(GO:2000017) |
| 0.3 | 12.0 | GO:0051310 | metaphase plate congression(GO:0051310) |
| 0.3 | 3.6 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.3 | 1.2 | GO:0043987 | histone H3-S10 phosphorylation(GO:0043987) |
| 0.3 | 2.7 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.2 | 2.5 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.2 | 1.7 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.2 | 0.7 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.2 | 10.8 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.2 | 1.8 | GO:0015074 | DNA integration(GO:0015074) |
| 0.2 | 0.9 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.2 | 3.0 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.2 | 0.6 | GO:1900045 | negative regulation of protein K63-linked ubiquitination(GO:1900045) negative regulation of protein polyubiquitination(GO:1902915) negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.2 | 1.4 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.2 | 2.1 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 3.5 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.2 | 1.6 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.1 | 1.9 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.1 | 2.3 | GO:0006490 | oligosaccharide-lipid intermediate biosynthetic process(GO:0006490) |
| 0.1 | 1.0 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.1 | 0.9 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.1 | 0.9 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 17.3 | GO:0007051 | spindle organization(GO:0007051) |
| 0.1 | 0.3 | GO:0060821 | inactivation of X chromosome by DNA methylation(GO:0060821) |
| 0.1 | 0.6 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.1 | 1.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.7 | GO:0045410 | positive regulation of interleukin-6 biosynthetic process(GO:0045410) |
| 0.1 | 1.6 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.1 | 0.8 | GO:0003352 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) |
| 0.1 | 1.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) |
| 0.1 | 0.6 | GO:0031087 | deadenylation-independent decapping of nuclear-transcribed mRNA(GO:0031087) |
| 0.1 | 0.9 | GO:0006241 | CTP biosynthetic process(GO:0006241) CTP metabolic process(GO:0046036) |
| 0.1 | 0.3 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.7 | GO:0090161 | Golgi ribbon formation(GO:0090161) |
| 0.0 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 2.1 | GO:0001825 | blastocyst formation(GO:0001825) |
| 0.0 | 1.1 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 4.2 | GO:0010212 | response to ionizing radiation(GO:0010212) |
| 0.0 | 0.6 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 2.0 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.0 | 0.3 | GO:0034508 | centromere complex assembly(GO:0034508) |
| 0.0 | 1.1 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 2.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 2.0 | GO:0006334 | nucleosome assembly(GO:0006334) |
| 0.0 | 2.4 | GO:0000956 | nuclear-transcribed mRNA catabolic process(GO:0000956) |
| 0.0 | 0.8 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 2.2 | GO:0008033 | tRNA processing(GO:0008033) |
| 0.0 | 0.8 | GO:0030521 | androgen receptor signaling pathway(GO:0030521) |
| 0.0 | 4.6 | GO:0042254 | ribosome biogenesis(GO:0042254) |
| 0.0 | 0.1 | GO:0021995 | anterior neuropore closure(GO:0021506) neuropore closure(GO:0021995) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.5 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 1.1 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.1 | GO:0071233 | response to leucine(GO:0043201) cellular response to leucine(GO:0071233) |
| 0.0 | 0.7 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.0 | GO:0003349 | epicardium-derived cardiac endothelial cell differentiation(GO:0003349) |
| 0.0 | 0.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.2 | 25.4 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 1.6 | 9.5 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 1.4 | 12.7 | GO:0008278 | cohesin complex(GO:0008278) |
| 1.2 | 3.5 | GO:0035101 | FACT complex(GO:0035101) |
| 1.1 | 12.4 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 1.1 | 5.5 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 1.1 | 12.0 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.0 | 5.0 | GO:0031523 | Myb complex(GO:0031523) |
| 1.0 | 8.9 | GO:0031390 | Ctf18 RFC-like complex(GO:0031390) |
| 0.9 | 6.6 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.9 | 10.4 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.8 | 5.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.7 | 3.5 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.7 | 10.7 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.7 | 3.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.6 | 11.9 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.5 | 2.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 3.5 | GO:0005638 | lamin filament(GO:0005638) |
| 0.5 | 5.9 | GO:0042555 | MCM complex(GO:0042555) |
| 0.4 | 2.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.4 | 3.0 | GO:0032300 | mismatch repair complex(GO:0032300) |
| 0.4 | 2.5 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.3 | 1.8 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.3 | 1.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.2 | 2.2 | GO:0034709 | methylosome(GO:0034709) |
| 0.2 | 2.4 | GO:0031595 | nuclear proteasome complex(GO:0031595) |
| 0.2 | 0.7 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.2 | 19.2 | GO:0000922 | spindle pole(GO:0000922) |
| 0.2 | 13.1 | GO:0000794 | condensed nuclear chromosome(GO:0000794) |
| 0.1 | 6.5 | GO:0000793 | condensed chromosome(GO:0000793) |
| 0.1 | 3.7 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 8.5 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.1 | 0.5 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.1 | 0.3 | GO:0070877 | microprocessor complex(GO:0070877) ribonuclease III complex(GO:1903095) |
| 0.1 | 4.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.1 | 4.7 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.1 | 0.8 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.1 | 0.2 | GO:0099631 | postsynaptic endocytic zone cytoplasmic component(GO:0099631) |
| 0.1 | 2.1 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 3.0 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 2.4 | GO:0015030 | Cajal body(GO:0015030) |
| 0.1 | 2.0 | GO:0044815 | nucleosome(GO:0000786) DNA packaging complex(GO:0044815) |
| 0.0 | 7.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 7.6 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 3.3 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 6.4 | GO:0000775 | chromosome, centromeric region(GO:0000775) |
| 0.0 | 0.7 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.6 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.3 | GO:0071014 | post-mRNA release spliceosomal complex(GO:0071014) |
| 0.0 | 3.8 | GO:0000781 | chromosome, telomeric region(GO:0000781) |
| 0.0 | 1.7 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 1.6 | GO:0030964 | mitochondrial respiratory chain complex I(GO:0005747) NADH dehydrogenase complex(GO:0030964) respiratory chain complex I(GO:0045271) |
| 0.0 | 1.8 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 1.2 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.3 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 12.6 | GO:0048471 | perinuclear region of cytoplasm(GO:0048471) |
| 0.0 | 0.8 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.9 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.5 | 20.7 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 2.2 | 10.8 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 1.0 | 3.0 | GO:0032142 | guanine/thymine mispair binding(GO:0032137) single guanine insertion binding(GO:0032142) |
| 0.8 | 2.5 | GO:0071207 | histone pre-mRNA stem-loop binding(GO:0071207) |
| 0.8 | 8.9 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.8 | 8.0 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.8 | 2.3 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.7 | 5.8 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.7 | 5.0 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.7 | 2.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.5 | 2.2 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.5 | 1.6 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.5 | 5.0 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.4 | 4.3 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.4 | 1.2 | GO:0019153 | protein-disulfide reductase (glutathione) activity(GO:0019153) |
| 0.3 | 1.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 2.7 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.3 | 12.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.3 | 2.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.3 | 9.9 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.3 | 13.1 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.3 | 6.3 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.2 | 0.9 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.2 | 5.5 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.2 | 1.2 | GO:0031493 | nucleosomal histone binding(GO:0031493) histone threonine kinase activity(GO:0035184) |
| 0.2 | 10.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.2 | 1.3 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.2 | 4.7 | GO:0070182 | DNA polymerase binding(GO:0070182) |
| 0.2 | 0.9 | GO:0038049 | glucocorticoid receptor activity(GO:0004883) transcription factor activity, ligand-activated RNA polymerase II transcription factor binding(GO:0038049) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 2.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.1 | 3.6 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.1 | 1.4 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.1 | 1.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.1 | 3.5 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.1 | 5.7 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.1 | 4.7 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.1 | 1.7 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.7 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.1 | 1.8 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 0.3 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 1.6 | GO:0008137 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.1 | 0.8 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 0.7 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 1.1 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 2.4 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 1.9 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.6 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.6 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 8.7 | GO:0004721 | phosphoprotein phosphatase activity(GO:0004721) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.6 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.8 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.5 | GO:0017081 | chloride channel regulator activity(GO:0017081) |
| 0.0 | 0.9 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 1.4 | GO:0019213 | deacetylase activity(GO:0019213) |
| 0.0 | 13.0 | GO:0004842 | ubiquitin-protein transferase activity(GO:0004842) |
| 0.0 | 1.1 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.6 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 1.7 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 10.3 | GO:0003682 | chromatin binding(GO:0003682) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 1.8 | GO:0004896 | cytokine receptor activity(GO:0004896) |
| 0.0 | 3.0 | GO:0042393 | histone binding(GO:0042393) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 1.8 | GO:0047485 | protein N-terminus binding(GO:0047485) |
| 0.0 | 1.4 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 0.3 | GO:0015114 | phosphate ion transmembrane transporter activity(GO:0015114) |
| 0.0 | 0.8 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 2.0 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.5 | GO:0003743 | translation initiation factor activity(GO:0003743) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 28.1 | PID ATM PATHWAY | ATM pathway |
| 0.5 | 10.4 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.4 | 17.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.3 | 11.9 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 1.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.3 | 10.4 | PID ATR PATHWAY | ATR signaling pathway |
| 0.2 | 19.6 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.1 | 7.2 | PID E2F PATHWAY | E2F transcription factor network |
| 0.1 | 3.5 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.1 | 2.0 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.1 | 2.7 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 3.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.9 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.0 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.9 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 1.0 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.4 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.5 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 13.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 1.1 | 24.6 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.6 | 5.5 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.6 | 14.2 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.5 | 7.2 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.4 | 13.8 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.4 | 5.9 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.3 | 5.7 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.3 | 20.7 | REACTOME LATE PHASE OF HIV LIFE CYCLE | Genes involved in Late Phase of HIV Life Cycle |
| 0.2 | 2.5 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.2 | 22.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 1.8 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.2 | 7.4 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.2 | 2.7 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 2.0 | REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | Genes involved in NRIF signals cell death from the nucleus |
| 0.1 | 2.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.1 | 3.5 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.1 | 2.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.1 | 1.9 | REACTOME DOUBLE STRAND BREAK REPAIR | Genes involved in Double-Strand Break Repair |
| 0.1 | 1.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.1 | 4.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.1 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 1.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 1.1 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.9 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 0.9 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.9 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 1.6 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.0 | 1.1 | REACTOME CDK MEDIATED PHOSPHORYLATION AND REMOVAL OF CDC6 | Genes involved in CDK-mediated phosphorylation and removal of Cdc6 |
| 0.0 | 1.8 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |