Project
avrg: 2D miR_HR1_12
Navigation
Downloads
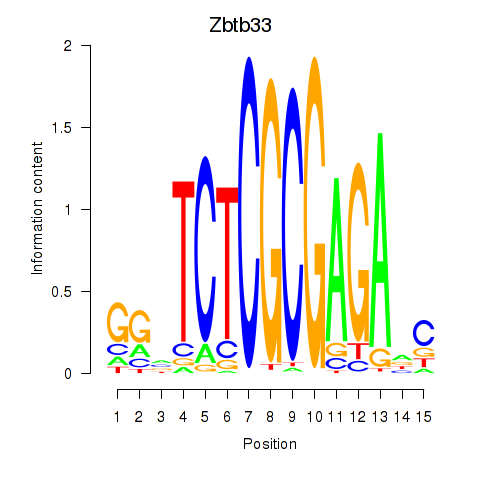
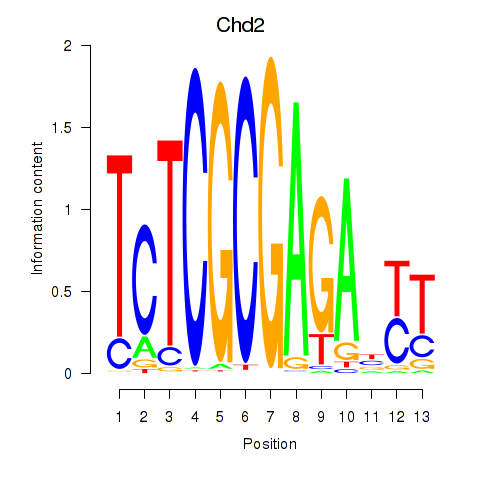
Results for Zbtb33_Chd2
Z-value: 6.81
Transcription factors associated with Zbtb33_Chd2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb33
|
ENSMUSG00000048047.3 | zinc finger and BTB domain containing 33 |
|
Chd2
|
ENSMUSG00000078671.4 | chromodomain helicase DNA binding protein 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb33 | mm10_v2_chrX_+_38189780_38189826 | 0.73 | 7.0e-03 | Click! |
| Chd2 | mm10_v2_chr7_-_73541738_73541758 | 0.72 | 8.1e-03 | Click! |
Activity profile of Zbtb33_Chd2 motif
Sorted Z-values of Zbtb33_Chd2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_34833631 | 16.44 |
ENSMUST00000093191.2
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr5_+_45669907 | 14.79 |
ENSMUST00000117396.1
|
Ncapg
|
non-SMC condensin I complex, subunit G |
| chr4_-_118437331 | 13.42 |
ENSMUST00000006565.6
|
Cdc20
|
cell division cycle 20 |
| chrX_+_151803642 | 12.97 |
ENSMUST00000156616.2
|
Huwe1
|
HECT, UBA and WWE domain containing 1 |
| chr14_-_57826128 | 12.28 |
ENSMUST00000022536.2
|
Ska3
|
spindle and kinetochore associated complex subunit 3 |
| chr19_+_38931008 | 11.73 |
ENSMUST00000145051.1
|
Hells
|
helicase, lymphoid specific |
| chr17_+_56040350 | 11.35 |
ENSMUST00000002914.8
|
Chaf1a
|
chromatin assembly factor 1, subunit A (p150) |
| chr4_+_24496434 | 11.13 |
ENSMUST00000108222.2
ENSMUST00000138567.2 ENSMUST00000050446.6 |
Mms22l
|
MMS22-like, DNA repair protein |
| chr19_+_38930909 | 10.43 |
ENSMUST00000025965.5
|
Hells
|
helicase, lymphoid specific |
| chr5_+_110286306 | 10.05 |
ENSMUST00000007296.5
ENSMUST00000112482.1 |
Pole
|
polymerase (DNA directed), epsilon |
| chr16_+_48994185 | 9.32 |
ENSMUST00000117994.1
ENSMUST00000048374.5 |
C330027C09Rik
|
RIKEN cDNA C330027C09 gene |
| chr15_+_99074968 | 8.72 |
ENSMUST00000039665.6
|
Troap
|
trophinin associated protein |
| chr3_+_69004969 | 7.79 |
ENSMUST00000136502.1
ENSMUST00000107803.1 |
Smc4
|
structural maintenance of chromosomes 4 |
| chr12_+_116405397 | 7.56 |
ENSMUST00000084828.3
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr3_+_69004711 | 7.49 |
ENSMUST00000042901.8
|
Smc4
|
structural maintenance of chromosomes 4 |
| chr5_-_110286159 | 6.59 |
ENSMUST00000031472.5
|
Pxmp2
|
peroxisomal membrane protein 2 |
| chr11_+_116671658 | 6.49 |
ENSMUST00000106378.1
ENSMUST00000144049.1 |
1810032O08Rik
|
RIKEN cDNA 1810032O08 gene |
| chr5_+_135778465 | 6.40 |
ENSMUST00000019323.6
|
Mdh2
|
malate dehydrogenase 2, NAD (mitochondrial) |
| chr12_-_72917760 | 6.20 |
ENSMUST00000110489.2
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr11_-_33163072 | 5.95 |
ENSMUST00000093201.6
ENSMUST00000101375.4 ENSMUST00000109354.3 ENSMUST00000075641.3 |
Npm1
|
nucleophosmin 1 |
| chr7_+_24884809 | 5.83 |
ENSMUST00000156372.1
ENSMUST00000124035.1 |
Rps19
|
ribosomal protein S19 |
| chr12_-_72917872 | 5.81 |
ENSMUST00000044000.5
|
4930447C04Rik
|
RIKEN cDNA 4930447C04 gene |
| chr10_-_88146867 | 5.77 |
ENSMUST00000164121.1
ENSMUST00000164803.1 ENSMUST00000168163.1 ENSMUST00000048518.9 |
Parpbp
|
PARP1 binding protein |
| chr3_+_105959369 | 5.66 |
ENSMUST00000010278.5
|
Wdr77
|
WD repeat domain 77 |
| chr5_+_30666886 | 5.47 |
ENSMUST00000144742.1
|
Cenpa
|
centromere protein A |
| chr11_-_73138245 | 5.39 |
ENSMUST00000052140.2
|
Gsg2
|
germ cell-specific gene 2 |
| chr6_+_91515928 | 4.93 |
ENSMUST00000040607.4
|
Lsm3
|
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr9_+_92542223 | 4.85 |
ENSMUST00000070522.7
ENSMUST00000160359.1 |
Plod2
|
procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr17_+_28328471 | 4.83 |
ENSMUST00000042334.8
|
Rpl10a
|
ribosomal protein L10A |
| chr7_+_24884611 | 4.65 |
ENSMUST00000108428.1
|
Rps19
|
ribosomal protein S19 |
| chr12_-_28635914 | 4.63 |
ENSMUST00000074267.3
|
Rps7
|
ribosomal protein S7 |
| chr9_-_64172879 | 4.59 |
ENSMUST00000176299.1
ENSMUST00000130127.1 ENSMUST00000176794.1 ENSMUST00000177045.1 |
Zwilch
|
zwilch kinetochore protein |
| chr3_-_54735001 | 4.50 |
ENSMUST00000153224.1
|
Exosc8
|
exosome component 8 |
| chr7_+_24884651 | 4.35 |
ENSMUST00000153451.2
ENSMUST00000108429.1 |
Rps19
|
ribosomal protein S19 |
| chr1_+_179803376 | 4.34 |
ENSMUST00000097454.2
|
Gm10518
|
predicted gene 10518 |
| chr11_+_98203059 | 4.29 |
ENSMUST00000107539.1
|
Cdk12
|
cyclin-dependent kinase 12 |
| chr5_+_33658567 | 4.26 |
ENSMUST00000114426.3
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr2_+_32961559 | 4.26 |
ENSMUST00000126610.1
|
Rpl12
|
ribosomal protein L12 |
| chr1_-_86359455 | 4.17 |
ENSMUST00000027438.6
|
Ncl
|
nucleolin |
| chr3_-_86142684 | 4.06 |
ENSMUST00000029722.6
|
Rps3a1
|
ribosomal protein S3A1 |
| chr6_+_86371489 | 4.01 |
ENSMUST00000089558.5
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr5_-_140321524 | 3.82 |
ENSMUST00000031534.6
|
Mad1l1
|
MAD1 mitotic arrest deficient 1-like 1 |
| chr3_-_88410295 | 3.75 |
ENSMUST00000056370.7
|
Pmf1
|
polyamine-modulated factor 1 |
| chr4_+_152039315 | 3.65 |
ENSMUST00000084116.6
ENSMUST00000105663.1 ENSMUST00000103197.3 |
Nol9
|
nucleolar protein 9 |
| chr7_-_118116171 | 3.58 |
ENSMUST00000131374.1
|
Rps15a
|
ribosomal protein S15A |
| chr9_+_64281575 | 3.56 |
ENSMUST00000034964.6
|
Tipin
|
timeless interacting protein |
| chr1_+_134962553 | 3.55 |
ENSMUST00000027687.7
|
Ube2t
|
ubiquitin-conjugating enzyme E2T (putative) |
| chr11_+_74770822 | 3.52 |
ENSMUST00000141755.1
ENSMUST00000010698.6 |
Mettl16
|
methyltransferase like 16 |
| chr18_+_75000469 | 3.52 |
ENSMUST00000079716.5
|
Rpl17
|
ribosomal protein L17 |
| chr13_+_63815240 | 3.52 |
ENSMUST00000021926.5
ENSMUST00000095724.3 ENSMUST00000143449.1 ENSMUST00000067821.5 |
Ercc6l2
|
excision repair cross-complementing rodent repair deficiency, complementation group 6 like 2 |
| chr7_-_48881032 | 3.50 |
ENSMUST00000058745.8
|
E2f8
|
E2F transcription factor 8 |
| chr7_-_118116128 | 3.48 |
ENSMUST00000128482.1
ENSMUST00000131840.1 |
Rps15a
|
ribosomal protein S15A |
| chr17_-_46247968 | 3.45 |
ENSMUST00000142706.2
ENSMUST00000173349.1 ENSMUST00000087026.6 |
Polr1c
|
polymerase (RNA) I polypeptide C |
| chr4_-_41464816 | 3.43 |
ENSMUST00000108055.2
ENSMUST00000154535.1 ENSMUST00000030148.5 |
Kif24
|
kinesin family member 24 |
| chr2_+_3336159 | 3.36 |
ENSMUST00000115089.1
|
Acbd7
|
acyl-Coenzyme A binding domain containing 7 |
| chr5_+_123907175 | 3.33 |
ENSMUST00000023869.8
|
Denr
|
density-regulated protein |
| chr16_-_4559720 | 3.29 |
ENSMUST00000005862.7
|
Tfap4
|
transcription factor AP4 |
| chr18_+_57878620 | 3.21 |
ENSMUST00000115366.2
|
Slc12a2
|
solute carrier family 12, member 2 |
| chr7_-_80901220 | 3.21 |
ENSMUST00000146402.1
ENSMUST00000026816.8 |
Wdr73
|
WD repeat domain 73 |
| chr1_-_179803625 | 3.18 |
ENSMUST00000027768.7
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr8_-_105851981 | 3.15 |
ENSMUST00000040776.4
|
Cenpt
|
centromere protein T |
| chr2_+_69897255 | 3.13 |
ENSMUST00000131553.1
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr4_-_9669068 | 3.08 |
ENSMUST00000078139.6
ENSMUST00000108340.2 ENSMUST00000084915.4 ENSMUST00000108337.1 ENSMUST00000084912.5 ENSMUST00000038564.6 ENSMUST00000146441.1 ENSMUST00000098275.2 |
Asph
|
aspartate-beta-hydroxylase |
| chr2_-_125123618 | 3.05 |
ENSMUST00000142718.1
ENSMUST00000152367.1 ENSMUST00000067780.3 ENSMUST00000147105.1 |
Myef2
|
myelin basic protein expression factor 2, repressor |
| chr9_+_44773191 | 2.91 |
ENSMUST00000147559.1
|
Ift46
|
intraflagellar transport 46 |
| chr2_-_39005574 | 2.89 |
ENSMUST00000080861.5
|
Rpl35
|
ribosomal protein L35 |
| chr8_+_123518835 | 2.88 |
ENSMUST00000093043.5
|
Gas8
|
growth arrest specific 8 |
| chr5_-_34660068 | 2.87 |
ENSMUST00000041364.9
|
Nop14
|
NOP14 nucleolar protein |
| chr16_-_17125106 | 2.83 |
ENSMUST00000093336.6
|
2610318N02Rik
|
RIKEN cDNA 2610318N02 gene |
| chr11_-_78183551 | 2.79 |
ENSMUST00000102483.4
|
Rpl23a
|
ribosomal protein L23A |
| chr14_+_57826210 | 2.77 |
ENSMUST00000022538.3
|
Mrp63
|
mitochondrial ribosomal protein 63 |
| chrX_+_106015699 | 2.77 |
ENSMUST00000033582.4
|
Cox7b
|
cytochrome c oxidase subunit VIIb |
| chr6_+_83326071 | 2.72 |
ENSMUST00000038658.8
ENSMUST00000101245.2 |
Mob1a
|
MOB kinase activator 1A |
| chr1_+_130717320 | 2.70 |
ENSMUST00000049813.4
|
Yod1
|
YOD1 OTU deubiquitinating enzyme 1 homologue (S. cerevisiae) |
| chr4_+_116685544 | 2.70 |
ENSMUST00000135573.1
ENSMUST00000151129.1 |
Prdx1
|
peroxiredoxin 1 |
| chr4_-_86857365 | 2.69 |
ENSMUST00000102814.4
|
Rps6
|
ribosomal protein S6 |
| chr5_+_33658123 | 2.69 |
ENSMUST00000074849.6
ENSMUST00000079534.4 |
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chrX_-_8074720 | 2.69 |
ENSMUST00000115636.3
ENSMUST00000115638.3 |
Suv39h1
|
suppressor of variegation 3-9 homolog 1 (Drosophila) |
| chr6_+_83326016 | 2.67 |
ENSMUST00000055261.4
|
Mob1a
|
MOB kinase activator 1A |
| chr17_-_33824346 | 2.64 |
ENSMUST00000173879.1
ENSMUST00000166693.2 ENSMUST00000173019.1 ENSMUST00000087342.6 ENSMUST00000173844.1 |
Rps28
|
ribosomal protein S28 |
| chr17_+_28691419 | 2.61 |
ENSMUST00000124886.1
|
Mapk14
|
mitogen-activated protein kinase 14 |
| chr9_+_14784638 | 2.57 |
ENSMUST00000034405.4
|
Mre11a
|
meiotic recombination 11 homolog A (S. cerevisiae) |
| chr2_+_26910747 | 2.57 |
ENSMUST00000102898.4
|
Rpl7a
|
ribosomal protein L7A |
| chr8_+_25754492 | 2.52 |
ENSMUST00000167899.1
|
Gm17484
|
predicted gene, 17484 |
| chr9_+_107950952 | 2.51 |
ENSMUST00000049348.3
|
Traip
|
TRAF-interacting protein |
| chr5_+_33658550 | 2.49 |
ENSMUST00000152847.1
|
Tacc3
|
transforming, acidic coiled-coil containing protein 3 |
| chr4_+_126677630 | 2.44 |
ENSMUST00000030642.2
|
Psmb2
|
proteasome (prosome, macropain) subunit, beta type 2 |
| chr14_-_99099701 | 2.41 |
ENSMUST00000042471.9
|
Dis3
|
DIS3 mitotic control homolog (S. cerevisiae) |
| chr4_+_116685859 | 2.40 |
ENSMUST00000129315.1
ENSMUST00000106470.1 |
Prdx1
|
peroxiredoxin 1 |
| chr7_+_81762947 | 2.39 |
ENSMUST00000133034.1
|
Fam103a1
|
family with sequence similarity 103, member A1 |
| chr4_+_107434617 | 2.39 |
ENSMUST00000135835.1
ENSMUST00000046005.2 |
Glis1
|
GLIS family zinc finger 1 |
| chr15_-_102624922 | 2.38 |
ENSMUST00000183765.1
|
ATF7
|
Cyclic AMP-dependent transcription factor ATF-7 |
| chrX_+_13071470 | 2.33 |
ENSMUST00000169594.2
|
Usp9x
|
ubiquitin specific peptidase 9, X chromosome |
| chr10_+_88146992 | 2.30 |
ENSMUST00000052355.7
|
Nup37
|
nucleoporin 37 |
| chr7_+_13278778 | 2.27 |
ENSMUST00000098814.4
ENSMUST00000146998.1 ENSMUST00000185145.1 |
Lig1
|
ligase I, DNA, ATP-dependent |
| chr15_-_102625061 | 2.27 |
ENSMUST00000184077.1
ENSMUST00000184906.1 ENSMUST00000169033.1 |
ATF7
|
Cyclic AMP-dependent transcription factor ATF-7 |
| chr3_-_69044697 | 2.26 |
ENSMUST00000136512.1
ENSMUST00000143454.1 ENSMUST00000107802.1 |
Trim59
|
tripartite motif-containing 59 |
| chr10_+_11149449 | 2.25 |
ENSMUST00000054814.7
ENSMUST00000159541.1 |
Shprh
|
SNF2 histone linker PHD RING helicase |
| chr1_-_119837613 | 2.25 |
ENSMUST00000064091.5
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr1_-_119837338 | 2.24 |
ENSMUST00000163435.1
|
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr15_+_76343504 | 2.23 |
ENSMUST00000023210.6
|
Cyc1
|
cytochrome c-1 |
| chr16_-_87495704 | 2.22 |
ENSMUST00000176750.1
ENSMUST00000175977.1 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr8_-_46211284 | 2.19 |
ENSMUST00000034049.4
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4 |
| chr1_-_128417352 | 2.15 |
ENSMUST00000027602.8
ENSMUST00000064309.7 |
Dars
|
aspartyl-tRNA synthetase |
| chr11_+_115603920 | 2.14 |
ENSMUST00000058109.8
|
Mrps7
|
mitchondrial ribosomal protein S7 |
| chr7_-_45124355 | 2.14 |
ENSMUST00000003521.8
|
Rps11
|
ribosomal protein S11 |
| chr5_-_77310049 | 2.11 |
ENSMUST00000047860.8
|
Noa1
|
nitric oxide associated 1 |
| chr10_+_88147061 | 2.10 |
ENSMUST00000169309.1
|
Nup37
|
nucleoporin 37 |
| chr16_-_48993931 | 2.10 |
ENSMUST00000114516.1
|
Dzip3
|
DAZ interacting protein 3, zinc finger |
| chr3_+_137864487 | 2.10 |
ENSMUST00000041045.7
|
H2afz
|
H2A histone family, member Z |
| chr11_+_72689997 | 2.07 |
ENSMUST00000155998.1
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chrX_-_164076482 | 2.07 |
ENSMUST00000134272.1
|
Siah1b
|
seven in absentia 1B |
| chrX_-_164076100 | 2.05 |
ENSMUST00000037928.2
ENSMUST00000071667.2 |
Siah1b
|
seven in absentia 1B |
| chr16_-_87495823 | 2.01 |
ENSMUST00000176041.1
ENSMUST00000026704.7 |
Cct8
|
chaperonin containing Tcp1, subunit 8 (theta) |
| chr15_-_31601506 | 1.99 |
ENSMUST00000161266.1
|
Cct5
|
chaperonin containing Tcp1, subunit 5 (epsilon) |
| chr12_-_91384403 | 1.98 |
ENSMUST00000141429.1
|
Cep128
|
centrosomal protein 128 |
| chr9_+_121719403 | 1.98 |
ENSMUST00000182225.1
|
Nktr
|
natural killer tumor recognition sequence |
| chr6_+_120666388 | 1.98 |
ENSMUST00000112686.1
|
Cecr2
|
cat eye syndrome chromosome region, candidate 2 |
| chr3_+_152210458 | 1.98 |
ENSMUST00000166984.1
ENSMUST00000106121.1 |
Fubp1
|
far upstream element (FUSE) binding protein 1 |
| chr7_+_96210107 | 1.97 |
ENSMUST00000138760.1
|
Tenm4
|
teneurin transmembrane protein 4 |
| chr9_-_44251464 | 1.94 |
ENSMUST00000034618.4
|
Pdzd3
|
PDZ domain containing 3 |
| chr2_-_119477613 | 1.93 |
ENSMUST00000110808.1
ENSMUST00000049920.7 |
Ino80
|
INO80 homolog (S. cerevisiae) |
| chr3_+_88336256 | 1.92 |
ENSMUST00000001451.5
|
Smg5
|
Smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans) |
| chr8_+_70315759 | 1.92 |
ENSMUST00000165819.2
ENSMUST00000140239.1 |
Gdf1
Cers1
|
growth differentiation factor 1 ceramide synthase 1 |
| chr11_-_98193260 | 1.92 |
ENSMUST00000092735.5
ENSMUST00000107545.2 |
Med1
|
mediator complex subunit 1 |
| chr3_-_122984404 | 1.90 |
ENSMUST00000090379.2
|
Usp53
|
ubiquitin specific peptidase 53 |
| chr8_-_31168699 | 1.88 |
ENSMUST00000033983.4
|
Mak16
|
MAK16 homolog (S. cerevisiae) |
| chr17_-_83631892 | 1.85 |
ENSMUST00000051482.1
|
Kcng3
|
potassium voltage-gated channel, subfamily G, member 3 |
| chr9_+_14784660 | 1.85 |
ENSMUST00000115632.3
ENSMUST00000147305.1 |
Mre11a
|
meiotic recombination 11 homolog A (S. cerevisiae) |
| chr1_+_160195215 | 1.84 |
ENSMUST00000135680.1
ENSMUST00000097193.2 |
Mrps14
|
mitochondrial ribosomal protein S14 |
| chr3_-_107333289 | 1.83 |
ENSMUST00000061772.9
|
Rbm15
|
RNA binding motif protein 15 |
| chr1_+_59684949 | 1.83 |
ENSMUST00000027174.3
|
Nop58
|
NOP58 ribonucleoprotein |
| chr19_+_30030589 | 1.81 |
ENSMUST00000112552.1
|
Uhrf2
|
ubiquitin-like, containing PHD and RING finger domains 2 |
| chrX_-_7947553 | 1.80 |
ENSMUST00000133349.1
|
Hdac6
|
histone deacetylase 6 |
| chr1_-_119836999 | 1.79 |
ENSMUST00000163621.1
ENSMUST00000168303.1 |
Ptpn4
|
protein tyrosine phosphatase, non-receptor type 4 |
| chr3_+_69721985 | 1.78 |
ENSMUST00000029358.8
|
Nmd3
|
NMD3 homolog (S. cerevisiae) |
| chr9_+_99575776 | 1.78 |
ENSMUST00000066650.5
ENSMUST00000148987.1 |
Dbr1
|
debranching enzyme homolog 1 (S. cerevisiae) |
| chr2_+_144270900 | 1.77 |
ENSMUST00000028910.2
ENSMUST00000110027.1 |
Mgme1
|
mitochondrial genome maintainance exonuclease 1 |
| chr14_+_30549131 | 1.76 |
ENSMUST00000022529.6
|
Tkt
|
transketolase |
| chr7_+_109519139 | 1.75 |
ENSMUST00000143107.1
|
Rpl27a
|
ribosomal protein L27A |
| chr14_-_105176860 | 1.73 |
ENSMUST00000163545.1
|
Rbm26
|
RNA binding motif protein 26 |
| chr9_+_44773027 | 1.72 |
ENSMUST00000125877.1
|
Ift46
|
intraflagellar transport 46 |
| chrX_-_7947763 | 1.71 |
ENSMUST00000154244.1
|
Hdac6
|
histone deacetylase 6 |
| chr7_-_131410325 | 1.71 |
ENSMUST00000154602.1
|
Ikzf5
|
IKAROS family zinc finger 5 |
| chr4_-_155761042 | 1.70 |
ENSMUST00000030903.5
|
Atad3a
|
ATPase family, AAA domain containing 3A |
| chr3_+_137864573 | 1.69 |
ENSMUST00000174561.1
ENSMUST00000173790.1 |
H2afz
|
H2A histone family, member Z |
| chr11_-_107189325 | 1.68 |
ENSMUST00000018577.7
ENSMUST00000106757.1 |
Nol11
|
nucleolar protein 11 |
| chr2_+_60209887 | 1.66 |
ENSMUST00000102748.4
ENSMUST00000102747.1 |
March7
|
membrane-associated ring finger (C3HC4) 7 |
| chr8_-_94838255 | 1.66 |
ENSMUST00000161762.1
ENSMUST00000162538.1 |
Ciapin1
|
cytokine induced apoptosis inhibitor 1 |
| chr7_-_46710642 | 1.65 |
ENSMUST00000143082.1
|
Saal1
|
serum amyloid A-like 1 |
| chr9_+_64173364 | 1.65 |
ENSMUST00000034966.7
|
Rpl4
|
ribosomal protein L4 |
| chr5_+_129787390 | 1.65 |
ENSMUST00000031402.8
|
Cct6a
|
chaperonin containing Tcp1, subunit 6a (zeta) |
| chr14_-_60251473 | 1.62 |
ENSMUST00000041905.6
|
Nupl1
|
nucleoporin like 1 |
| chr15_-_85131949 | 1.62 |
ENSMUST00000023068.6
|
Smc1b
|
structural maintenance of chromosomes 1B |
| chr9_+_66350465 | 1.60 |
ENSMUST00000042824.6
|
Herc1
|
hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 |
| chr3_-_105687552 | 1.60 |
ENSMUST00000090680.6
|
Ddx20
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 |
| chr5_-_117389029 | 1.59 |
ENSMUST00000111953.1
ENSMUST00000086461.6 |
Rfc5
|
replication factor C (activator 1) 5 |
| chr15_-_31601786 | 1.57 |
ENSMUST00000022842.8
|
Cct5
|
chaperonin containing Tcp1, subunit 5 (epsilon) |
| chr5_-_100429503 | 1.56 |
ENSMUST00000181873.1
ENSMUST00000180779.1 |
5430416N02Rik
|
RIKEN cDNA 5430416N02 gene |
| chr11_+_90638127 | 1.56 |
ENSMUST00000020851.8
|
Cox11
|
cytochrome c oxidase assembly protein 11 |
| chr19_+_44333092 | 1.55 |
ENSMUST00000058856.8
|
Scd4
|
stearoyl-coenzyme A desaturase 4 |
| chr6_-_30509706 | 1.53 |
ENSMUST00000064330.6
ENSMUST00000102991.2 ENSMUST00000115157.1 ENSMUST00000148638.1 |
Tmem209
|
transmembrane protein 209 |
| chr13_-_111490028 | 1.52 |
ENSMUST00000091236.4
|
Gpbp1
|
GC-rich promoter binding protein 1 |
| chr3_-_69004475 | 1.51 |
ENSMUST00000154741.1
ENSMUST00000148031.1 |
Ift80
|
intraflagellar transport 80 |
| chr8_-_84969412 | 1.48 |
ENSMUST00000147812.1
|
Rnaseh2a
|
ribonuclease H2, large subunit |
| chr10_-_85957775 | 1.48 |
ENSMUST00000001834.3
|
Rtcb
|
RNA 2',3'-cyclic phosphate and 5'-OH ligase |
| chr19_+_58943413 | 1.47 |
ENSMUST00000054280.6
|
Eno4
|
enolase 4 |
| chr16_+_13256481 | 1.46 |
ENSMUST00000009713.7
ENSMUST00000115809.1 |
Mkl2
|
MKL/myocardin-like 2 |
| chr14_-_105177263 | 1.45 |
ENSMUST00000163499.1
|
Rbm26
|
RNA binding motif protein 26 |
| chr11_-_88864534 | 1.44 |
ENSMUST00000018572.4
|
Akap1
|
A kinase (PRKA) anchor protein 1 |
| chr11_+_78032346 | 1.42 |
ENSMUST00000122342.1
ENSMUST00000092881.3 |
Dhrs13
|
dehydrogenase/reductase (SDR family) member 13 |
| chr10_-_93589621 | 1.40 |
ENSMUST00000020203.6
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr17_-_42876417 | 1.39 |
ENSMUST00000024709.7
|
Cd2ap
|
CD2-associated protein |
| chr2_-_6130117 | 1.36 |
ENSMUST00000126551.1
ENSMUST00000054254.5 ENSMUST00000114942.2 |
Proser2
|
proline and serine rich 2 |
| chrX_+_42151002 | 1.36 |
ENSMUST00000123245.1
|
Stag2
|
stromal antigen 2 |
| chr14_-_105177280 | 1.35 |
ENSMUST00000100327.3
ENSMUST00000022715.7 |
Rbm26
|
RNA binding motif protein 26 |
| chr5_+_88764983 | 1.34 |
ENSMUST00000031311.9
|
Dck
|
deoxycytidine kinase |
| chr11_-_119300070 | 1.33 |
ENSMUST00000026667.8
|
Eif4a3
|
eukaryotic translation initiation factor 4A3 |
| chr14_+_57999305 | 1.32 |
ENSMUST00000180534.1
|
3110083C13Rik
|
RIKEN cDNA 3110083C13 gene |
| chr2_-_151009364 | 1.31 |
ENSMUST00000109896.1
|
Ninl
|
ninein-like |
| chr2_-_151039363 | 1.29 |
ENSMUST00000128627.1
ENSMUST00000066640.4 |
Ninl
Nanp
|
ninein-like N-acetylneuraminic acid phosphatase |
| chr2_+_24949747 | 1.28 |
ENSMUST00000028350.3
|
Zmynd19
|
zinc finger, MYND domain containing 19 |
| chr2_-_10080322 | 1.28 |
ENSMUST00000145530.1
ENSMUST00000026887.7 ENSMUST00000114896.1 ENSMUST00000114897.2 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr9_-_53667429 | 1.28 |
ENSMUST00000166367.1
ENSMUST00000034529.7 |
Cul5
|
cullin 5 |
| chr4_-_62525036 | 1.27 |
ENSMUST00000030091.3
|
Pole3
|
polymerase (DNA directed), epsilon 3 (p17 subunit) |
| chr19_-_27429807 | 1.27 |
ENSMUST00000076219.4
|
D19Bwg1357e
|
DNA segment, Chr 19, Brigham & Women's Genetics 1357 expressed |
| chr5_+_53590215 | 1.27 |
ENSMUST00000037618.6
|
Rbpj
|
recombination signal binding protein for immunoglobulin kappa J region |
| chr10_-_67285180 | 1.26 |
ENSMUST00000159002.1
ENSMUST00000077839.6 |
Nrbf2
|
nuclear receptor binding factor 2 |
| chr2_-_34870921 | 1.23 |
ENSMUST00000028225.5
|
Psmd5
|
proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 |
| chr5_-_136135989 | 1.22 |
ENSMUST00000150406.1
ENSMUST00000006301.4 |
Lrwd1
|
leucine-rich repeats and WD repeat domain containing 1 |
| chr6_-_125166463 | 1.21 |
ENSMUST00000117757.2
ENSMUST00000073605.8 |
Gapdh
|
glyceraldehyde-3-phosphate dehydrogenase |
| chr1_+_58393119 | 1.21 |
ENSMUST00000050552.8
|
Bzw1
|
basic leucine zipper and W2 domains 1 |
| chr10_-_80039674 | 1.18 |
ENSMUST00000004786.9
|
Polr2e
|
polymerase (RNA) II (DNA directed) polypeptide E |
| chr2_-_10080055 | 1.17 |
ENSMUST00000130067.1
ENSMUST00000139810.1 |
Atp5c1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 |
| chr5_-_124425572 | 1.17 |
ENSMUST00000168651.1
|
Sbno1
|
sno, strawberry notch homolog 1 (Drosophila) |
| chr8_-_121652895 | 1.17 |
ENSMUST00000046386.4
|
Zcchc14
|
zinc finger, CCHC domain containing 14 |
| chr9_-_62811592 | 1.17 |
ENSMUST00000034775.8
|
Fem1b
|
feminization 1 homolog b (C. elegans) |
| chr7_+_127511976 | 1.15 |
ENSMUST00000098025.4
|
Srcap
|
Snf2-related CREBBP activator protein |
| chr11_-_53430779 | 1.14 |
ENSMUST00000061326.4
ENSMUST00000109021.3 |
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr15_-_98567630 | 1.14 |
ENSMUST00000012104.6
|
Ccnt1
|
cyclin T1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb33_Chd2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 4.0 | 12.0 | GO:0010705 | meiotic DNA double-strand break processing involved in reciprocal meiotic recombination(GO:0010705) |
| 3.7 | 14.8 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 3.4 | 13.4 | GO:0098763 | mitotic cell cycle phase(GO:0098763) |
| 3.4 | 10.1 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 3.2 | 9.6 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 3.2 | 22.2 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 2.0 | 30.3 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 1.5 | 4.5 | GO:0070846 | misfolded protein transport(GO:0070843) polyubiquitinated protein transport(GO:0070844) polyubiquitinated misfolded protein transport(GO:0070845) Hsp90 deacetylation(GO:0070846) |
| 1.5 | 4.5 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 1.5 | 25.1 | GO:0071459 | protein localization to chromosome, centromeric region(GO:0071459) |
| 1.2 | 4.8 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 1.2 | 3.5 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 1.2 | 3.5 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 1.1 | 3.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 1.0 | 3.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 1.0 | 3.8 | GO:0090235 | regulation of metaphase plate congression(GO:0090235) |
| 0.9 | 3.7 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.9 | 6.4 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.9 | 2.7 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.9 | 4.4 | GO:0031860 | telomeric 3' overhang formation(GO:0031860) |
| 0.9 | 1.7 | GO:1901189 | regulation of cell adhesion involved in heart morphogenesis(GO:0061344) positive regulation of ephrin receptor signaling pathway(GO:1901189) |
| 0.9 | 9.4 | GO:1903405 | protein localization to nuclear body(GO:1903405) protein localization to Cajal body(GO:1904867) regulation of protein localization to Cajal body(GO:1904869) positive regulation of protein localization to Cajal body(GO:1904871) protein localization to nucleoplasm(GO:1990173) |
| 0.8 | 3.3 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.8 | 2.3 | GO:1904569 | regulation of selenocysteine incorporation(GO:1904569) |
| 0.7 | 2.2 | GO:0015866 | ADP transport(GO:0015866) |
| 0.7 | 5.7 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.7 | 2.7 | GO:0071043 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.7 | 2.0 | GO:0060912 | cardiac cell fate specification(GO:0060912) |
| 0.7 | 4.6 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) |
| 0.6 | 1.9 | GO:0072720 | cellular response to UV-A(GO:0071492) response to dithiothreitol(GO:0072720) |
| 0.6 | 9.4 | GO:0030953 | astral microtubule organization(GO:0030953) |
| 0.6 | 1.8 | GO:0046166 | glyceraldehyde-3-phosphate biosynthetic process(GO:0046166) |
| 0.6 | 6.4 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.6 | 2.3 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.5 | 2.7 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.5 | 2.6 | GO:0033140 | negative regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033140) |
| 0.5 | 4.2 | GO:2000232 | regulation of rRNA processing(GO:2000232) |
| 0.5 | 1.0 | GO:0090666 | scaRNA localization to Cajal body(GO:0090666) |
| 0.5 | 3.6 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.5 | 7.6 | GO:0030261 | chromosome condensation(GO:0030261) |
| 0.5 | 12.0 | GO:0045005 | replication fork processing(GO:0031297) DNA-dependent DNA replication maintenance of fidelity(GO:0045005) |
| 0.5 | 1.9 | GO:0021698 | cerebellar cortex structural organization(GO:0021698) |
| 0.5 | 1.9 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.5 | 4.3 | GO:0070475 | rRNA base methylation(GO:0070475) |
| 0.4 | 2.7 | GO:2001184 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.4 | 1.8 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.4 | 5.8 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.4 | 1.8 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.4 | 12.2 | GO:0000470 | maturation of LSU-rRNA(GO:0000470) |
| 0.4 | 2.1 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.4 | 3.7 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.4 | 1.2 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.4 | 1.5 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.4 | 1.9 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.4 | 2.7 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.4 | 2.2 | GO:0033762 | response to glucagon(GO:0033762) mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.4 | 6.5 | GO:0070816 | phosphorylation of RNA polymerase II C-terminal domain(GO:0070816) |
| 0.4 | 2.9 | GO:1904526 | regulation of microtubule binding(GO:1904526) |
| 0.4 | 11.4 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.4 | 1.1 | GO:2000819 | regulation of nucleotide-excision repair(GO:2000819) |
| 0.3 | 7.0 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.3 | 2.1 | GO:0032439 | endosome localization(GO:0032439) positive regulation of pinocytosis(GO:0048549) |
| 0.3 | 1.0 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.3 | 1.3 | GO:0046125 | pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.3 | 0.7 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.3 | 1.0 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.3 | 1.9 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.3 | 0.9 | GO:0060220 | eye photoreceptor cell fate commitment(GO:0042706) photoreceptor cell fate commitment(GO:0046552) camera-type eye photoreceptor cell fate commitment(GO:0060220) |
| 0.3 | 3.4 | GO:0007019 | microtubule depolymerization(GO:0007019) |
| 0.3 | 1.2 | GO:0071169 | establishment of protein localization to chromatin(GO:0071169) |
| 0.3 | 1.8 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.3 | 3.7 | GO:0036297 | interstrand cross-link repair(GO:0036297) |
| 0.3 | 3.3 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.3 | 11.4 | GO:0006284 | base-excision repair(GO:0006284) |
| 0.3 | 1.5 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.3 | 6.6 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.3 | 0.3 | GO:0072402 | response to cell cycle checkpoint signaling(GO:0072396) response to DNA integrity checkpoint signaling(GO:0072402) response to DNA damage checkpoint signaling(GO:0072423) |
| 0.3 | 1.3 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.3 | 1.8 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.3 | 7.0 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.2 | 1.0 | GO:0036367 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.2 | 1.4 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.2 | 3.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.2 | 1.6 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.2 | 1.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.2 | 2.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.2 | 1.5 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.2 | 0.8 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.2 | 4.7 | GO:0060285 | cilium or flagellum-dependent cell motility(GO:0001539) cilium-dependent cell motility(GO:0060285) |
| 0.2 | 1.4 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.2 | 1.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.2 | 5.9 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.2 | 1.6 | GO:1900262 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.2 | 1.4 | GO:0032876 | negative regulation of DNA endoreduplication(GO:0032876) |
| 0.2 | 1.9 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.2 | 1.1 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.2 | 1.3 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.2 | 0.6 | GO:0001887 | selenium compound metabolic process(GO:0001887) selenocysteine metabolic process(GO:0016259) |
| 0.2 | 0.5 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.2 | 0.9 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.2 | 1.9 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.2 | 2.5 | GO:0010804 | negative regulation of tumor necrosis factor-mediated signaling pathway(GO:0010804) |
| 0.2 | 2.0 | GO:0001842 | neural fold formation(GO:0001842) |
| 0.2 | 1.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.2 | 0.5 | GO:0036090 | cleavage furrow ingression(GO:0036090) |
| 0.2 | 0.6 | GO:0000460 | maturation of 5.8S rRNA(GO:0000460) |
| 0.1 | 1.8 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.1 | 1.2 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.1 | 0.7 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.1 | 0.4 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.1 | 7.5 | GO:0002181 | cytoplasmic translation(GO:0002181) |
| 0.1 | 2.5 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.1 | 5.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.1 | 0.4 | GO:0043396 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) positive regulation of corticotropin-releasing hormone secretion(GO:0051466) |
| 0.1 | 9.4 | GO:2000179 | positive regulation of neural precursor cell proliferation(GO:2000179) |
| 0.1 | 12.3 | GO:0031110 | regulation of microtubule polymerization or depolymerization(GO:0031110) |
| 0.1 | 2.3 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.1 | 1.5 | GO:0006388 | tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.1 | 1.6 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.7 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 1.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.1 | 1.3 | GO:0021942 | radial glia guided migration of Purkinje cell(GO:0021942) |
| 0.1 | 0.4 | GO:1903011 | sequestering of BMP in extracellular matrix(GO:0035582) negative regulation of bone development(GO:1903011) |
| 0.1 | 1.6 | GO:0021702 | cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.1 | 0.7 | GO:0050861 | positive regulation of B cell receptor signaling pathway(GO:0050861) |
| 0.1 | 0.6 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.1 | 0.3 | GO:0002946 | tRNA C5-cytosine methylation(GO:0002946) |
| 0.1 | 3.5 | GO:1900087 | positive regulation of G1/S transition of mitotic cell cycle(GO:1900087) |
| 0.1 | 3.6 | GO:0030490 | maturation of SSU-rRNA(GO:0030490) |
| 0.1 | 0.6 | GO:0006265 | resolution of meiotic recombination intermediates(GO:0000712) DNA topological change(GO:0006265) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.9 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.1 | 1.7 | GO:0002643 | regulation of tolerance induction(GO:0002643) |
| 0.1 | 0.8 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 3.0 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.1 | 0.9 | GO:0006337 | nucleosome disassembly(GO:0006337) |
| 0.1 | 1.7 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.4 | GO:0072710 | response to hydroxyurea(GO:0072710) cellular response to hydroxyurea(GO:0072711) |
| 0.1 | 0.5 | GO:0032377 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.1 | 0.8 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.1 | 0.3 | GO:0010286 | heat acclimation(GO:0010286) |
| 0.1 | 2.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.1 | 0.5 | GO:0051415 | interphase microtubule nucleation by interphase microtubule organizing center(GO:0051415) microtubule nucleation by microtubule organizing center(GO:0051418) |
| 0.1 | 0.5 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.1 | 2.2 | GO:0000184 | nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:0000184) |
| 0.1 | 0.5 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.1 | 0.7 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.4 | GO:0061034 | olfactory bulb mitral cell layer development(GO:0061034) |
| 0.1 | 2.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.1 | 0.7 | GO:0061032 | visceral serous pericardium development(GO:0061032) |
| 0.1 | 0.5 | GO:1900364 | regulation of mRNA polyadenylation(GO:1900363) negative regulation of mRNA polyadenylation(GO:1900364) |
| 0.1 | 1.9 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.7 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.1 | 0.3 | GO:0051096 | positive regulation of helicase activity(GO:0051096) |
| 0.1 | 3.9 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.1 | 1.0 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.5 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 2.2 | GO:0006418 | tRNA aminoacylation for protein translation(GO:0006418) |
| 0.0 | 0.3 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.7 | GO:0019430 | removal of superoxide radicals(GO:0019430) |
| 0.0 | 0.8 | GO:0006744 | ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.5 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 3.6 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 1.3 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.2 | GO:0031145 | anaphase-promoting complex-dependent catabolic process(GO:0031145) |
| 0.0 | 0.8 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.9 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.5 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.6 | GO:0007250 | activation of NF-kappaB-inducing kinase activity(GO:0007250) |
| 0.0 | 0.1 | GO:1904742 | regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.5 | GO:0060416 | response to growth hormone(GO:0060416) |
| 0.0 | 0.3 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.2 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) |
| 0.0 | 0.8 | GO:0051567 | histone H3-K9 methylation(GO:0051567) |
| 0.0 | 0.5 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 1.2 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 3.6 | GO:0042787 | protein ubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0042787) |
| 0.0 | 1.5 | GO:0045844 | positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.0 | 5.3 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 1.9 | GO:0010923 | negative regulation of phosphatase activity(GO:0010923) |
| 0.0 | 0.1 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.5 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 1.3 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.3 | GO:0034453 | microtubule anchoring(GO:0034453) |
| 0.0 | 0.5 | GO:0003170 | heart valve development(GO:0003170) |
| 0.0 | 0.3 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 1.0 | GO:0042990 | regulation of transcription factor import into nucleus(GO:0042990) |
| 0.0 | 0.3 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.2 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 1.5 | GO:0007093 | mitotic cell cycle checkpoint(GO:0007093) |
| 0.0 | 0.9 | GO:0030317 | sperm motility(GO:0030317) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.3 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.2 | GO:0042407 | cristae formation(GO:0042407) |
| 0.0 | 0.8 | GO:0032436 | positive regulation of proteasomal ubiquitin-dependent protein catabolic process(GO:0032436) |
| 0.0 | 2.4 | GO:0000377 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:0072395 | signal transduction involved in cell cycle checkpoint(GO:0072395) signal transduction involved in DNA integrity checkpoint(GO:0072401) signal transduction involved in DNA damage checkpoint(GO:0072422) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.8 | 11.4 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 3.8 | 37.6 | GO:0000796 | condensin complex(GO:0000796) |
| 3.7 | 11.1 | GO:0035101 | FACT complex(GO:0035101) |
| 2.3 | 11.3 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 2.2 | 28.7 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 1.5 | 4.6 | GO:1990423 | RZZ complex(GO:1990423) |
| 1.4 | 4.3 | GO:0002944 | cyclin K-CDK12 complex(GO:0002944) |
| 1.4 | 5.5 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 1.2 | 10.5 | GO:0000801 | central element(GO:0000801) |
| 1.1 | 3.2 | GO:0031533 | mRNA cap methyltransferase complex(GO:0031533) |
| 0.9 | 3.7 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.9 | 6.0 | GO:0001652 | granular component(GO:0001652) |
| 0.8 | 4.2 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.8 | 39.9 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.8 | 7.6 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.7 | 24.6 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.7 | 4.4 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.7 | 7.8 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.7 | 7.1 | GO:0034709 | methylosome(GO:0034709) |
| 0.7 | 4.9 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.7 | 9.4 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.6 | 3.8 | GO:0001740 | Barr body(GO:0001740) |
| 0.6 | 3.6 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.6 | 3.6 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.6 | 2.9 | GO:0030689 | Noc complex(GO:0030689) |
| 0.6 | 4.0 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.5 | 13.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.5 | 2.7 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.5 | 2.0 | GO:0090537 | CERF complex(GO:0090537) |
| 0.5 | 2.3 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.4 | 1.2 | GO:0031251 | PAN complex(GO:0031251) |
| 0.4 | 4.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) |
| 0.4 | 2.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.4 | 3.1 | GO:0000836 | ER ubiquitin ligase complex(GO:0000835) Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.4 | 3.6 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.3 | 1.7 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.3 | 25.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.3 | 2.4 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.3 | 2.5 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.3 | 1.8 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.3 | 2.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.3 | 4.6 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.3 | 8.4 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.3 | 1.6 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.2 | 6.6 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.2 | 1.3 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.2 | 5.1 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.2 | 0.8 | GO:0017133 | mitochondrial electron transfer flavoprotein complex(GO:0017133) electron transfer flavoprotein complex(GO:0045251) |
| 0.2 | 2.2 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.2 | 1.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.2 | 1.6 | GO:0034719 | SMN-Sm protein complex(GO:0034719) |
| 0.2 | 3.3 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.2 | 1.5 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 1.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.2 | 3.7 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.1 | 1.8 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.1 | 1.8 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.1 | 1.5 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.1 | 4.0 | GO:0015935 | small ribosomal subunit(GO:0015935) |
| 0.1 | 1.4 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.1 | 0.9 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.1 | 1.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 1.4 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.1 | 0.7 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.1 | 2.4 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.1 | 3.2 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.1 | 1.2 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.1 | 0.4 | GO:0097059 | CRLF-CLCF1 complex(GO:0097058) CNTFR-CLCF1 complex(GO:0097059) |
| 0.1 | 3.8 | GO:0016592 | mediator complex(GO:0016592) |
| 0.1 | 1.2 | GO:0031414 | N-terminal protein acetyltransferase complex(GO:0031414) |
| 0.1 | 7.1 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.1 | 0.7 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.1 | 0.4 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) |
| 0.1 | 4.5 | GO:0016235 | aggresome(GO:0016235) |
| 0.1 | 0.8 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.1 | 0.6 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.1 | 2.5 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.1 | 2.0 | GO:0030687 | preribosome, large subunit precursor(GO:0030687) |
| 0.1 | 0.5 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.1 | 1.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.1 | 0.2 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 1.0 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 2.7 | GO:0044815 | DNA packaging complex(GO:0044815) |
| 0.1 | 0.4 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.1 | 3.0 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.1 | 3.4 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.1 | 0.3 | GO:0097361 | CIA complex(GO:0097361) |
| 0.1 | 0.6 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.1 | 0.9 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.4 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 2.0 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 9.9 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.9 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:1990716 | axonemal central apparatus(GO:1990716) |
| 0.0 | 0.5 | GO:0005845 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0000923 | equatorial microtubule organizing center(GO:0000923) |
| 0.0 | 0.5 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.7 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.4 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 2.9 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 2.7 | GO:0032993 | protein-DNA complex(GO:0032993) |
| 0.0 | 0.6 | GO:0035267 | NuA4 histone acetyltransferase complex(GO:0035267) H4/H2A histone acetyltransferase complex(GO:0043189) H4 histone acetyltransferase complex(GO:1902562) |
| 0.0 | 0.4 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 2.8 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 2.7 | GO:0005840 | ribosome(GO:0005840) |
| 0.0 | 1.4 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0000815 | ESCRT III complex(GO:0000815) |
| 0.0 | 0.2 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.0 | 0.4 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 3.5 | GO:0043296 | apical junction complex(GO:0043296) |
| 0.0 | 2.2 | GO:0005814 | centriole(GO:0005814) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 1.1 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 1.2 | GO:0000792 | heterochromatin(GO:0000792) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 1.3 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.2 | GO:0071011 | precatalytic spliceosome(GO:0071011) |
| 0.0 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 3.3 | 16.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 3.1 | 9.2 | GO:0030629 | U6 snRNA 3'-end binding(GO:0030629) |
| 2.2 | 13.4 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 2.1 | 6.4 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 1.4 | 11.4 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 1.3 | 10.6 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 1.3 | 10.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 1.2 | 4.8 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 1.1 | 4.5 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 1.1 | 3.2 | GO:0004482 | mRNA (guanine-N7-)-methyltransferase activity(GO:0004482) |
| 0.9 | 6.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.9 | 7.9 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.8 | 4.2 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.8 | 2.3 | GO:0035605 | peptidyl-cysteine S-nitrosylase activity(GO:0035605) |
| 0.7 | 2.2 | GO:0036033 | mediator complex binding(GO:0036033) |
| 0.7 | 4.6 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.7 | 6.6 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.6 | 1.9 | GO:0008297 | single-stranded DNA exodeoxyribonuclease activity(GO:0008297) |
| 0.5 | 3.6 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.5 | 2.1 | GO:0071987 | WD40-repeat domain binding(GO:0071987) |
| 0.4 | 4.0 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.4 | 2.6 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.4 | 3.5 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.4 | 14.8 | GO:0017134 | fibroblast growth factor binding(GO:0017134) |
| 0.4 | 1.5 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.3 | 1.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |
| 0.3 | 1.0 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.3 | 2.7 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.3 | 2.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.3 | 2.3 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.3 | 1.9 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.3 | 1.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.3 | 1.6 | GO:1990188 | euchromatin binding(GO:1990188) |
| 0.3 | 4.4 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.3 | 4.6 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.3 | 1.8 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.3 | 3.2 | GO:0015377 | cation:chloride symporter activity(GO:0015377) |
| 0.3 | 3.7 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.3 | 1.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.3 | 6.9 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 3.7 | GO:0070034 | telomerase RNA binding(GO:0070034) |
| 0.2 | 1.9 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.2 | 2.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.2 | 11.0 | GO:0031492 | nucleosomal DNA binding(GO:0031492) |
| 0.2 | 2.7 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.2 | 7.6 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.2 | 34.7 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.2 | 2.2 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.2 | 4.3 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.2 | 1.9 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.2 | 0.6 | GO:0033680 | ATP-dependent DNA/RNA helicase activity(GO:0033680) |
| 0.2 | 1.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.2 | 32.5 | GO:0004386 | helicase activity(GO:0004386) |
| 0.2 | 1.5 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.2 | 0.5 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.2 | 0.7 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.2 | 0.7 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.2 | 14.8 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.2 | 1.5 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.6 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.2 | 1.7 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.1 | 3.1 | GO:1990381 | ubiquitin-specific protease binding(GO:1990381) |
| 0.1 | 1.0 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 3.9 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.1 | 0.9 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.1 | 0.6 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 3.3 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.1 | 0.7 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.1 | 3.3 | GO:0070888 | E-box binding(GO:0070888) |
| 0.1 | 4.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 0.8 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 6.2 | GO:0043425 | bHLH transcription factor binding(GO:0043425) |
| 0.1 | 1.5 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.1 | 2.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.1 | 2.6 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.1 | 28.8 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.1 | 1.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 1.5 | GO:1990226 | histone methyltransferase binding(GO:1990226) |
| 0.1 | 3.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.1 | 1.9 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.1 | 0.3 | GO:0045131 | pre-mRNA branch point binding(GO:0045131) |
| 0.1 | 0.7 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.1 | 0.5 | GO:0000340 | RNA 7-methylguanosine cap binding(GO:0000340) |
| 0.1 | 0.2 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.1 | 3.8 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.1 | 0.3 | GO:0015226 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 1.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.1 | 1.4 | GO:0009055 | electron carrier activity(GO:0009055) |
| 0.1 | 1.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.1 | 1.7 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.1 | 3.6 | GO:0016706 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, 2-oxoglutarate as one donor, and incorporation of one atom each of oxygen into both donors(GO:0016706) |
| 0.1 | 0.7 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.1 | 0.8 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.1 | 1.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.1 | 2.0 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.1 | 0.2 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 1.7 | GO:0001103 | RNA polymerase II repressing transcription factor binding(GO:0001103) |
| 0.1 | 1.5 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.0 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.1 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.1 | 1.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.0 | 0.5 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 3.1 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.8 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.4 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 11.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0001055 | RNA polymerase II activity(GO:0001055) |
| 0.0 | 0.6 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 1.8 | GO:0016875 | aminoacyl-tRNA ligase activity(GO:0004812) ligase activity, forming carbon-oxygen bonds(GO:0016875) ligase activity, forming aminoacyl-tRNA and related compounds(GO:0016876) |
| 0.0 | 1.3 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.8 | GO:0016891 | endoribonuclease activity, producing 5'-phosphomonoesters(GO:0016891) |
| 0.0 | 0.3 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 2.6 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.5 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.8 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.0 | 0.7 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.2 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.4 | GO:0000993 | RNA polymerase II core binding(GO:0000993) |
| 0.0 | 1.1 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.3 | GO:0016929 | SUMO-specific protease activity(GO:0016929) |
| 0.0 | 3.0 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 5.9 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.7 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0003684 | damaged DNA binding(GO:0003684) |
| 0.0 | 0.5 | GO:0001104 | RNA polymerase II transcription cofactor activity(GO:0001104) |
| 0.0 | 0.1 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.3 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 45.1 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.4 | 24.1 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.3 | 9.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.3 | 17.5 | PID PLK1 PATHWAY | PLK1 signaling events |
| 0.2 | 13.6 | PID P53 REGULATION PATHWAY | p53 pathway |
| 0.2 | 5.2 | PID ATR PATHWAY | ATR signaling pathway |
| 0.1 | 13.1 | PID HDAC CLASSI PATHWAY | Signaling events mediated by HDAC Class I |
| 0.1 | 2.3 | PID FANCONI PATHWAY | Fanconi anemia pathway |
| 0.1 | 3.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.1 | 2.7 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.1 | 4.2 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.1 | 5.7 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.1 | 1.5 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 3.8 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.1 | 2.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.1 | 3.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 4.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.8 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 1.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.6 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 4.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.5 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.5 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 11.6 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.7 | 13.7 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.7 | 6.9 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.6 | 35.1 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.5 | 5.9 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.5 | 7.1 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.5 | 10.3 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.4 | 25.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.3 | 4.9 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.3 | 6.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.3 | 16.6 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.3 | 13.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.2 | 19.8 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.2 | 4.6 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.2 | 2.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.2 | 5.2 | REACTOME NONSENSE MEDIATED DECAY ENHANCED BY THE EXON JUNCTION COMPLEX | Genes involved in Nonsense Mediated Decay Enhanced by the Exon Junction Complex |
| 0.2 | 2.2 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.2 | 3.5 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.2 | 5.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.1 | 0.8 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.1 | 6.9 | REACTOME RESPIRATORY ELECTRON TRANSPORT | Genes involved in Respiratory electron transport |
| 0.1 | 7.9 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.1 | 2.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.1 | 4.9 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.1 | 1.9 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.1 | 1.0 | REACTOME EXTENSION OF TELOMERES | Genes involved in Extension of Telomeres |
| 0.1 | 1.1 | REACTOME TRANSCRIPTIONAL ACTIVITY OF SMAD2 SMAD3 SMAD4 HETEROTRIMER | Genes involved in Transcriptional activity of SMAD2/SMAD3:SMAD4 heterotrimer |
| 0.1 | 4.8 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.1 | 1.1 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.8 | REACTOME CYTOSOLIC TRNA AMINOACYLATION | Genes involved in Cytosolic tRNA aminoacylation |
| 0.1 | 1.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.1 | 12.2 | REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | Genes involved in Antigen processing: Ubiquitination & Proteasome degradation |
| 0.1 | 0.7 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 1.3 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.8 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 1.0 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 1.3 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.1 | REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | Genes involved in Antiviral mechanism by IFN-stimulated genes |
| 0.0 | 1.7 | REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | Genes involved in Loss of Nlp from mitotic centrosomes |
| 0.0 | 1.3 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 1.0 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 2.9 | REACTOME TRANSPORT OF INORGANIC CATIONS ANIONS AND AMINO ACIDS OLIGOPEPTIDES | Genes involved in Transport of inorganic cations/anions and amino acids/oligopeptides |
| 0.0 | 1.0 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.5 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.5 | REACTOME NUCLEAR SIGNALING BY ERBB4 | Genes involved in Nuclear signaling by ERBB4 |
| 0.0 | 1.0 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 1.1 | REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | Genes involved in Class B/2 (Secretin family receptors) |