Project
avrg: 2D miR_HR1_12
Navigation
Downloads
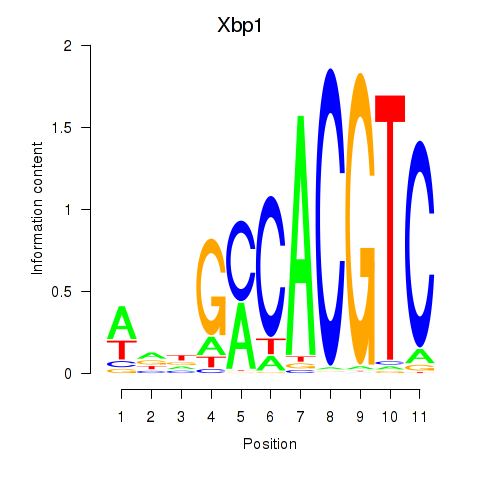
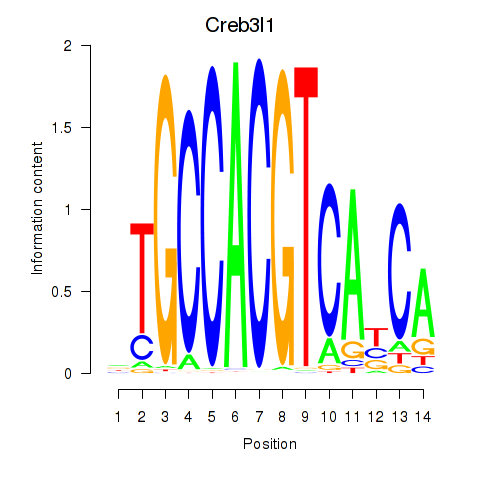
Results for Xbp1_Creb3l1
Z-value: 5.04
Transcription factors associated with Xbp1_Creb3l1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Xbp1
|
ENSMUSG00000020484.12 | X-box binding protein 1 |
|
Creb3l1
|
ENSMUSG00000027230.9 | cAMP responsive element binding protein 3-like 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Creb3l1 | mm10_v2_chr2_-_92024502_92024529 | 0.92 | 2.6e-05 | Click! |
| Xbp1 | mm10_v2_chr11_+_5520652_5520659 | 0.88 | 1.8e-04 | Click! |
Activity profile of Xbp1_Creb3l1 motif
Sorted Z-values of Xbp1_Creb3l1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_45092130 | 13.91 |
ENSMUST00000148175.1
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr7_-_45092198 | 13.30 |
ENSMUST00000140449.1
ENSMUST00000117546.1 ENSMUST00000019683.3 |
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr7_-_45091713 | 8.72 |
ENSMUST00000141576.1
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr15_+_79516396 | 6.00 |
ENSMUST00000010974.7
|
Kdelr3
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 3 |
| chr3_+_93520473 | 5.25 |
ENSMUST00000029515.4
|
S100a11
|
S100 calcium binding protein A11 (calgizzarin) |
| chr2_+_180725263 | 5.03 |
ENSMUST00000094218.3
|
Slc17a9
|
solute carrier family 17, member 9 |
| chr18_+_57354733 | 4.97 |
ENSMUST00000025490.8
|
Prrc1
|
proline-rich coiled-coil 1 |
| chr15_-_32244632 | 4.52 |
ENSMUST00000181536.1
|
0610007N19Rik
|
RIKEN cDNA 0610007N19 |
| chr9_+_72925622 | 3.77 |
ENSMUST00000038489.5
|
Pygo1
|
pygopus 1 |
| chr19_+_11965817 | 3.71 |
ENSMUST00000025590.9
|
Osbp
|
oxysterol binding protein |
| chr5_+_113735782 | 3.61 |
ENSMUST00000065698.5
|
Ficd
|
FIC domain containing |
| chr5_+_137288273 | 3.27 |
ENSMUST00000024099.4
ENSMUST00000085934.3 |
Ache
|
acetylcholinesterase |
| chr7_+_35555367 | 3.26 |
ENSMUST00000181932.1
|
B230322F03Rik
|
RIKEN cDNA B230322F03 gene |
| chr6_-_54593139 | 3.21 |
ENSMUST00000046520.6
|
Fkbp14
|
FK506 binding protein 14 |
| chr5_+_92137896 | 3.07 |
ENSMUST00000031355.6
|
Uso1
|
USO1 vesicle docking factor |
| chr10_-_128400448 | 3.03 |
ENSMUST00000167859.1
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr19_+_8741669 | 3.01 |
ENSMUST00000176314.1
ENSMUST00000073430.7 ENSMUST00000175901.1 |
Stx5a
|
syntaxin 5A |
| chr10_+_91082940 | 2.85 |
ENSMUST00000020150.3
|
Ikbip
|
IKBKB interacting protein |
| chr19_+_8741473 | 2.81 |
ENSMUST00000177373.1
ENSMUST00000010254.9 |
Stx5a
|
syntaxin 5A |
| chr4_-_118544010 | 2.78 |
ENSMUST00000128098.1
|
Tmem125
|
transmembrane protein 125 |
| chr3_-_27710413 | 2.71 |
ENSMUST00000046157.4
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr8_+_70302518 | 2.41 |
ENSMUST00000066469.7
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr19_+_8741413 | 2.38 |
ENSMUST00000176381.1
|
Stx5a
|
syntaxin 5A |
| chr11_+_69095217 | 2.13 |
ENSMUST00000101004.2
|
Per1
|
period circadian clock 1 |
| chr4_+_43562672 | 2.07 |
ENSMUST00000167751.1
ENSMUST00000132631.1 |
Creb3
|
cAMP responsive element binding protein 3 |
| chr2_-_168742100 | 2.05 |
ENSMUST00000109177.1
|
Atp9a
|
ATPase, class II, type 9A |
| chr6_-_126740151 | 2.05 |
ENSMUST00000112242.1
|
Kcna6
|
potassium voltage-gated channel, shaker-related, subfamily, member 6 |
| chr9_-_104063049 | 2.01 |
ENSMUST00000035166.5
|
Uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr2_+_150786735 | 2.00 |
ENSMUST00000045441.7
|
Pygb
|
brain glycogen phosphorylase |
| chr8_+_70302761 | 1.99 |
ENSMUST00000150968.1
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr10_+_91083036 | 1.95 |
ENSMUST00000020149.5
|
Ikbip
|
IKBKB interacting protein |
| chr4_-_129640691 | 1.92 |
ENSMUST00000084264.5
|
Txlna
|
taxilin alpha |
| chr4_-_114908892 | 1.87 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chrX_+_73787002 | 1.87 |
ENSMUST00000166518.1
|
Ssr4
|
signal sequence receptor, delta |
| chr6_+_70844499 | 1.86 |
ENSMUST00000034093.8
ENSMUST00000162950.1 |
Eif2ak3
|
eukaryotic translation initiation factor 2 alpha kinase 3 |
| chr4_-_155653184 | 1.86 |
ENSMUST00000030937.1
|
Mmp23
|
matrix metallopeptidase 23 |
| chr17_-_34031684 | 1.84 |
ENSMUST00000169397.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr12_+_51377560 | 1.81 |
ENSMUST00000021335.5
|
Scfd1
|
Sec1 family domain containing 1 |
| chr17_-_34031544 | 1.80 |
ENSMUST00000025186.8
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr13_-_37994111 | 1.79 |
ENSMUST00000021864.6
|
Ssr1
|
signal sequence receptor, alpha |
| chr6_-_52204415 | 1.77 |
ENSMUST00000048794.6
|
Hoxa5
|
homeobox A5 |
| chr10_+_61680302 | 1.76 |
ENSMUST00000020285.8
|
Sar1a
|
SAR1 gene homolog A (S. cerevisiae) |
| chr5_+_64970069 | 1.75 |
ENSMUST00000031080.8
|
Fam114a1
|
family with sequence similarity 114, member A1 |
| chrX_+_73787062 | 1.73 |
ENSMUST00000002090.2
|
Ssr4
|
signal sequence receptor, delta |
| chr15_-_57892358 | 1.68 |
ENSMUST00000022993.5
|
Derl1
|
Der1-like domain family, member 1 |
| chr17_-_34031644 | 1.66 |
ENSMUST00000171872.1
|
Slc39a7
|
solute carrier family 39 (zinc transporter), member 7 |
| chr11_-_108343917 | 1.66 |
ENSMUST00000059595.4
|
Prkca
|
protein kinase C, alpha |
| chr16_-_36874806 | 1.63 |
ENSMUST00000075946.5
|
Eaf2
|
ELL associated factor 2 |
| chr18_+_51117754 | 1.62 |
ENSMUST00000116639.2
|
Prr16
|
proline rich 16 |
| chr5_+_143403819 | 1.61 |
ENSMUST00000110731.2
|
Kdelr2
|
KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 2 |
| chr14_+_20674311 | 1.61 |
ENSMUST00000048657.8
|
Sec24c
|
Sec24 related gene family, member C (S. cerevisiae) |
| chr16_+_36875119 | 1.60 |
ENSMUST00000135406.1
ENSMUST00000114812.1 ENSMUST00000134616.1 ENSMUST00000023534.6 |
Golgb1
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr1_-_183369529 | 1.57 |
ENSMUST00000069922.5
|
Mia3
|
melanoma inhibitory activity 3 |
| chr13_-_55415166 | 1.57 |
ENSMUST00000054146.3
|
Pfn3
|
profilin 3 |
| chr5_+_30869623 | 1.54 |
ENSMUST00000114716.1
|
Tmem214
|
transmembrane protein 214 |
| chr17_-_26199008 | 1.53 |
ENSMUST00000142410.1
ENSMUST00000120333.1 ENSMUST00000039113.7 |
Pdia2
|
protein disulfide isomerase associated 2 |
| chr7_+_139212974 | 1.53 |
ENSMUST00000016124.8
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr19_+_5088534 | 1.51 |
ENSMUST00000025811.4
|
Yif1a
|
Yip1 interacting factor homolog A (S. cerevisiae) |
| chr9_-_103202113 | 1.50 |
ENSMUST00000035157.8
|
Srprb
|
signal recognition particle receptor, B subunit |
| chr10_+_42761483 | 1.48 |
ENSMUST00000019937.4
|
Sec63
|
SEC63-like (S. cerevisiae) |
| chr12_-_36042476 | 1.44 |
ENSMUST00000020896.8
|
Tspan13
|
tetraspanin 13 |
| chr11_-_86807624 | 1.43 |
ENSMUST00000018569.7
|
Dhx40
|
DEAH (Asp-Glu-Ala-His) box polypeptide 40 |
| chr12_+_13269111 | 1.42 |
ENSMUST00000042953.8
|
Nbas
|
neuroblastoma amplified sequence |
| chr2_+_70661556 | 1.40 |
ENSMUST00000112201.1
ENSMUST00000028509.4 ENSMUST00000133432.1 ENSMUST00000112205.1 |
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr5_+_30869579 | 1.39 |
ENSMUST00000046349.7
|
Tmem214
|
transmembrane protein 214 |
| chr3_-_97767916 | 1.37 |
ENSMUST00000045243.8
|
Pde4dip
|
phosphodiesterase 4D interacting protein (myomegalin) |
| chr1_-_13589717 | 1.36 |
ENSMUST00000027068.4
|
Tram1
|
translocating chain-associating membrane protein 1 |
| chr4_+_137594140 | 1.32 |
ENSMUST00000105840.1
ENSMUST00000055131.6 ENSMUST00000105839.1 ENSMUST00000105838.1 |
Usp48
|
ubiquitin specific peptidase 48 |
| chr3_+_96697100 | 1.32 |
ENSMUST00000107077.3
|
Pias3
|
protein inhibitor of activated STAT 3 |
| chrX_+_56454871 | 1.32 |
ENSMUST00000039374.2
ENSMUST00000101553.2 |
Ddx26b
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr6_-_4086914 | 1.32 |
ENSMUST00000049166.4
|
Bet1
|
blocked early in transport 1 homolog (S. cerevisiae) |
| chr6_-_88518760 | 1.31 |
ENSMUST00000032168.5
|
Sec61a1
|
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr3_-_108562153 | 1.29 |
ENSMUST00000106622.2
|
Tmem167b
|
transmembrane protein 167B |
| chr7_-_122067263 | 1.29 |
ENSMUST00000033159.3
|
Ears2
|
glutamyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr7_+_116504409 | 1.28 |
ENSMUST00000183175.1
|
Nucb2
|
nucleobindin 2 |
| chr17_-_64331817 | 1.28 |
ENSMUST00000172733.1
ENSMUST00000172818.1 |
Pja2
|
praja 2, RING-H2 motif containing |
| chr5_-_115158169 | 1.26 |
ENSMUST00000053271.5
ENSMUST00000112121.1 |
Mlec
|
malectin |
| chr7_+_116504363 | 1.26 |
ENSMUST00000032895.8
|
Nucb2
|
nucleobindin 2 |
| chr14_-_79301623 | 1.24 |
ENSMUST00000022595.7
|
Rgcc
|
regulator of cell cycle |
| chr6_+_88084473 | 1.24 |
ENSMUST00000032143.6
|
Rpn1
|
ribophorin I |
| chr9_+_57560934 | 1.23 |
ENSMUST00000045791.9
|
Scamp2
|
secretory carrier membrane protein 2 |
| chr3_-_108086590 | 1.21 |
ENSMUST00000102638.1
ENSMUST00000102637.1 |
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr15_-_102136225 | 1.20 |
ENSMUST00000154032.1
|
Spryd3
|
SPRY domain containing 3 |
| chr4_-_132144486 | 1.20 |
ENSMUST00000056336.1
|
Oprd1
|
opioid receptor, delta 1 |
| chr16_-_11134624 | 1.15 |
ENSMUST00000038424.7
|
Txndc11
|
thioredoxin domain containing 11 |
| chr9_+_104063376 | 1.14 |
ENSMUST00000120854.1
|
Acad11
|
acyl-Coenzyme A dehydrogenase family, member 11 |
| chr10_+_41519493 | 1.11 |
ENSMUST00000019962.8
|
Cd164
|
CD164 antigen |
| chr5_+_110176640 | 1.11 |
ENSMUST00000112512.1
|
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr7_+_139213003 | 1.11 |
ENSMUST00000156768.1
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr11_-_96916448 | 1.10 |
ENSMUST00000103152.4
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr2_-_91070180 | 1.09 |
ENSMUST00000153367.1
ENSMUST00000079976.3 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chr2_-_144011202 | 1.08 |
ENSMUST00000016072.5
ENSMUST00000037875.5 |
Rrbp1
|
ribosome binding protein 1 |
| chr6_+_87042838 | 1.08 |
ENSMUST00000113658.1
ENSMUST00000113657.1 ENSMUST00000113655.1 ENSMUST00000032057.7 |
Gfpt1
|
glutamine fructose-6-phosphate transaminase 1 |
| chr11_-_20741580 | 1.07 |
ENSMUST00000035350.5
|
Aftph
|
aftiphilin |
| chr14_-_60086832 | 1.07 |
ENSMUST00000080368.5
|
Atp8a2
|
ATPase, aminophospholipid transporter-like, class I, type 8A, member 2 |
| chr17_+_29490812 | 1.04 |
ENSMUST00000024811.6
|
Pim1
|
proviral integration site 1 |
| chr4_+_43441939 | 1.04 |
ENSMUST00000060864.6
|
Tesk1
|
testis specific protein kinase 1 |
| chr2_-_91070283 | 1.00 |
ENSMUST00000111436.2
ENSMUST00000073575.5 |
Slc39a13
|
solute carrier family 39 (metal ion transporter), member 13 |
| chrX_-_155623325 | 1.00 |
ENSMUST00000038665.5
|
Ptchd1
|
patched domain containing 1 |
| chr10_-_24101951 | 0.99 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr11_-_72796028 | 0.95 |
ENSMUST00000156294.1
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr4_-_151861698 | 0.94 |
ENSMUST00000049790.7
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr11_-_68853119 | 0.94 |
ENSMUST00000018880.7
|
Ndel1
|
nuclear distribution gene E-like homolog 1 (A. nidulans) |
| chr4_-_151861762 | 0.92 |
ENSMUST00000097774.2
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr6_+_29348069 | 0.92 |
ENSMUST00000173216.1
ENSMUST00000031779.10 ENSMUST00000090481.7 |
Calu
|
calumenin |
| chr5_-_35679416 | 0.91 |
ENSMUST00000114233.2
|
Htra3
|
HtrA serine peptidase 3 |
| chr2_+_155133501 | 0.91 |
ENSMUST00000029126.8
ENSMUST00000109685.1 |
Itch
|
itchy, E3 ubiquitin protein ligase |
| chr3_+_96697076 | 0.91 |
ENSMUST00000162778.2
ENSMUST00000064900.9 |
Pias3
|
protein inhibitor of activated STAT 3 |
| chr4_-_134000857 | 0.90 |
ENSMUST00000105887.1
ENSMUST00000012262.5 ENSMUST00000144668.1 ENSMUST00000105889.3 |
Dhdds
|
dehydrodolichyl diphosphate synthase |
| chr11_-_96916366 | 0.89 |
ENSMUST00000144731.1
ENSMUST00000127048.1 |
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr13_-_55321928 | 0.89 |
ENSMUST00000035242.7
|
Rab24
|
RAB24, member RAS oncogene family |
| chr3_+_123267445 | 0.88 |
ENSMUST00000047923.7
|
Sec24d
|
Sec24 related gene family, member D (S. cerevisiae) |
| chr11_-_96916407 | 0.88 |
ENSMUST00000130774.1
|
Cdk5rap3
|
CDK5 regulatory subunit associated protein 3 |
| chr3_-_65392579 | 0.88 |
ENSMUST00000029414.5
|
Ssr3
|
signal sequence receptor, gamma |
| chr4_-_129641060 | 0.87 |
ENSMUST00000046425.9
ENSMUST00000133803.1 |
Txlna
|
taxilin alpha |
| chr19_+_8898090 | 0.87 |
ENSMUST00000096246.3
|
Ganab
|
alpha glucosidase 2 alpha neutral subunit |
| chr2_-_155692317 | 0.87 |
ENSMUST00000103140.4
|
Trpc4ap
|
transient receptor potential cation channel, subfamily C, member 4 associated protein |
| chr9_-_44767792 | 0.85 |
ENSMUST00000034607.9
|
Arcn1
|
archain 1 |
| chr4_-_43000451 | 0.84 |
ENSMUST00000030164.7
|
Vcp
|
valosin containing protein |
| chr1_-_93478785 | 0.83 |
ENSMUST00000170883.1
|
Hdlbp
|
high density lipoprotein (HDL) binding protein |
| chr12_-_59011996 | 0.81 |
ENSMUST00000021375.5
|
Sec23a
|
SEC23A (S. cerevisiae) |
| chr7_-_121035096 | 0.80 |
ENSMUST00000065740.2
|
Gm9905
|
predicted gene 9905 |
| chr14_-_75754475 | 0.79 |
ENSMUST00000049168.7
|
Cog3
|
component of oligomeric golgi complex 3 |
| chr3_-_108562349 | 0.78 |
ENSMUST00000090546.5
|
Tmem167b
|
transmembrane protein 167B |
| chr1_+_119526125 | 0.77 |
ENSMUST00000183952.1
|
TMEM185B
|
Transmembrane protein 185B |
| chr5_+_110176868 | 0.77 |
ENSMUST00000139611.1
ENSMUST00000031477.8 |
Golga3
|
golgi autoantigen, golgin subfamily a, 3 |
| chr7_+_122067164 | 0.77 |
ENSMUST00000033158.4
|
Ubfd1
|
ubiquitin family domain containing 1 |
| chr11_-_72795801 | 0.76 |
ENSMUST00000079681.5
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr6_-_82774448 | 0.73 |
ENSMUST00000000642.4
|
Hk2
|
hexokinase 2 |
| chr10_+_85102627 | 0.73 |
ENSMUST00000095383.4
|
AI597468
|
expressed sequence AI597468 |
| chr2_-_39065505 | 0.72 |
ENSMUST00000039165.8
|
Golga1
|
golgi autoantigen, golgin subfamily a, 1 |
| chr11_+_54100924 | 0.69 |
ENSMUST00000093107.5
ENSMUST00000019050.5 ENSMUST00000174616.1 ENSMUST00000129499.1 ENSMUST00000126840.1 |
P4ha2
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha II polypeptide |
| chr7_-_127393604 | 0.68 |
ENSMUST00000165495.1
ENSMUST00000106303.2 ENSMUST00000074249.6 |
E430018J23Rik
|
RIKEN cDNA E430018J23 gene |
| chr12_+_59130767 | 0.66 |
ENSMUST00000175877.1
|
Ctage5
|
CTAGE family, member 5 |
| chrX_+_56346390 | 0.65 |
ENSMUST00000101560.3
|
Zfp449
|
zinc finger protein 449 |
| chr15_+_23036449 | 0.65 |
ENSMUST00000164787.1
|
Cdh18
|
cadherin 18 |
| chr12_+_69184158 | 0.63 |
ENSMUST00000060579.8
|
Mgat2
|
mannoside acetylglucosaminyltransferase 2 |
| chr19_+_46761578 | 0.63 |
ENSMUST00000077666.4
ENSMUST00000099373.4 |
Cnnm2
|
cyclin M2 |
| chr3_-_65392546 | 0.62 |
ENSMUST00000119896.1
|
Ssr3
|
signal sequence receptor, gamma |
| chr16_-_11134601 | 0.60 |
ENSMUST00000118362.1
ENSMUST00000118679.1 |
Txndc11
|
thioredoxin domain containing 11 |
| chr5_+_124540695 | 0.59 |
ENSMUST00000060226.4
|
Tmed2
|
transmembrane emp24 domain trafficking protein 2 |
| chr5_-_72559599 | 0.58 |
ENSMUST00000074948.4
ENSMUST00000087216.5 |
Nfxl1
|
nuclear transcription factor, X-box binding-like 1 |
| chr5_+_144190284 | 0.57 |
ENSMUST00000060747.7
|
Bhlha15
|
basic helix-loop-helix family, member a15 |
| chr4_-_11386679 | 0.56 |
ENSMUST00000043781.7
ENSMUST00000108310.1 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr7_+_28693032 | 0.56 |
ENSMUST00000151227.1
ENSMUST00000108281.1 |
Fbxo27
|
F-box protein 27 |
| chr8_+_25720054 | 0.55 |
ENSMUST00000068916.8
ENSMUST00000139836.1 |
Ppapdc1b
|
phosphatidic acid phosphatase type 2 domain containing 1B |
| chr10_-_78753046 | 0.55 |
ENSMUST00000105383.2
|
Ccdc105
|
coiled-coil domain containing 105 |
| chr13_+_73937799 | 0.54 |
ENSMUST00000099384.2
|
Brd9
|
bromodomain containing 9 |
| chr11_-_20741447 | 0.54 |
ENSMUST00000177543.1
|
Aftph
|
aftiphilin |
| chr13_+_81657732 | 0.53 |
ENSMUST00000049055.6
|
Lysmd3
|
LysM, putative peptidoglycan-binding, domain containing 3 |
| chr19_-_6909599 | 0.51 |
ENSMUST00000173091.1
|
Prdx5
|
peroxiredoxin 5 |
| chr16_+_52031549 | 0.51 |
ENSMUST00000114471.1
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr2_+_121140407 | 0.49 |
ENSMUST00000028702.3
ENSMUST00000110665.1 |
Adal
|
adenosine deaminase-like |
| chr17_-_47833169 | 0.48 |
ENSMUST00000131971.1
ENSMUST00000129360.1 ENSMUST00000113280.1 ENSMUST00000132125.1 |
Mdfi
|
MyoD family inhibitor |
| chr4_-_151861667 | 0.48 |
ENSMUST00000169423.2
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr4_-_43562397 | 0.48 |
ENSMUST00000030187.7
|
Tln1
|
talin 1 |
| chr4_-_11386757 | 0.47 |
ENSMUST00000108313.1
ENSMUST00000108311.2 |
Esrp1
|
epithelial splicing regulatory protein 1 |
| chr10_-_24092320 | 0.47 |
ENSMUST00000092654.2
|
Taar8b
|
trace amine-associated receptor 8B |
| chr17_-_51831884 | 0.47 |
ENSMUST00000124222.1
|
Satb1
|
special AT-rich sequence binding protein 1 |
| chr2_-_121140371 | 0.46 |
ENSMUST00000099486.2
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr8_+_108714644 | 0.46 |
ENSMUST00000043896.8
|
Zfhx3
|
zinc finger homeobox 3 |
| chr11_-_69948145 | 0.46 |
ENSMUST00000179298.1
ENSMUST00000018710.6 ENSMUST00000135437.1 ENSMUST00000141837.2 ENSMUST00000142500.1 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr19_-_41263931 | 0.45 |
ENSMUST00000025989.8
|
Tm9sf3
|
transmembrane 9 superfamily member 3 |
| chr7_+_100706623 | 0.45 |
ENSMUST00000107042.1
|
Fam168a
|
family with sequence similarity 168, member A |
| chr17_-_47833256 | 0.44 |
ENSMUST00000152455.1
ENSMUST00000035375.7 |
Mdfi
|
MyoD family inhibitor |
| chr11_-_68853019 | 0.44 |
ENSMUST00000108672.1
|
Ndel1
|
nuclear distribution gene E-like homolog 1 (A. nidulans) |
| chr17_-_25861456 | 0.43 |
ENSMUST00000079461.8
ENSMUST00000176923.1 |
Wdr90
|
WD repeat domain 90 |
| chr6_-_72958465 | 0.43 |
ENSMUST00000114050.1
|
Tmsb10
|
thymosin, beta 10 |
| chr8_+_71367210 | 0.43 |
ENSMUST00000019169.7
|
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr19_+_6276725 | 0.42 |
ENSMUST00000025684.3
|
Ehd1
|
EH-domain containing 1 |
| chr13_-_100833369 | 0.41 |
ENSMUST00000067246.4
|
Slc30a5
|
solute carrier family 30 (zinc transporter), member 5 |
| chr7_-_126799134 | 0.40 |
ENSMUST00000087566.4
|
Aldoa
|
aldolase A, fructose-bisphosphate |
| chr5_+_76974683 | 0.40 |
ENSMUST00000101087.3
ENSMUST00000120550.1 |
Srp72
|
signal recognition particle 72 |
| chr7_-_127387113 | 0.40 |
ENSMUST00000164345.1
ENSMUST00000049052.3 |
9130019O22Rik
|
RIKEN cDNA 9130019O22 gene |
| chr17_+_34032071 | 0.39 |
ENSMUST00000174299.1
ENSMUST00000173554.1 |
Rxrb
|
retinoid X receptor beta |
| chr7_-_81992277 | 0.38 |
ENSMUST00000026096.7
|
Bnc1
|
basonuclin 1 |
| chr9_+_44379490 | 0.37 |
ENSMUST00000066601.6
|
Hyou1
|
hypoxia up-regulated 1 |
| chr1_+_172082509 | 0.36 |
ENSMUST00000135192.1
|
Copa
|
coatomer protein complex subunit alpha |
| chr2_+_121140498 | 0.36 |
ENSMUST00000119031.1
ENSMUST00000110662.2 |
Adal
|
adenosine deaminase-like |
| chr17_-_35201996 | 0.36 |
ENSMUST00000167924.1
ENSMUST00000025263.8 |
Tnf
|
tumor necrosis factor |
| chr5_+_124541296 | 0.36 |
ENSMUST00000124529.1
|
Tmed2
|
transmembrane emp24 domain trafficking protein 2 |
| chr1_-_172082757 | 0.36 |
ENSMUST00000003550.4
|
Ncstn
|
nicastrin |
| chr9_-_21091974 | 0.34 |
ENSMUST00000115487.1
|
Raver1
|
ribonucleoprotein, PTB-binding 1 |
| chr17_-_56183887 | 0.34 |
ENSMUST00000019723.7
|
D17Wsu104e
|
DNA segment, Chr 17, Wayne State University 104, expressed |
| chr4_+_44756553 | 0.33 |
ENSMUST00000107824.2
|
Zcchc7
|
zinc finger, CCHC domain containing 7 |
| chr11_-_100970887 | 0.32 |
ENSMUST00000060792.5
|
Ptrf
|
polymerase I and transcript release factor |
| chr4_-_129640959 | 0.32 |
ENSMUST00000132217.1
ENSMUST00000130017.1 ENSMUST00000154105.1 |
Txlna
|
taxilin alpha |
| chr14_+_26638237 | 0.32 |
ENSMUST00000112318.3
|
Arf4
|
ADP-ribosylation factor 4 |
| chr2_+_164497518 | 0.32 |
ENSMUST00000103101.4
ENSMUST00000117066.1 |
Pigt
|
phosphatidylinositol glycan anchor biosynthesis, class T |
| chr11_-_59228162 | 0.31 |
ENSMUST00000163300.1
ENSMUST00000061242.7 |
Arf1
|
ADP-ribosylation factor 1 |
| chr8_+_71366848 | 0.30 |
ENSMUST00000110053.2
ENSMUST00000110054.1 ENSMUST00000139541.1 |
Use1
|
unconventional SNARE in the ER 1 homolog (S. cerevisiae) |
| chr10_+_24076500 | 0.30 |
ENSMUST00000051133.5
|
Taar8a
|
trace amine-associated receptor 8A |
| chr3_+_89246397 | 0.29 |
ENSMUST00000168900.1
|
Krtcap2
|
keratinocyte associated protein 2 |
| chr6_-_72958097 | 0.28 |
ENSMUST00000114049.1
|
Tmsb10
|
thymosin, beta 10 |
| chr19_-_5771376 | 0.26 |
ENSMUST00000025890.8
|
Scyl1
|
SCY1-like 1 (S. cerevisiae) |
| chr1_+_92910758 | 0.26 |
ENSMUST00000027487.8
|
Rnpepl1
|
arginyl aminopeptidase (aminopeptidase B)-like 1 |
| chr19_+_8819401 | 0.26 |
ENSMUST00000096753.3
|
Hnrnpul2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr5_+_120431770 | 0.26 |
ENSMUST00000031591.7
|
Lhx5
|
LIM homeobox protein 5 |
| chr9_+_44379536 | 0.26 |
ENSMUST00000161318.1
ENSMUST00000160902.1 |
Hyou1
|
hypoxia up-regulated 1 |
| chrX_+_20658326 | 0.25 |
ENSMUST00000089217.4
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Xbp1_Creb3l1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.6 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 1.0 | 3.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 1.0 | 12.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.8 | 2.5 | GO:0070093 | negative regulation of glucagon secretion(GO:0070093) positive regulation of testosterone secretion(GO:2000845) |
| 0.7 | 2.1 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) establishment of viral latency(GO:0019043) release from viral latency(GO:0019046) |
| 0.7 | 3.3 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.6 | 1.8 | GO:0060574 | bronchiole development(GO:0060435) intestinal epithelial cell maturation(GO:0060574) |
| 0.5 | 4.9 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.5 | 6.6 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.4 | 2.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.4 | 1.2 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.4 | 1.7 | GO:0010360 | negative regulation of anion channel activity(GO:0010360) regulation of dense core granule biogenesis(GO:2000705) |
| 0.4 | 7.4 | GO:0071577 | zinc II ion transmembrane transport(GO:0071577) |
| 0.3 | 1.9 | GO:0070417 | negative regulation of translation in response to stress(GO:0032055) cellular response to cold(GO:0070417) |
| 0.3 | 0.9 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.3 | 1.4 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.3 | 0.9 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.3 | 0.8 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.3 | 0.8 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.2 | 1.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.2 | 2.8 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.2 | 0.7 | GO:1905053 | regulation of base-excision repair(GO:1905051) positive regulation of base-excision repair(GO:1905053) |
| 0.2 | 1.3 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.2 | 1.1 | GO:0006041 | glucosamine metabolic process(GO:0006041) |
| 0.2 | 0.6 | GO:0031590 | wybutosine metabolic process(GO:0031590) wybutosine biosynthetic process(GO:0031591) |
| 0.2 | 8.6 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.2 | 3.8 | GO:0007289 | spermatid nucleus differentiation(GO:0007289) |
| 0.2 | 4.1 | GO:0044144 | regulation of growth of symbiont in host(GO:0044126) modulation of growth of symbiont involved in interaction with host(GO:0044144) |
| 0.2 | 1.3 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.2 | 2.7 | GO:0060510 | Type II pneumocyte differentiation(GO:0060510) |
| 0.2 | 0.5 | GO:0060785 | regulation of apoptosis involved in tissue homeostasis(GO:0060785) |
| 0.2 | 2.4 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.2 | 0.5 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 1.2 | GO:0032261 | purine nucleotide salvage(GO:0032261) IMP salvage(GO:0032264) |
| 0.1 | 1.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.7 | GO:1904925 | negative regulation of mitochondrial membrane permeability(GO:0035795) positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.1 | 0.4 | GO:2001137 | positive regulation of endocytic recycling(GO:2001137) |
| 0.1 | 5.0 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.1 | 1.0 | GO:2000138 | positive regulation of cell proliferation involved in heart morphogenesis(GO:2000138) |
| 0.1 | 0.5 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 2.2 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.1 | 1.7 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.1 | 1.0 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.1 | 0.7 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.1 | 0.8 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.1 | 0.6 | GO:1903297 | regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903297) negative regulation of hypoxia-induced intrinsic apoptotic signaling pathway(GO:1903298) intrinsic apoptotic signaling pathway in response to hypoxia(GO:1990144) |
| 0.1 | 1.6 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.1 | 1.5 | GO:1902175 | regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902175) |
| 0.1 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.1 | 0.2 | GO:0072272 | proximal/distal pattern formation involved in metanephric nephron development(GO:0072272) |
| 0.1 | 1.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.1 | 0.3 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 1.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 1.2 | GO:0018279 | protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.9 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.4 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.3 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.9 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 1.5 | GO:0010259 | multicellular organism aging(GO:0010259) |
| 0.0 | 0.4 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.6 | GO:0048312 | mitochondrial calcium ion transport(GO:0006851) intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.2 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.5 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 1.5 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.4 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.0 | 0.5 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 1.9 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 1.6 | GO:0032233 | positive regulation of actin filament bundle assembly(GO:0032233) |
| 0.0 | 0.5 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.3 | GO:0018195 | peptidyl-arginine modification(GO:0018195) |
| 0.0 | 0.2 | GO:0003085 | negative regulation of systemic arterial blood pressure(GO:0003085) |
| 0.0 | 0.2 | GO:2000653 | regulation of genetic imprinting(GO:2000653) |
| 0.0 | 0.9 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 1.4 | GO:0007030 | Golgi organization(GO:0007030) |
| 0.0 | 0.1 | GO:2000680 | regulation of rubidium ion transport(GO:2000680) |
| 0.0 | 0.4 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 1.3 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.0 | 5.5 | GO:0007283 | spermatogenesis(GO:0007283) |
| 0.0 | 0.1 | GO:0043982 | histone H4-K5 acetylation(GO:0043981) histone H4-K8 acetylation(GO:0043982) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.5 | 1.4 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.4 | 6.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.4 | 1.6 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.4 | 3.2 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.4 | 8.1 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.4 | 1.9 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.3 | 0.8 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.3 | 3.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 0.4 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.2 | 1.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.2 | 2.6 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.2 | 9.5 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.2 | 3.9 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.2 | 0.7 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 1.4 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.1 | 1.3 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.1 | 1.5 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.1 | 1.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.1 | 3.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.1 | 1.5 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.1 | 0.9 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.3 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.4 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 2.9 | GO:0005881 | cytoplasmic microtubule(GO:0005881) |
| 0.0 | 1.2 | GO:0032590 | dendrite membrane(GO:0032590) |
| 0.0 | 1.3 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 65.3 | GO:0005783 | endoplasmic reticulum(GO:0005783) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.6 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.0 | 0.1 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.0 | 0.5 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.4 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 2.4 | GO:0055037 | recycling endosome(GO:0055037) |
| 0.0 | 0.9 | GO:0005776 | autophagosome(GO:0005776) |
| 0.0 | 2.0 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 3.6 | GO:0016323 | basolateral plasma membrane(GO:0016323) |
| 0.0 | 12.8 | GO:0005794 | Golgi apparatus(GO:0005794) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.9 | 7.6 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.8 | 3.3 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.7 | 2.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.6 | 1.7 | GO:0004698 | calcium-dependent protein kinase C activity(GO:0004698) |
| 0.5 | 1.6 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.5 | 2.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.5 | 1.9 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.4 | 1.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.3 | 10.8 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.3 | 1.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.3 | 1.4 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.2 | 2.2 | GO:0031545 | peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.2 | 1.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 0.8 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 8.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.2 | 0.5 | GO:0072541 | peroxynitrite reductase activity(GO:0072541) |
| 0.2 | 2.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.2 | 0.6 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.2 | 1.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.1 | 3.7 | GO:0015248 | sterol transporter activity(GO:0015248) |
| 0.1 | 1.5 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.1 | 3.6 | GO:0070566 | adenylyltransferase activity(GO:0070566) |
| 0.1 | 3.1 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.1 | 2.4 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.1 | 2.0 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 0.9 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.1 | 1.9 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.1 | 10.2 | GO:0000149 | SNARE binding(GO:0000149) |
| 0.1 | 0.7 | GO:0008865 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 1.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.1 | 1.7 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.1 | 1.5 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 0.6 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.1 | 2.2 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.1 | 0.9 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 1.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 2.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.1 | 0.8 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.1 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 4.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.1 | 0.9 | GO:0004659 | prenyltransferase activity(GO:0004659) |
| 0.1 | 0.3 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.1 | 33.0 | GO:0005509 | calcium ion binding(GO:0005509) |
| 0.1 | 0.4 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.4 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 1.5 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 1.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 1.6 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 2.0 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.5 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.9 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 1.2 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0016004 | phospholipase activator activity(GO:0016004) lipase activator activity(GO:0060229) |
| 0.0 | 2.8 | GO:0008565 | protein transporter activity(GO:0008565) |
| 0.0 | 1.1 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 1.4 | GO:0003724 | RNA helicase activity(GO:0003724) ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 1.9 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 1.7 | GO:0020037 | heme binding(GO:0020037) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0043996 | histone acetyltransferase activity (H4-K5 specific)(GO:0043995) histone acetyltransferase activity (H4-K8 specific)(GO:0043996) histone acetyltransferase activity (H4-K16 specific)(GO:0046972) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 1.3 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.3 | GO:0070006 | metalloaminopeptidase activity(GO:0070006) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 2.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.1 | 3.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.1 | 2.1 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.1 | 2.1 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.1 | 1.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 1.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 2.9 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 3.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 1.8 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.7 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.5 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 4.7 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.0 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.4 | 8.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.3 | 15.2 | REACTOME ACTIVATION OF CHAPERONE GENES BY XBP1S | Genes involved in Activation of Chaperone Genes by XBP1(S) |
| 0.2 | 3.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.2 | 1.7 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 2.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.1 | 0.9 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.1 | 3.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.1 | 1.7 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.1 | 3.7 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.9 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.1 | 3.1 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.1 | 1.1 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 1.0 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 2.0 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 2.1 | REACTOME BMAL1 CLOCK NPAS2 ACTIVATES CIRCADIAN EXPRESSION | Genes involved in BMAL1:CLOCK/NPAS2 Activates Circadian Expression |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 4.7 | REACTOME SRP DEPENDENT COTRANSLATIONAL PROTEIN TARGETING TO MEMBRANE | Genes involved in SRP-dependent cotranslational protein targeting to membrane |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.5 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |