Project
avrg: 2D miR_HR1_12
Navigation
Downloads
Results for Nfe2_Bach1_Mafk
Z-value: 0.33
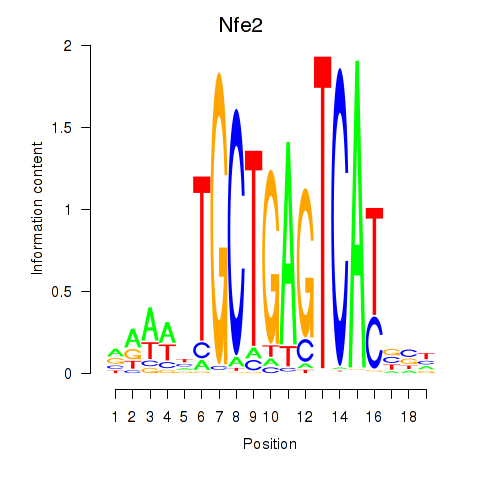
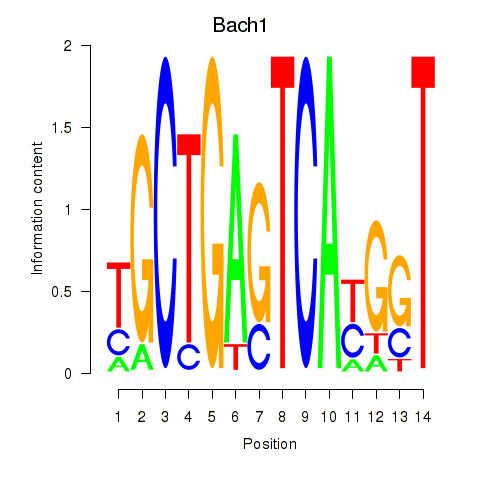
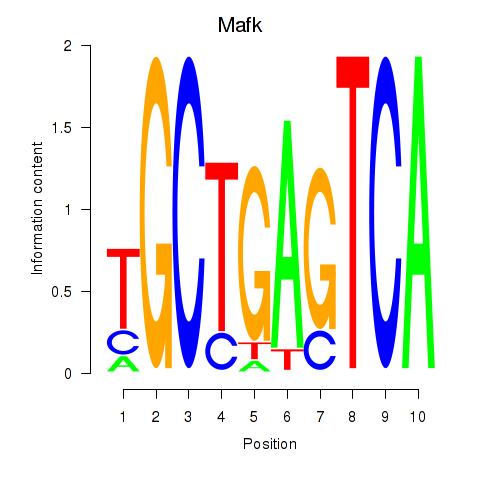
Transcription factors associated with Nfe2_Bach1_Mafk
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Nfe2
|
ENSMUSG00000058794.6 | nuclear factor, erythroid derived 2 |
|
Bach1
|
ENSMUSG00000025612.5 | BTB and CNC homology 1, basic leucine zipper transcription factor 1 |
|
Mafk
|
ENSMUSG00000018143.4 | v-maf musculoaponeurotic fibrosarcoma oncogene family, protein K (avian) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfe2 | mm10_v2_chr15_-_103255433_103255445 | 0.37 | 2.3e-01 | Click! |
| Mafk | mm10_v2_chr5_+_139791513_139791539 | 0.16 | 6.2e-01 | Click! |
| Bach1 | mm10_v2_chr16_+_87698904_87698959 | -0.06 | 8.6e-01 | Click! |
Activity profile of Nfe2_Bach1_Mafk motif
Sorted Z-values of Nfe2_Bach1_Mafk motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_164269371 | 0.60 |
ENSMUST00000145412.1
ENSMUST00000033749.7 |
Pir
|
pirin |
| chr9_+_86695542 | 0.57 |
ENSMUST00000150367.2
|
A330041J22Rik
|
RIKEN cDNA A330041J22 gene |
| chr18_+_61639542 | 0.48 |
ENSMUST00000183083.1
ENSMUST00000183087.1 |
Gm20748
|
predicted gene, 20748 |
| chr7_+_126766397 | 0.44 |
ENSMUST00000032944.7
|
Gdpd3
|
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr6_-_129533267 | 0.42 |
ENSMUST00000181594.1
|
1700101I11Rik
|
RIKEN cDNA 1700101I11 gene |
| chr5_-_139484420 | 0.40 |
ENSMUST00000150992.1
|
Zfand2a
|
zinc finger, AN1-type domain 2A |
| chr2_+_167777467 | 0.36 |
ENSMUST00000139927.1
ENSMUST00000127441.1 |
Gm14321
|
predicted gene 14321 |
| chr2_-_160313616 | 0.36 |
ENSMUST00000109475.2
|
Gm826
|
predicted gene 826 |
| chr5_-_108675569 | 0.36 |
ENSMUST00000051757.7
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chr6_-_16898441 | 0.35 |
ENSMUST00000031533.7
|
Tfec
|
transcription factor EC |
| chr19_+_42052228 | 0.30 |
ENSMUST00000164518.2
|
4933411K16Rik
|
RIKEN cDNA 4933411K16 gene |
| chr2_+_11705712 | 0.30 |
ENSMUST00000138856.1
ENSMUST00000078834.5 ENSMUST00000114834.3 ENSMUST00000114833.3 ENSMUST00000114831.2 ENSMUST00000114832.2 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr2_-_30415389 | 0.28 |
ENSMUST00000142096.1
|
Crat
|
carnitine acetyltransferase |
| chr5_+_112255813 | 0.25 |
ENSMUST00000031286.6
ENSMUST00000131673.1 ENSMUST00000112375.1 |
Crybb1
|
crystallin, beta B1 |
| chr1_+_165769392 | 0.25 |
ENSMUST00000040298.4
|
Creg1
|
cellular repressor of E1A-stimulated genes 1 |
| chr2_+_11705437 | 0.24 |
ENSMUST00000148748.1
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr6_+_129397297 | 0.24 |
ENSMUST00000032262.7
|
Clec1b
|
C-type lectin domain family 1, member b |
| chr4_-_43558386 | 0.21 |
ENSMUST00000130353.1
|
Tln1
|
talin 1 |
| chr11_-_30649510 | 0.21 |
ENSMUST00000074613.3
|
Acyp2
|
acylphosphatase 2, muscle type |
| chr7_-_126799163 | 0.21 |
ENSMUST00000032934.5
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr4_-_140774196 | 0.21 |
ENSMUST00000026381.6
|
Padi4
|
peptidyl arginine deiminase, type IV |
| chr9_+_64117147 | 0.20 |
ENSMUST00000034969.7
|
Lctl
|
lactase-like |
| chr10_+_4611971 | 0.20 |
ENSMUST00000105590.1
ENSMUST00000067086.7 |
Esr1
|
estrogen receptor 1 (alpha) |
| chr11_-_120630516 | 0.20 |
ENSMUST00000106181.1
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr9_+_7272514 | 0.20 |
ENSMUST00000015394.8
|
Mmp13
|
matrix metallopeptidase 13 |
| chr6_+_5725639 | 0.20 |
ENSMUST00000115556.1
ENSMUST00000115555.1 ENSMUST00000115559.3 |
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1 |
| chr2_+_11705459 | 0.19 |
ENSMUST00000126394.1
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr17_-_45573253 | 0.19 |
ENSMUST00000165127.1
ENSMUST00000166469.1 ENSMUST00000024739.7 |
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr6_+_129397478 | 0.18 |
ENSMUST00000112081.2
ENSMUST00000112079.2 |
Clec1b
|
C-type lectin domain family 1, member b |
| chr19_-_8839181 | 0.18 |
ENSMUST00000096259.4
|
Gng3
|
guanine nucleotide binding protein (G protein), gamma 3 |
| chr11_-_120630126 | 0.17 |
ENSMUST00000106180.1
|
Mafg
|
v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G (avian) |
| chr12_-_40248073 | 0.16 |
ENSMUST00000169926.1
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr2_+_11705355 | 0.16 |
ENSMUST00000128156.2
|
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr7_+_120917744 | 0.16 |
ENSMUST00000033173.7
ENSMUST00000106483.2 |
Polr3e
|
polymerase (RNA) III (DNA directed) polypeptide E |
| chr10_+_116143881 | 0.16 |
ENSMUST00000105271.2
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr19_+_8839298 | 0.15 |
ENSMUST00000160556.1
|
Bscl2
|
Bernardinelli-Seip congenital lipodystrophy 2 homolog (human) |
| chr8_-_107588392 | 0.15 |
ENSMUST00000044106.4
|
Psmd7
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 7 |
| chr2_-_20943413 | 0.15 |
ENSMUST00000140230.1
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr12_-_108275409 | 0.15 |
ENSMUST00000136175.1
|
Ccdc85c
|
coiled-coil domain containing 85C |
| chr12_-_85288419 | 0.15 |
ENSMUST00000121930.1
|
Acyp1
|
acylphosphatase 1, erythrocyte (common) type |
| chr1_-_63686904 | 0.15 |
ENSMUST00000090313.4
|
Dytn
|
dystrotelin |
| chr18_-_10030017 | 0.14 |
ENSMUST00000116669.1
ENSMUST00000092096.6 |
Usp14
|
ubiquitin specific peptidase 14 |
| chr2_+_164948219 | 0.14 |
ENSMUST00000017881.2
|
Mmp9
|
matrix metallopeptidase 9 |
| chr17_-_45572495 | 0.14 |
ENSMUST00000130406.1
|
Hsp90ab1
|
heat shock protein 90 alpha (cytosolic), class B member 1 |
| chr7_-_126799134 | 0.14 |
ENSMUST00000087566.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr7_+_84528964 | 0.14 |
ENSMUST00000180387.1
|
Gm2115
|
predicted gene 2115 |
| chr2_+_11705287 | 0.14 |
ENSMUST00000135341.1
ENSMUST00000138349.1 ENSMUST00000123600.2 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr12_-_76795489 | 0.13 |
ENSMUST00000082431.3
|
Gpx2
|
glutathione peroxidase 2 |
| chrX_+_53724826 | 0.13 |
ENSMUST00000069209.1
|
4930502E18Rik
|
RIKEN cDNA 4930502E18 gene |
| chr16_+_90220742 | 0.13 |
ENSMUST00000023707.9
|
Sod1
|
superoxide dismutase 1, soluble |
| chr16_+_11405648 | 0.13 |
ENSMUST00000096273.2
|
Snx29
|
sorting nexin 29 |
| chr8_+_123332676 | 0.13 |
ENSMUST00000010298.6
|
Spire2
|
spire homolog 2 (Drosophila) |
| chr12_-_84400851 | 0.13 |
ENSMUST00000117286.1
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr15_+_102503722 | 0.12 |
ENSMUST00000096145.4
|
Gm10337
|
predicted gene 10337 |
| chr16_+_20651652 | 0.12 |
ENSMUST00000007212.8
|
Psmd2
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 |
| chr9_-_86695897 | 0.11 |
ENSMUST00000034989.8
|
Me1
|
malic enzyme 1, NADP(+)-dependent, cytosolic |
| chr12_-_84400929 | 0.11 |
ENSMUST00000122194.1
|
Entpd5
|
ectonucleoside triphosphate diphosphohydrolase 5 |
| chr12_+_70974621 | 0.11 |
ENSMUST00000160027.1
ENSMUST00000160864.1 |
Psma3
|
proteasome (prosome, macropain) subunit, alpha type 3 |
| chr2_+_30078213 | 0.11 |
ENSMUST00000150770.1
|
Pkn3
|
protein kinase N3 |
| chr1_+_86064619 | 0.11 |
ENSMUST00000027432.8
|
Psmd1
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 1 |
| chr1_+_107589997 | 0.11 |
ENSMUST00000112706.2
ENSMUST00000000514.4 |
Serpinb8
|
serine (or cysteine) peptidase inhibitor, clade B, member 8 |
| chr17_+_35841668 | 0.11 |
ENSMUST00000174124.1
|
Mdc1
|
mediator of DNA damage checkpoint 1 |
| chr11_-_49187037 | 0.10 |
ENSMUST00000153999.1
ENSMUST00000066531.6 |
Btnl9
|
butyrophilin-like 9 |
| chr8_+_105827721 | 0.10 |
ENSMUST00000034365.4
|
Tsnaxip1
|
translin-associated factor X (Tsnax) interacting protein 1 |
| chr4_+_116685544 | 0.10 |
ENSMUST00000135573.1
ENSMUST00000151129.1 |
Prdx1
|
peroxiredoxin 1 |
| chr13_+_29014399 | 0.10 |
ENSMUST00000146336.1
ENSMUST00000130109.1 |
A330102I10Rik
|
RIKEN cDNA A330102I10 gene |
| chr14_+_70077375 | 0.10 |
ENSMUST00000035908.1
|
Egr3
|
early growth response 3 |
| chr9_-_122310921 | 0.10 |
ENSMUST00000180685.1
|
Gm26797
|
predicted gene, 26797 |
| chr17_-_45592262 | 0.10 |
ENSMUST00000164769.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chr1_+_174172738 | 0.10 |
ENSMUST00000027817.7
|
Spta1
|
spectrin alpha, erythrocytic 1 |
| chr16_-_36990449 | 0.09 |
ENSMUST00000075869.6
|
Fbxo40
|
F-box protein 40 |
| chr12_+_89812467 | 0.09 |
ENSMUST00000110133.2
ENSMUST00000110130.2 |
Nrxn3
|
neurexin III |
| chr11_-_48871408 | 0.09 |
ENSMUST00000097271.2
|
Irgm1
|
immunity-related GTPase family M member 1 |
| chr9_+_65214690 | 0.09 |
ENSMUST00000069000.7
|
Parp16
|
poly (ADP-ribose) polymerase family, member 16 |
| chr16_-_18811972 | 0.09 |
ENSMUST00000000028.7
ENSMUST00000115585.1 |
Cdc45
|
cell division cycle 45 |
| chr2_+_155775333 | 0.09 |
ENSMUST00000029141.5
|
Mmp24
|
matrix metallopeptidase 24 |
| chr14_-_21848924 | 0.09 |
ENSMUST00000124549.1
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr6_+_5725812 | 0.09 |
ENSMUST00000115554.1
ENSMUST00000153942.1 |
Dync1i1
|
dynein cytoplasmic 1 intermediate chain 1 |
| chr13_-_23710714 | 0.09 |
ENSMUST00000091707.6
ENSMUST00000006787.7 ENSMUST00000091706.6 |
Hfe
|
hemochromatosis |
| chr1_-_172219715 | 0.08 |
ENSMUST00000170700.1
ENSMUST00000003554.4 |
Casq1
|
calsequestrin 1 |
| chr17_-_45592485 | 0.08 |
ENSMUST00000166119.1
|
Slc29a1
|
solute carrier family 29 (nucleoside transporters), member 1 |
| chrX_-_136868537 | 0.08 |
ENSMUST00000058814.6
|
Rab9b
|
RAB9B, member RAS oncogene family |
| chr1_-_33907721 | 0.08 |
ENSMUST00000115161.1
ENSMUST00000062289.8 |
Bend6
|
BEN domain containing 6 |
| chr13_-_54611332 | 0.08 |
ENSMUST00000091609.4
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr4_+_116685859 | 0.08 |
ENSMUST00000129315.1
ENSMUST00000106470.1 |
Prdx1
|
peroxiredoxin 1 |
| chrX_+_93675088 | 0.08 |
ENSMUST00000045898.3
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta isoform |
| chr10_+_118204942 | 0.08 |
ENSMUST00000096691.4
|
Il22
|
interleukin 22 |
| chr4_+_109406623 | 0.08 |
ENSMUST00000124209.1
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr1_-_120074023 | 0.08 |
ENSMUST00000056089.7
|
Tmem37
|
transmembrane protein 37 |
| chr13_-_54611274 | 0.08 |
ENSMUST00000049575.7
|
Cltb
|
clathrin, light polypeptide (Lcb) |
| chr13_-_85127514 | 0.08 |
ENSMUST00000179230.1
|
Gm4076
|
predicted gene 4076 |
| chr2_-_30415302 | 0.07 |
ENSMUST00000132981.2
ENSMUST00000129494.1 |
Crat
|
carnitine acetyltransferase |
| chr4_-_19922599 | 0.07 |
ENSMUST00000029900.5
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr14_+_67716262 | 0.07 |
ENSMUST00000150768.1
|
Kctd9
|
potassium channel tetramerisation domain containing 9 |
| chr11_+_80428598 | 0.07 |
ENSMUST00000173938.1
ENSMUST00000017572.7 |
Psmd11
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr9_+_32224457 | 0.07 |
ENSMUST00000183121.1
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr6_-_120357422 | 0.07 |
ENSMUST00000032283.5
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr3_+_28805436 | 0.07 |
ENSMUST00000043867.5
|
Rpl22l1
|
ribosomal protein L22 like 1 |
| chr2_+_163547148 | 0.07 |
ENSMUST00000109411.1
ENSMUST00000018094.6 |
Hnf4a
|
hepatic nuclear factor 4, alpha |
| chr1_-_167285110 | 0.07 |
ENSMUST00000027839.8
|
Uck2
|
uridine-cytidine kinase 2 |
| chr8_+_119437118 | 0.07 |
ENSMUST00000152420.1
ENSMUST00000098365.3 |
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr8_-_107403197 | 0.07 |
ENSMUST00000003947.8
|
Nqo1
|
NAD(P)H dehydrogenase, quinone 1 |
| chr15_-_79834261 | 0.07 |
ENSMUST00000148358.1
|
Cbx6
|
chromobox 6 |
| chr7_-_44670820 | 0.07 |
ENSMUST00000048102.7
|
Myh14
|
myosin, heavy polypeptide 14 |
| chr1_-_10719898 | 0.07 |
ENSMUST00000035577.6
|
Cpa6
|
carboxypeptidase A6 |
| chr6_-_120357342 | 0.07 |
ENSMUST00000163827.1
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr13_-_14613017 | 0.07 |
ENSMUST00000015816.3
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr11_+_78178105 | 0.07 |
ENSMUST00000147819.1
|
Tlcd1
|
TLC domain containing 1 |
| chr5_-_21785115 | 0.07 |
ENSMUST00000115193.1
ENSMUST00000115192.1 ENSMUST00000115195.1 ENSMUST00000030771.5 |
Dnajc2
|
DnaJ (Hsp40) homolog, subfamily C, member 2 |
| chr14_+_79515618 | 0.07 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr10_+_128933782 | 0.07 |
ENSMUST00000099112.2
|
Itga7
|
integrin alpha 7 |
| chrX_+_56779699 | 0.07 |
ENSMUST00000114772.2
ENSMUST00000114768.3 ENSMUST00000155882.1 |
Fhl1
|
four and a half LIM domains 1 |
| chr18_+_61045139 | 0.07 |
ENSMUST00000025522.4
ENSMUST00000115274.1 |
Pdgfrb
|
platelet derived growth factor receptor, beta polypeptide |
| chrX_+_56779437 | 0.06 |
ENSMUST00000114773.3
|
Fhl1
|
four and a half LIM domains 1 |
| chr5_-_124187150 | 0.06 |
ENSMUST00000161938.1
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr14_+_54476100 | 0.06 |
ENSMUST00000164766.1
ENSMUST00000164697.1 |
Rem2
|
rad and gem related GTP binding protein 2 |
| chr10_-_43901712 | 0.06 |
ENSMUST00000020012.6
|
Qrsl1
|
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr15_+_3270767 | 0.06 |
ENSMUST00000082424.4
ENSMUST00000159158.1 ENSMUST00000159216.1 ENSMUST00000160311.1 |
Sepp1
|
selenoprotein P, plasma, 1 |
| chr2_+_112261926 | 0.06 |
ENSMUST00000028553.3
|
Nop10
|
NOP10 ribonucleoprotein |
| chr1_-_75046639 | 0.06 |
ENSMUST00000152855.1
|
Nhej1
|
nonhomologous end-joining factor 1 |
| chr7_+_127800604 | 0.06 |
ENSMUST00000046863.5
ENSMUST00000106272.1 ENSMUST00000139068.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chr5_+_25247344 | 0.06 |
ENSMUST00000114950.1
|
Galnt11
|
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 11 |
| chr3_+_123267445 | 0.06 |
ENSMUST00000047923.7
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr11_-_118342500 | 0.06 |
ENSMUST00000103024.3
|
BC100451
|
cDNA sequence BC100451 |
| chr10_-_116473418 | 0.06 |
ENSMUST00000087965.4
ENSMUST00000164271.1 |
Kcnmb4
|
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr4_-_57956283 | 0.06 |
ENSMUST00000030051.5
|
Txn1
|
thioredoxin 1 |
| chr11_+_78178651 | 0.06 |
ENSMUST00000092880.7
ENSMUST00000127587.1 ENSMUST00000108338.1 |
Tlcd1
|
TLC domain containing 1 |
| chr7_+_19083842 | 0.06 |
ENSMUST00000032568.7
ENSMUST00000122999.1 ENSMUST00000108473.3 ENSMUST00000108474.1 |
Dmpk
|
dystrophia myotonica-protein kinase |
| chr1_+_127306706 | 0.06 |
ENSMUST00000171405.1
|
Mgat5
|
mannoside acetylglucosaminyltransferase 5 |
| chrX_-_97377190 | 0.06 |
ENSMUST00000037353.3
|
Eda2r
|
ectodysplasin A2 receptor |
| chrX_+_163911401 | 0.06 |
ENSMUST00000140845.1
|
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr2_-_84886692 | 0.05 |
ENSMUST00000054514.5
ENSMUST00000151799.1 |
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr15_-_76195710 | 0.05 |
ENSMUST00000023226.6
|
Plec
|
plectin |
| chr9_-_43239816 | 0.05 |
ENSMUST00000034512.5
|
Oaf
|
OAF homolog (Drosophila) |
| chr5_-_65428354 | 0.05 |
ENSMUST00000131263.1
|
Ugdh
|
UDP-glucose dehydrogenase |
| chr6_-_120357440 | 0.05 |
ENSMUST00000112703.1
|
Ccdc77
|
coiled-coil domain containing 77 |
| chr16_-_22439570 | 0.05 |
ENSMUST00000170393.1
|
Etv5
|
ets variant gene 5 |
| chr13_+_14613242 | 0.05 |
ENSMUST00000170836.2
|
Psma2
|
proteasome (prosome, macropain) subunit, alpha type 2 |
| chr5_+_34525797 | 0.05 |
ENSMUST00000125817.1
ENSMUST00000067638.7 |
Sh3bp2
|
SH3-domain binding protein 2 |
| chr15_-_79605084 | 0.05 |
ENSMUST00000023065.6
|
Dmc1
|
DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination |
| chr12_+_51690966 | 0.05 |
ENSMUST00000021338.8
|
Ap4s1
|
adaptor-related protein complex AP-4, sigma 1 |
| chr7_-_34230281 | 0.05 |
ENSMUST00000038027.4
|
Gpi1
|
glucose phosphate isomerase 1 |
| chr17_+_27839974 | 0.05 |
ENSMUST00000071006.7
|
Snrpc
|
U1 small nuclear ribonucleoprotein C |
| chr2_-_156004147 | 0.05 |
ENSMUST00000156993.1
ENSMUST00000141437.1 |
6430550D23Rik
|
RIKEN cDNA 6430550D23 gene |
| chr9_-_21239310 | 0.05 |
ENSMUST00000164812.1
ENSMUST00000049567.4 |
Keap1
|
kelch-like ECH-associated protein 1 |
| chr15_-_76206309 | 0.05 |
ENSMUST00000073418.6
ENSMUST00000171634.1 ENSMUST00000076442.5 |
Plec
|
plectin |
| chr14_-_20393473 | 0.05 |
ENSMUST00000061444.3
|
Mrps16
|
mitochondrial ribosomal protein S16 |
| chr16_+_48431237 | 0.05 |
ENSMUST00000023330.6
|
Morc1
|
microrchidia 1 |
| chr17_+_35049966 | 0.05 |
ENSMUST00000007257.9
|
Clic1
|
chloride intracellular channel 1 |
| chr1_+_157526127 | 0.05 |
ENSMUST00000111700.1
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr3_-_116968969 | 0.05 |
ENSMUST00000143611.1
ENSMUST00000040097.7 |
Palmd
|
palmdelphin |
| chr19_+_26623419 | 0.05 |
ENSMUST00000176584.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr19_-_6080311 | 0.05 |
ENSMUST00000159832.1
|
Vps51
|
vacuolar protein sorting 51 homolog (S. cerevisiae) |
| chr2_+_181319714 | 0.04 |
ENSMUST00000098971.4
ENSMUST00000054622.8 ENSMUST00000108814.1 ENSMUST00000048608.9 ENSMUST00000108815.1 |
Rtel1
|
regulator of telomere elongation helicase 1 |
| chr17_+_35016576 | 0.04 |
ENSMUST00000007245.1
ENSMUST00000172499.1 |
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr16_-_18343882 | 0.04 |
ENSMUST00000130752.1
ENSMUST00000115628.2 |
Tango2
|
transport and golgi organization 2 |
| chr5_-_100500592 | 0.04 |
ENSMUST00000149714.1
ENSMUST00000046154.5 |
Lin54
|
lin-54 homolog (C. elegans) |
| chr17_-_24644933 | 0.04 |
ENSMUST00000019684.5
|
Slc9a3r2
|
solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 |
| chr7_-_113347273 | 0.04 |
ENSMUST00000117577.1
|
Btbd10
|
BTB (POZ) domain containing 10 |
| chr5_+_147860615 | 0.04 |
ENSMUST00000031654.6
|
Pomp
|
proteasome maturation protein |
| chr10_+_69212634 | 0.04 |
ENSMUST00000020101.5
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr10_+_18845071 | 0.04 |
ENSMUST00000019998.7
|
Perp
|
PERP, TP53 apoptosis effector |
| chr3_+_81999461 | 0.04 |
ENSMUST00000107736.1
|
Asic5
|
acid-sensing (proton-gated) ion channel family member 5 |
| chr5_+_32247351 | 0.04 |
ENSMUST00000101376.2
|
Plb1
|
phospholipase B1 |
| chr9_+_54950808 | 0.04 |
ENSMUST00000172407.1
|
Psma4
|
proteasome (prosome, macropain) subunit, alpha type 4 |
| chr7_+_127244511 | 0.04 |
ENSMUST00000052509.4
|
Zfp771
|
zinc finger protein 771 |
| chr11_-_109363654 | 0.04 |
ENSMUST00000070956.3
|
Gm11696
|
predicted gene 11696 |
| chr11_-_49187159 | 0.04 |
ENSMUST00000046522.6
|
Btnl9
|
butyrophilin-like 9 |
| chr11_+_107479478 | 0.04 |
ENSMUST00000021063.6
ENSMUST00000106752.3 |
Psmd12
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
| chr2_+_156475803 | 0.04 |
ENSMUST00000029155.8
|
Epb4.1l1
|
erythrocyte protein band 4.1-like 1 |
| chr8_+_94984399 | 0.04 |
ENSMUST00000093271.6
|
Gpr56
|
G protein-coupled receptor 56 |
| chr3_+_95929325 | 0.04 |
ENSMUST00000171368.1
ENSMUST00000168106.1 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr17_+_28692568 | 0.04 |
ENSMUST00000114752.1
|
Mapk14
|
mitogen-activated protein kinase 14 |
| chr7_+_127800844 | 0.04 |
ENSMUST00000106271.1
ENSMUST00000138432.1 |
Hsd3b7
|
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 |
| chrX_+_162901226 | 0.04 |
ENSMUST00000101095.2
|
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chrX_-_97377150 | 0.04 |
ENSMUST00000113832.1
|
Eda2r
|
ectodysplasin A2 receptor |
| chr2_+_61711694 | 0.04 |
ENSMUST00000028278.7
|
Psmd14
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 |
| chr9_+_58823512 | 0.04 |
ENSMUST00000034889.8
|
Hcn4
|
hyperpolarization-activated, cyclic nucleotide-gated K+ 4 |
| chr9_+_54950782 | 0.04 |
ENSMUST00000034848.7
|
Psma4
|
proteasome (prosome, macropain) subunit, alpha type 4 |
| chr11_+_87582201 | 0.04 |
ENSMUST00000133202.1
|
Sept4
|
septin 4 |
| chr7_+_49759100 | 0.04 |
ENSMUST00000085272.5
|
Htatip2
|
HIV-1 tat interactive protein 2, homolog (human) |
| chr17_+_78937124 | 0.04 |
ENSMUST00000024887.4
|
Ndufaf7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex assembly factor 7 |
| chr7_-_75308373 | 0.04 |
ENSMUST00000085164.5
|
Sv2b
|
synaptic vesicle glycoprotein 2 b |
| chr1_+_72307413 | 0.04 |
ENSMUST00000027379.8
|
Xrcc5
|
X-ray repair complementing defective repair in Chinese hamster cells 5 |
| chr11_-_106256045 | 0.04 |
ENSMUST00000021048.6
|
Ftsj3
|
FtsJ homolog 3 (E. coli) |
| chr11_+_62077018 | 0.04 |
ENSMUST00000092415.5
|
Specc1
|
sperm antigen with calponin homology and coiled-coil domains 1 |
| chr17_-_56133817 | 0.04 |
ENSMUST00000167545.1
|
Sema6b
|
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6B |
| chr5_-_136565432 | 0.04 |
ENSMUST00000176172.1
|
Cux1
|
cut-like homeobox 1 |
| chr11_-_43747963 | 0.04 |
ENSMUST00000048578.2
ENSMUST00000109278.1 |
Ttc1
|
tetratricopeptide repeat domain 1 |
| chr3_-_89393629 | 0.04 |
ENSMUST00000124783.1
ENSMUST00000126027.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr6_-_67037399 | 0.04 |
ENSMUST00000043098.6
|
Gadd45a
|
growth arrest and DNA-damage-inducible 45 alpha |
| chr7_-_17056669 | 0.04 |
ENSMUST00000037762.4
|
Hif3a
|
hypoxia inducible factor 3, alpha subunit |
| chr19_+_26753588 | 0.04 |
ENSMUST00000177116.1
|
Smarca2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr3_+_95929246 | 0.04 |
ENSMUST00000165307.1
ENSMUST00000015893.6 |
Anp32e
|
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrX_+_162901567 | 0.04 |
ENSMUST00000112303.1
ENSMUST00000033727.7 |
Ctps2
|
cytidine 5'-triphosphate synthase 2 |
| chr11_+_120673359 | 0.04 |
ENSMUST00000135346.1
ENSMUST00000127269.1 ENSMUST00000131727.2 ENSMUST00000149389.1 ENSMUST00000153346.1 |
Aspscr1
|
alveolar soft part sarcoma chromosome region, candidate 1 (human) |
| chr11_+_76945719 | 0.04 |
ENSMUST00000125145.1
|
Blmh
|
bleomycin hydrolase |
| chr7_-_114276107 | 0.04 |
ENSMUST00000033008.9
|
Psma1
|
proteasome (prosome, macropain) subunit, alpha type 1 |
| chrX_-_122397351 | 0.04 |
ENSMUST00000079490.4
|
Nap1l3
|
nucleosome assembly protein 1-like 3 |
| chr12_+_111758848 | 0.04 |
ENSMUST00000084941.5
|
Klc1
|
kinesin light chain 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Nfe2_Bach1_Mafk
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903659 | transforming growth factor beta activation(GO:0036363) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.2 | GO:0036414 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 1.0 | GO:0032825 | positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0070649 | formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.2 | GO:0060523 | Sertoli cell proliferation(GO:0060011) prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:1902299 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) pre-replicative complex assembly(GO:0036388) DNA replication preinitiation complex assembly(GO:0071163) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.2 | GO:0019254 | carnitine metabolic process, CoA-linked(GO:0019254) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.3 | GO:2000582 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0042977 | regulation of activation of JAK2 kinase activity(GO:0010534) activation of JAK2 kinase activity(GO:0042977) negative regulation of activation of JAK2 kinase activity(GO:1902569) |
| 0.0 | 0.1 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0072277 | cell migration involved in vasculogenesis(GO:0035441) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:1904430 | negative regulation of t-circle formation(GO:1904430) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.4 | GO:0030220 | platelet formation(GO:0030220) |
| 0.0 | 0.6 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.0 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0035754 | B cell chemotaxis(GO:0035754) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.0 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.0 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:2000304 | positive regulation of sphingolipid biosynthetic process(GO:0090154) positive regulation of arachidonic acid secretion(GO:0090238) positive regulation of ceramide biosynthetic process(GO:2000304) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.1 | GO:0060335 | positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0061718 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.0 | GO:0086046 | membrane depolarization during SA node cell action potential(GO:0086046) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.1 | 0.3 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.1 | GO:0005656 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.2 | GO:0097550 | transcriptional preinitiation complex(GO:0097550) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.3 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.6 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.0 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0003998 | acylphosphatase activity(GO:0003998) |
| 0.1 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.2 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.1 | 0.3 | GO:0002135 | CTP binding(GO:0002135) |
| 0.1 | 0.2 | GO:0004092 | carnitine O-acetyltransferase activity(GO:0004092) |
| 0.0 | 0.2 | GO:0004382 | guanosine-diphosphatase activity(GO:0004382) |
| 0.0 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.2 | GO:0004784 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.0 | 0.1 | GO:0004471 | malic enzyme activity(GO:0004470) malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.6 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.4 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0070540 | stearic acid binding(GO:0070540) |
| 0.0 | 0.5 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0003979 | UDP-glucose 6-dehydrogenase activity(GO:0003979) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0032050 | clathrin heavy chain binding(GO:0032050) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.0 | 0.2 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.0 | GO:1990450 | linear polyubiquitin binding(GO:1990450) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.9 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |