Project
2D miR_HR1_12
Navigation
Downloads
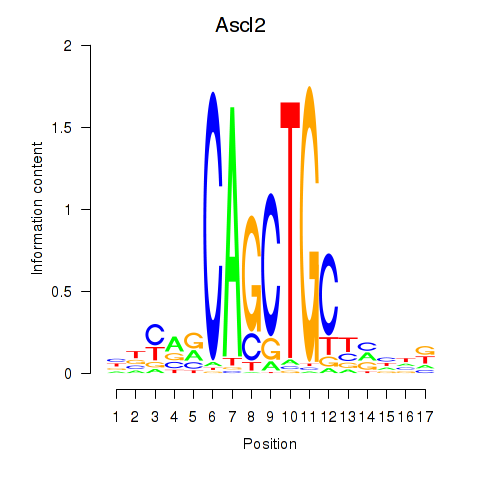
Results for Ascl2
Z-value: 2.04
Transcription factors associated with Ascl2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ascl2
|
ENSMUSG00000009248.5 | achaete-scute family bHLH transcription factor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ascl2 | mm10_v2_chr7_-_142969238_142969264 | -0.60 | 4.1e-02 | Click! |
Activity profile of Ascl2 motif
Sorted Z-values of Ascl2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_128206346 | 5.87 |
ENSMUST00000033049.7
|
Cox6a2
|
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr14_+_33923582 | 4.43 |
ENSMUST00000168727.1
|
Gdf10
|
growth differentiation factor 10 |
| chr11_-_120648104 | 4.28 |
ENSMUST00000026134.2
|
Myadml2
|
myeloid-associated differentiation marker-like 2 |
| chr11_+_69965396 | 3.92 |
ENSMUST00000018713.6
|
Cldn7
|
claudin 7 |
| chr12_-_17176888 | 3.82 |
ENSMUST00000170580.1
|
Kcnf1
|
potassium voltage-gated channel, subfamily F, member 1 |
| chr10_+_127866457 | 3.59 |
ENSMUST00000092058.3
|
BC089597
|
cDNA sequence BC089597 |
| chr1_+_120340569 | 3.32 |
ENSMUST00000037286.8
|
C1ql2
|
complement component 1, q subcomponent-like 2 |
| chr7_-_143074037 | 2.98 |
ENSMUST00000136602.1
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr2_-_25196759 | 2.84 |
ENSMUST00000081869.6
|
Tor4a
|
torsin family 4, member A |
| chr4_-_138367966 | 2.74 |
ENSMUST00000030535.3
|
Cda
|
cytidine deaminase |
| chr11_+_98664341 | 2.65 |
ENSMUST00000017348.2
|
Gsdma
|
gasdermin A |
| chr12_-_103457195 | 2.64 |
ENSMUST00000044687.6
|
Ifi27l2b
|
interferon, alpha-inducible protein 27 like 2B |
| chr16_-_22439570 | 2.63 |
ENSMUST00000170393.1
|
Etv5
|
ets variant gene 5 |
| chr2_+_164403194 | 2.59 |
ENSMUST00000017151.1
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr4_-_130275542 | 2.52 |
ENSMUST00000154846.1
ENSMUST00000105996.1 |
Serinc2
|
serine incorporator 2 |
| chr15_-_71727815 | 2.48 |
ENSMUST00000022953.8
|
Fam135b
|
family with sequence similarity 135, member B |
| chr15_-_101850778 | 2.41 |
ENSMUST00000023790.3
|
Krt1
|
keratin 1 |
| chr1_+_171419027 | 2.40 |
ENSMUST00000171362.1
|
Tstd1
|
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
| chr7_+_44225430 | 2.40 |
ENSMUST00000075162.3
|
Klk1
|
kallikrein 1 |
| chr1_-_84696182 | 2.35 |
ENSMUST00000049126.6
|
Dner
|
delta/notch-like EGF-related receptor |
| chr7_+_44198191 | 2.28 |
ENSMUST00000085450.2
|
Klk1b3
|
kallikrein 1-related peptidase b3 |
| chr19_-_5349574 | 2.26 |
ENSMUST00000025764.5
|
Cst6
|
cystatin E/M |
| chr4_-_130275213 | 2.23 |
ENSMUST00000122374.1
|
Serinc2
|
serine incorporator 2 |
| chr4_-_141846277 | 2.23 |
ENSMUST00000105781.1
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr14_+_30879257 | 2.18 |
ENSMUST00000040715.6
|
Mustn1
|
musculoskeletal, embryonic nuclear protein 1 |
| chr12_+_108334341 | 2.12 |
ENSMUST00000021684.4
|
Cyp46a1
|
cytochrome P450, family 46, subfamily a, polypeptide 1 |
| chr10_-_128401218 | 2.09 |
ENSMUST00000042666.5
|
Slc39a5
|
solute carrier family 39 (metal ion transporter), member 5 |
| chr11_-_59182810 | 2.08 |
ENSMUST00000108793.2
|
Gjc2
|
gap junction protein, gamma 2 |
| chr6_+_107529717 | 2.05 |
ENSMUST00000049285.8
|
Lrrn1
|
leucine rich repeat protein 1, neuronal |
| chr15_-_100599983 | 2.01 |
ENSMUST00000073837.6
|
Pou6f1
|
POU domain, class 6, transcription factor 1 |
| chr4_-_141846359 | 2.01 |
ENSMUST00000037059.10
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr6_+_125349699 | 1.98 |
ENSMUST00000032491.8
|
Tnfrsf1a
|
tumor necrosis factor receptor superfamily, member 1a |
| chr15_-_32244632 | 1.98 |
ENSMUST00000181536.1
|
0610007N19Rik
|
RIKEN cDNA 0610007N19 |
| chr11_+_53519920 | 1.92 |
ENSMUST00000147912.1
|
Sept8
|
septin 8 |
| chr7_-_131322292 | 1.92 |
ENSMUST00000046611.7
|
Cuzd1
|
CUB and zona pellucida-like domains 1 |
| chr10_+_34483400 | 1.89 |
ENSMUST00000019913.7
ENSMUST00000170771.1 |
Frk
|
fyn-related kinase |
| chr9_-_45204083 | 1.89 |
ENSMUST00000034599.8
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr9_-_62537036 | 1.88 |
ENSMUST00000048043.5
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr18_-_82406777 | 1.87 |
ENSMUST00000065224.6
|
Galr1
|
galanin receptor 1 |
| chr2_+_174760619 | 1.85 |
ENSMUST00000029030.2
|
Edn3
|
endothelin 3 |
| chr4_-_8239034 | 1.85 |
ENSMUST00000066674.7
|
Car8
|
carbonic anhydrase 8 |
| chr8_-_111691002 | 1.85 |
ENSMUST00000034435.5
|
Ctrb1
|
chymotrypsinogen B1 |
| chr17_-_57228003 | 1.75 |
ENSMUST00000177046.1
ENSMUST00000024988.8 |
C3
|
complement component 3 |
| chr11_-_119086221 | 1.75 |
ENSMUST00000026665.7
|
Cbx4
|
chromobox 4 |
| chr12_+_79130777 | 1.74 |
ENSMUST00000021550.6
|
Arg2
|
arginase type II |
| chr4_+_58943575 | 1.74 |
ENSMUST00000107554.1
|
Zkscan16
|
zinc finger with KRAB and SCAN domains 16 |
| chr10_+_69212634 | 1.70 |
ENSMUST00000020101.5
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr10_-_128673896 | 1.69 |
ENSMUST00000054764.7
|
Suox
|
sulfite oxidase |
| chr4_-_148149684 | 1.69 |
ENSMUST00000126615.1
|
Fbxo6
|
F-box protein 6 |
| chr15_-_58364148 | 1.68 |
ENSMUST00000068515.7
|
Anxa13
|
annexin A13 |
| chr5_+_35757875 | 1.67 |
ENSMUST00000101280.3
ENSMUST00000054598.5 ENSMUST00000114205.1 ENSMUST00000114206.2 |
Ablim2
|
actin-binding LIM protein 2 |
| chr5_+_135168283 | 1.65 |
ENSMUST00000031692.5
|
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr7_-_66427469 | 1.65 |
ENSMUST00000015278.7
|
Aldh1a3
|
aldehyde dehydrogenase family 1, subfamily A3 |
| chr16_-_31314804 | 1.64 |
ENSMUST00000115230.1
ENSMUST00000130560.1 |
Apod
|
apolipoprotein D |
| chr11_+_96929260 | 1.61 |
ENSMUST00000054311.5
ENSMUST00000107636.3 |
Prr15l
|
proline rich 15-like |
| chr11_+_53519871 | 1.60 |
ENSMUST00000120878.2
|
Sept8
|
septin 8 |
| chr7_+_44188205 | 1.60 |
ENSMUST00000073713.6
|
Klk1b24
|
kallikrein 1-related peptidase b24 |
| chr6_-_148444336 | 1.57 |
ENSMUST00000060095.8
ENSMUST00000100772.3 |
Tmtc1
|
transmembrane and tetratricopeptide repeat containing 1 |
| chr4_-_137430517 | 1.57 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr12_+_112678803 | 1.54 |
ENSMUST00000174780.1
ENSMUST00000169593.1 ENSMUST00000173942.1 |
Zbtb42
|
zinc finger and BTB domain containing 42 |
| chr9_-_62510498 | 1.54 |
ENSMUST00000164246.2
|
Coro2b
|
coronin, actin binding protein, 2B |
| chr5_+_102845007 | 1.53 |
ENSMUST00000070000.4
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr11_+_96929367 | 1.51 |
ENSMUST00000062172.5
|
Prr15l
|
proline rich 15-like |
| chr10_-_75560330 | 1.51 |
ENSMUST00000051129.9
|
Fam211b
|
family with sequence similarity 211, member B |
| chr11_+_45980309 | 1.51 |
ENSMUST00000049038.3
|
Sox30
|
SRY-box containing gene 30 |
| chr5_+_135168382 | 1.50 |
ENSMUST00000111187.3
ENSMUST00000111188.1 |
Bcl7b
|
B cell CLL/lymphoma 7B |
| chr5_+_33721724 | 1.48 |
ENSMUST00000067150.7
ENSMUST00000169212.2 ENSMUST00000114411.2 ENSMUST00000164207.3 |
Fgfr3
|
fibroblast growth factor receptor 3 |
| chr15_+_99591028 | 1.47 |
ENSMUST00000169082.1
|
Aqp5
|
aquaporin 5 |
| chr11_-_114795888 | 1.44 |
ENSMUST00000000206.3
|
Btbd17
|
BTB (POZ) domain containing 17 |
| chr2_+_125136692 | 1.43 |
ENSMUST00000099452.2
|
Ctxn2
|
cortexin 2 |
| chr3_-_129970152 | 1.43 |
ENSMUST00000029624.8
|
Ccdc109b
|
coiled-coil domain containing 109B |
| chr1_-_88205674 | 1.42 |
ENSMUST00000119972.2
|
Dnajb3
|
DnaJ (Hsp40) homolog, subfamily B, member 3 |
| chr6_+_56017489 | 1.42 |
ENSMUST00000052827.4
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr13_-_62888282 | 1.42 |
ENSMUST00000092888.4
|
Fbp1
|
fructose bisphosphatase 1 |
| chr8_-_41374602 | 1.42 |
ENSMUST00000110417.1
ENSMUST00000034000.8 ENSMUST00000143057.1 |
Asah1
|
N-acylsphingosine amidohydrolase 1 |
| chr7_+_44207307 | 1.41 |
ENSMUST00000077354.4
|
Klk1b4
|
kallikrein 1-related pepidase b4 |
| chr7_-_127993831 | 1.41 |
ENSMUST00000033056.3
|
Pycard
|
PYD and CARD domain containing |
| chr7_+_80246375 | 1.40 |
ENSMUST00000058266.6
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr2_-_77170592 | 1.39 |
ENSMUST00000164114.2
ENSMUST00000049544.7 |
Ccdc141
|
coiled-coil domain containing 141 |
| chr9_-_96437434 | 1.39 |
ENSMUST00000070500.2
|
BC043934
|
cDNA sequence BC043934 |
| chr1_+_74854954 | 1.38 |
ENSMUST00000160379.2
|
Cdk5r2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39) |
| chr2_+_127854628 | 1.38 |
ENSMUST00000028859.1
|
Acoxl
|
acyl-Coenzyme A oxidase-like |
| chr15_+_84720052 | 1.37 |
ENSMUST00000006029.4
ENSMUST00000172307.2 |
Arhgap8
|
Rho GTPase activating protein 8 |
| chr13_-_95444827 | 1.36 |
ENSMUST00000045583.7
|
Crhbp
|
corticotropin releasing hormone binding protein |
| chr7_-_31126945 | 1.36 |
ENSMUST00000098548.4
|
Scn1b
|
sodium channel, voltage-gated, type I, beta |
| chr16_-_32797413 | 1.36 |
ENSMUST00000115116.1
ENSMUST00000041123.8 |
Muc20
|
mucin 20 |
| chr9_-_53975246 | 1.36 |
ENSMUST00000048409.7
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr5_+_149411749 | 1.36 |
ENSMUST00000093110.5
|
Medag
|
mesenteric estrogen dependent adipogenesis |
| chr4_+_133553370 | 1.35 |
ENSMUST00000042706.2
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr7_+_141461728 | 1.35 |
ENSMUST00000167491.1
ENSMUST00000165194.1 |
Efcab4a
|
EF-hand calcium binding domain 4A |
| chr2_+_35282380 | 1.34 |
ENSMUST00000028239.6
|
Gsn
|
gelsolin |
| chr2_-_168741752 | 1.34 |
ENSMUST00000029060.4
|
Atp9a
|
ATPase, class II, type 9A |
| chr3_+_102469918 | 1.33 |
ENSMUST00000106925.2
ENSMUST00000035952.3 |
Ngf
|
nerve growth factor |
| chr7_+_45216671 | 1.33 |
ENSMUST00000134420.1
|
Tead2
|
TEA domain family member 2 |
| chr3_-_75270073 | 1.33 |
ENSMUST00000039047.4
|
Serpini2
|
serine (or cysteine) peptidase inhibitor, clade I, member 2 |
| chr4_-_130275523 | 1.33 |
ENSMUST00000146478.1
|
Serinc2
|
serine incorporator 2 |
| chr7_-_104315455 | 1.32 |
ENSMUST00000106837.1
ENSMUST00000106839.2 ENSMUST00000070943.6 |
Trim12a
|
tripartite motif-containing 12A |
| chr9_-_57836706 | 1.30 |
ENSMUST00000164010.1
ENSMUST00000171444.1 ENSMUST00000098686.3 |
Arid3b
|
AT rich interactive domain 3B (BRIGHT-like) |
| chr12_+_26469204 | 1.30 |
ENSMUST00000020969.3
|
Cmpk2
|
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr6_-_29179584 | 1.29 |
ENSMUST00000159200.1
|
Prrt4
|
proline-rich transmembrane protein 4 |
| chr18_-_44662251 | 1.29 |
ENSMUST00000164666.1
|
Mcc
|
mutated in colorectal cancers |
| chr8_+_62951195 | 1.28 |
ENSMUST00000118003.1
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chrX_+_99042581 | 1.28 |
ENSMUST00000036606.7
|
Stard8
|
START domain containing 8 |
| chr17_-_57247632 | 1.28 |
ENSMUST00000005975.6
|
Gpr108
|
G protein-coupled receptor 108 |
| chr7_+_80246529 | 1.27 |
ENSMUST00000107381.1
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr5_+_24428208 | 1.27 |
ENSMUST00000115049.2
|
Slc4a2
|
solute carrier family 4 (anion exchanger), member 2 |
| chr4_+_41755210 | 1.26 |
ENSMUST00000108038.1
ENSMUST00000084695.4 |
Galt
|
galactose-1-phosphate uridyl transferase |
| chr5_+_35757951 | 1.26 |
ENSMUST00000114204.1
ENSMUST00000129347.1 |
Ablim2
|
actin-binding LIM protein 2 |
| chr1_-_156674290 | 1.26 |
ENSMUST00000079625.4
|
Tor3a
|
torsin family 3, member A |
| chr15_+_102144362 | 1.25 |
ENSMUST00000023807.6
|
Igfbp6
|
insulin-like growth factor binding protein 6 |
| chr10_-_61147625 | 1.23 |
ENSMUST00000122259.1
|
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr5_+_117841839 | 1.23 |
ENSMUST00000142742.2
|
Nos1
|
nitric oxide synthase 1, neuronal |
| chr2_+_174760781 | 1.22 |
ENSMUST00000140908.1
|
Edn3
|
endothelin 3 |
| chr16_+_17276291 | 1.22 |
ENSMUST00000164950.1
ENSMUST00000159242.1 |
Tmem191c
|
transmembrane protein 191C |
| chr2_+_84734050 | 1.22 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chr1_-_14918862 | 1.22 |
ENSMUST00000041447.4
|
Trpa1
|
transient receptor potential cation channel, subfamily A, member 1 |
| chr4_-_141933080 | 1.17 |
ENSMUST00000036701.7
|
Fhad1
|
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr6_+_112273758 | 1.16 |
ENSMUST00000032376.5
|
Lmcd1
|
LIM and cysteine-rich domains 1 |
| chr10_+_11343387 | 1.16 |
ENSMUST00000069106.4
|
Epm2a
|
epilepsy, progressive myoclonic epilepsy, type 2 gene alpha |
| chr19_-_45591820 | 1.16 |
ENSMUST00000160003.1
ENSMUST00000162879.1 |
Fbxw4
|
F-box and WD-40 domain protein 4 |
| chr3_-_129969989 | 1.16 |
ENSMUST00000146340.1
|
Ccdc109b
|
coiled-coil domain containing 109B |
| chr10_-_61147659 | 1.15 |
ENSMUST00000092498.5
ENSMUST00000137833.1 ENSMUST00000155919.1 |
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr17_-_87282793 | 1.15 |
ENSMUST00000146560.2
|
4833418N02Rik
|
RIKEN cDNA 4833418N02 gene |
| chr1_-_130715734 | 1.14 |
ENSMUST00000066863.6
ENSMUST00000050406.4 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr16_+_17276337 | 1.13 |
ENSMUST00000159065.1
ENSMUST00000159494.1 ENSMUST00000159811.1 |
Tmem191c
|
transmembrane protein 191C |
| chr4_-_118544010 | 1.13 |
ENSMUST00000128098.1
|
Tmem125
|
transmembrane protein 125 |
| chr11_-_79146407 | 1.13 |
ENSMUST00000018478.4
ENSMUST00000108264.1 |
Ksr1
|
kinase suppressor of ras 1 |
| chr9_-_108567336 | 1.10 |
ENSMUST00000074208.4
|
Ndufaf3
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 3 |
| chr7_+_139213239 | 1.08 |
ENSMUST00000106104.1
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr16_+_17276662 | 1.08 |
ENSMUST00000069420.4
|
Tmem191c
|
transmembrane protein 191C |
| chr9_+_74848437 | 1.08 |
ENSMUST00000161862.1
ENSMUST00000162089.1 ENSMUST00000160017.1 ENSMUST00000160950.1 |
Gm16551
Gm20649
|
predicted gene 16551 predicted gene 20649 |
| chr10_+_128270546 | 1.07 |
ENSMUST00000105238.3
ENSMUST00000085708.2 |
Stat2
|
signal transducer and activator of transcription 2 |
| chr3_+_132085281 | 1.06 |
ENSMUST00000029665.5
|
Dkk2
|
dickkopf homolog 2 (Xenopus laevis) |
| chr12_-_103443653 | 1.05 |
ENSMUST00000055071.8
|
Ifi27l2a
|
interferon, alpha-inducible protein 27 like 2A |
| chr5_-_98030727 | 1.04 |
ENSMUST00000031281.9
|
Antxr2
|
anthrax toxin receptor 2 |
| chr10_-_120201558 | 1.03 |
ENSMUST00000020448.4
|
Irak3
|
interleukin-1 receptor-associated kinase 3 |
| chr9_+_107975529 | 1.03 |
ENSMUST00000035216.4
|
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chr11_+_112782182 | 1.01 |
ENSMUST00000000579.2
|
Sox9
|
SRY-box containing gene 9 |
| chr7_+_139212974 | 1.00 |
ENSMUST00000016124.8
|
Lrrc27
|
leucine rich repeat containing 27 |
| chr5_+_91139591 | 0.99 |
ENSMUST00000031325.4
|
Areg
|
amphiregulin |
| chrX_+_143518576 | 0.99 |
ENSMUST00000033640.7
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3 |
| chr5_-_137212389 | 0.98 |
ENSMUST00000179412.1
|
A630081J09Rik
|
RIKEN cDNA A630081J09 gene |
| chr8_-_91134027 | 0.96 |
ENSMUST00000125257.1
|
Aktip
|
thymoma viral proto-oncogene 1 interacting protein |
| chr4_-_155785864 | 0.96 |
ENSMUST00000097742.2
|
Tmem88b
|
transmembrane protein 88B |
| chr6_-_52191695 | 0.96 |
ENSMUST00000101395.2
|
Hoxa4
|
homeobox A4 |
| chr4_-_147642496 | 0.96 |
ENSMUST00000133006.1
ENSMUST00000037565.7 ENSMUST00000105720.1 |
2610305D13Rik
|
RIKEN cDNA 2610305D13 gene |
| chr11_+_121434913 | 0.96 |
ENSMUST00000026175.2
ENSMUST00000092302.4 ENSMUST00000103014.3 |
Fn3k
|
fructosamine 3 kinase |
| chr6_+_118066356 | 0.95 |
ENSMUST00000164960.1
|
Rasgef1a
|
RasGEF domain family, member 1A |
| chr8_+_3665747 | 0.95 |
ENSMUST00000014118.2
|
1810033B17Rik
|
RIKEN cDNA 1810033B17 gene |
| chr9_-_39604124 | 0.94 |
ENSMUST00000042485.4
ENSMUST00000141370.1 |
AW551984
|
expressed sequence AW551984 |
| chr8_-_91133942 | 0.94 |
ENSMUST00000120213.1
ENSMUST00000109609.2 |
Aktip
|
thymoma viral proto-oncogene 1 interacting protein |
| chr5_+_136967859 | 0.94 |
ENSMUST00000001790.5
|
Cldn15
|
claudin 15 |
| chr7_-_114562945 | 0.94 |
ENSMUST00000119712.1
ENSMUST00000032908.8 |
Cyp2r1
|
cytochrome P450, family 2, subfamily r, polypeptide 1 |
| chr11_+_69059750 | 0.92 |
ENSMUST00000051888.2
|
2310047M10Rik
|
RIKEN cDNA 2310047M10 gene |
| chr1_-_167393826 | 0.92 |
ENSMUST00000028005.2
|
Mgst3
|
microsomal glutathione S-transferase 3 |
| chr11_+_83116809 | 0.92 |
ENSMUST00000037994.7
|
Slfn1
|
schlafen 1 |
| chr7_+_16944645 | 0.92 |
ENSMUST00000094807.5
|
Pnmal2
|
PNMA-like 2 |
| chr7_-_16874845 | 0.91 |
ENSMUST00000181501.1
|
9330104G04Rik
|
RIKEN cDNA 9330104G04 gene |
| chr4_-_147809788 | 0.91 |
ENSMUST00000105734.3
ENSMUST00000176201.1 |
Gm13157
Gm20707
|
predicted gene 13157 predicted gene 20707 |
| chr5_+_137641334 | 0.91 |
ENSMUST00000177466.1
ENSMUST00000166099.2 |
Sap25
|
sin3 associated polypeptide |
| chr19_-_29523159 | 0.90 |
ENSMUST00000180986.1
|
A930007I19Rik
|
RIKEN cDNA A930007I19 gene |
| chr1_-_162866502 | 0.90 |
ENSMUST00000046049.7
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr1_+_127868773 | 0.90 |
ENSMUST00000037649.5
|
Rab3gap1
|
RAB3 GTPase activating protein subunit 1 |
| chr5_+_81021202 | 0.89 |
ENSMUST00000117253.1
ENSMUST00000120128.1 |
Lphn3
|
latrophilin 3 |
| chr4_-_154160632 | 0.89 |
ENSMUST00000105639.3
ENSMUST00000030896.8 |
Tprgl
|
transformation related protein 63 regulated like |
| chrX_+_101449078 | 0.89 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr9_-_71592346 | 0.88 |
ENSMUST00000093823.1
|
Myzap
|
myocardial zonula adherens protein |
| chr4_-_133263042 | 0.87 |
ENSMUST00000105908.3
ENSMUST00000030674.7 |
Sytl1
|
synaptotagmin-like 1 |
| chr2_-_26933781 | 0.87 |
ENSMUST00000154651.1
ENSMUST00000015011.3 |
Surf4
|
surfeit gene 4 |
| chr3_-_148989316 | 0.87 |
ENSMUST00000098518.2
|
Lphn2
|
latrophilin 2 |
| chrX_-_7572843 | 0.86 |
ENSMUST00000132788.1
|
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr3_-_104511812 | 0.86 |
ENSMUST00000046316.6
|
Lrig2
|
leucine-rich repeats and immunoglobulin-like domains 2 |
| chr17_-_56716788 | 0.86 |
ENSMUST00000067931.5
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chr17_+_5492558 | 0.86 |
ENSMUST00000089185.4
|
Zdhhc14
|
zinc finger, DHHC domain containing 14 |
| chr11_-_120098673 | 0.85 |
ENSMUST00000093901.5
ENSMUST00000026442.4 ENSMUST00000106225.3 |
Enthd2
|
ENTH domain containing 2 |
| chr19_+_40612392 | 0.85 |
ENSMUST00000134063.1
|
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr13_-_119408985 | 0.84 |
ENSMUST00000099149.3
ENSMUST00000069902.6 ENSMUST00000109204.1 |
Nnt
|
nicotinamide nucleotide transhydrogenase |
| chr6_-_124733121 | 0.84 |
ENSMUST00000112484.3
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr6_-_72235559 | 0.84 |
ENSMUST00000042646.7
|
Atoh8
|
atonal homolog 8 (Drosophila) |
| chr16_-_17576206 | 0.84 |
ENSMUST00000090165.4
ENSMUST00000164623.1 |
Slc7a4
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 4 |
| chr19_-_6015152 | 0.82 |
ENSMUST00000025891.8
|
Capn1
|
calpain 1 |
| chr17_-_22166190 | 0.82 |
ENSMUST00000080249.5
|
Zfp947
|
zinc finger protein 947 |
| chr7_+_83584910 | 0.82 |
ENSMUST00000039317.7
ENSMUST00000164944.1 |
Tmc3
|
transmembrane channel-like gene family 3 |
| chr4_-_156050785 | 0.82 |
ENSMUST00000051509.8
|
Ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr7_+_28071230 | 0.82 |
ENSMUST00000138392.1
ENSMUST00000076648.7 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr16_+_21794384 | 0.82 |
ENSMUST00000180830.1
|
1300002E11Rik
|
RIKEN cDNA 1300002E11 gene |
| chr12_+_17690793 | 0.81 |
ENSMUST00000071858.3
|
Hpcal1
|
hippocalcin-like 1 |
| chr2_-_170406501 | 0.81 |
ENSMUST00000154650.1
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chrX_-_112698642 | 0.81 |
ENSMUST00000039887.3
|
Pof1b
|
premature ovarian failure 1B |
| chr1_+_162639148 | 0.81 |
ENSMUST00000028020.9
|
Myoc
|
myocilin |
| chr17_-_25433263 | 0.81 |
ENSMUST00000159623.1
|
Cacna1h
|
calcium channel, voltage-dependent, T type, alpha 1H subunit |
| chr3_+_108284089 | 0.79 |
ENSMUST00000102632.4
|
Sort1
|
sortilin 1 |
| chr2_-_172940299 | 0.79 |
ENSMUST00000009143.7
|
Bmp7
|
bone morphogenetic protein 7 |
| chr19_-_56822161 | 0.79 |
ENSMUST00000118592.1
|
A630007B06Rik
|
RIKEN cDNA A630007B06 gene |
| chr17_+_55952623 | 0.78 |
ENSMUST00000003274.6
|
Ebi3
|
Epstein-Barr virus induced gene 3 |
| chr9_+_57560934 | 0.77 |
ENSMUST00000045791.9
|
Scamp2
|
secretory carrier membrane protein 2 |
| chr2_-_152933202 | 0.77 |
ENSMUST00000099200.2
|
Foxs1
|
forkhead box S1 |
| chr4_-_156050465 | 0.77 |
ENSMUST00000184684.1
|
Ttll10
|
tubulin tyrosine ligase-like family, member 10 |
| chr13_-_62858364 | 0.77 |
ENSMUST00000021907.7
|
Fbp2
|
fructose bisphosphatase 2 |
| chr6_-_72617000 | 0.77 |
ENSMUST00000070524.4
|
Tgoln1
|
trans-golgi network protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ascl2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.7 | GO:0006216 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.9 | 6.1 | GO:1904219 | regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904217) positive regulation of CDP-diacylglycerol-serine O-phosphatidyltransferase activity(GO:1904219) positive regulation of serine C-palmitoyltransferase activity(GO:1904222) |
| 0.7 | 2.2 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.7 | 2.1 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.6 | 1.9 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.5 | 1.6 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.5 | 1.6 | GO:0034442 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.5 | 3.2 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.5 | 1.5 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.5 | 1.4 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) positive regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002582) positive regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002588) |
| 0.5 | 1.4 | GO:0071314 | cellular response to cocaine(GO:0071314) |
| 0.5 | 1.4 | GO:0086047 | corticospinal neuron axon guidance(GO:0021966) membrane depolarization during Purkinje myocyte cell action potential(GO:0086047) |
| 0.5 | 5.9 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.4 | 1.3 | GO:1903903 | positive regulation of keratinocyte apoptotic process(GO:1902174) regulation of establishment of T cell polarity(GO:1903903) |
| 0.4 | 2.6 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.4 | 1.7 | GO:0090467 | L-arginine import(GO:0043091) arginine import(GO:0090467) |
| 0.4 | 1.3 | GO:0009197 | dUDP biosynthetic process(GO:0006227) pyrimidine nucleoside diphosphate metabolic process(GO:0009138) pyrimidine nucleoside diphosphate biosynthetic process(GO:0009139) pyrimidine deoxyribonucleoside diphosphate metabolic process(GO:0009196) pyrimidine deoxyribonucleoside diphosphate biosynthetic process(GO:0009197) dUDP metabolic process(GO:0046077) |
| 0.4 | 1.3 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.4 | 1.2 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by nitric oxide(GO:0099548) |
| 0.4 | 1.1 | GO:0007439 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.4 | 1.5 | GO:0072190 | ureter urothelium development(GO:0072190) |
| 0.4 | 2.2 | GO:1902732 | positive regulation of chondrocyte proliferation(GO:1902732) |
| 0.4 | 1.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.3 | 1.7 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.3 | 1.4 | GO:0021586 | pons maturation(GO:0021586) |
| 0.3 | 0.9 | GO:1900477 | negative regulation of G1/S transition of mitotic cell cycle by negative regulation of transcription from RNA polymerase II promoter(GO:1900477) |
| 0.3 | 2.4 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.3 | 0.9 | GO:0097051 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) |
| 0.3 | 1.4 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.3 | 0.8 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.3 | 1.3 | GO:0014042 | positive regulation of neuron maturation(GO:0014042) |
| 0.3 | 1.6 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.3 | 0.8 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.2 | 1.0 | GO:0060598 | dichotomous subdivision of terminal units involved in mammary gland duct morphogenesis(GO:0060598) |
| 0.2 | 1.2 | GO:1904058 | positive regulation of sensory perception of pain(GO:1904058) |
| 0.2 | 0.6 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.2 | 1.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.2 | 3.9 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.2 | 1.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.2 | 0.8 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.2 | 0.4 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.2 | 0.8 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.2 | 1.5 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.5 | GO:0042939 | glutathione transport(GO:0034635) tripeptide transport(GO:0042939) |
| 0.2 | 0.5 | GO:0021577 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.2 | 1.5 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.2 | 1.5 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.2 | 0.2 | GO:1905072 | cardiac jelly development(GO:1905072) |
| 0.2 | 0.8 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 0.5 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.2 | 1.3 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.2 | 0.8 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.2 | 2.3 | GO:0007220 | Notch receptor processing(GO:0007220) |
| 0.2 | 0.6 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.1 | 2.4 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.1 | 0.7 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 1.6 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.4 | GO:2001293 | malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.8 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.1 | 1.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.1 | 0.4 | GO:0039521 | lysosomal microautophagy(GO:0016237) piecemeal microautophagy of nucleus(GO:0034727) suppression by virus of host autophagy(GO:0039521) negative regulation of sphingolipid biosynthesis involved in cellular sphingolipid homeostasis(GO:0090157) |
| 0.1 | 1.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.1 | 0.2 | GO:0014735 | regulation of muscle atrophy(GO:0014735) |
| 0.1 | 0.8 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.1 | 0.4 | GO:0032470 | positive regulation of endoplasmic reticulum calcium ion concentration(GO:0032470) |
| 0.1 | 1.8 | GO:1901550 | regulation of endothelial cell development(GO:1901550) regulation of establishment of endothelial barrier(GO:1903140) |
| 0.1 | 0.5 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.1 | 0.5 | GO:0014894 | response to inactivity(GO:0014854) response to muscle inactivity(GO:0014870) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 1.2 | GO:0046959 | habituation(GO:0046959) |
| 0.1 | 0.3 | GO:0070948 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.5 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 1.0 | GO:1903800 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.1 | 0.9 | GO:1903874 | ferrous iron transmembrane transport(GO:1903874) |
| 0.1 | 0.7 | GO:1903817 | negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.1 | 0.3 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.1 | GO:1904192 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 0.3 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 1.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 1.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.1 | 1.4 | GO:0048012 | hepatocyte growth factor receptor signaling pathway(GO:0048012) |
| 0.1 | 0.8 | GO:2000344 | cortisol metabolic process(GO:0034650) cortisol biosynthetic process(GO:0034651) positive regulation of acrosome reaction(GO:2000344) |
| 0.1 | 0.9 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.1 | 0.3 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.1 | 0.9 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 0.4 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.1 | 0.3 | GO:0003199 | endocardial cushion to mesenchymal transition involved in heart valve formation(GO:0003199) |
| 0.1 | 1.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.1 | 0.3 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) |
| 0.1 | 0.6 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 4.2 | GO:0010862 | positive regulation of pathway-restricted SMAD protein phosphorylation(GO:0010862) |
| 0.1 | 0.7 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.7 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.5 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.1 | 0.4 | GO:0070070 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.1 | 1.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.9 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.4 | GO:0061314 | Notch signaling involved in heart development(GO:0061314) |
| 0.1 | 0.4 | GO:1902004 | positive regulation of beta-amyloid formation(GO:1902004) |
| 0.1 | 0.9 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.1 | 0.4 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.1 | 0.5 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.2 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.1 | 3.0 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.1 | 0.8 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.1 | 0.8 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.1 | 0.4 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.1 | 0.6 | GO:0061727 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.1 | 1.0 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.1 | 0.3 | GO:0090527 | actin filament reorganization(GO:0090527) |
| 0.1 | 0.5 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.1 | 0.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.1 | 0.2 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.1 | 0.3 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.1 | 0.7 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.1 | 0.3 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.1 | 0.2 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) |
| 0.1 | 0.4 | GO:0040031 | snRNA modification(GO:0040031) |
| 0.1 | 1.4 | GO:0043552 | positive regulation of phosphatidylinositol 3-kinase activity(GO:0043552) |
| 0.1 | 0.2 | GO:0014719 | skeletal muscle satellite cell activation(GO:0014719) |
| 0.1 | 0.4 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.1 | 2.0 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.3 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 2.0 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.6 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 1.2 | GO:0010763 | positive regulation of fibroblast migration(GO:0010763) |
| 0.0 | 1.9 | GO:0003333 | amino acid transmembrane transport(GO:0003333) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.5 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 1.9 | GO:0071300 | cellular response to retinoic acid(GO:0071300) |
| 0.0 | 0.6 | GO:0034312 | diol biosynthetic process(GO:0034312) |
| 0.0 | 0.9 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.5 | GO:1990118 | sodium ion import(GO:0097369) sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 1.2 | GO:0015985 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) energy coupled proton transmembrane transport, against electrochemical gradient(GO:0015988) ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 1.5 | GO:0010644 | cell communication by electrical coupling(GO:0010644) |
| 0.0 | 1.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.7 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 1.1 | GO:0006739 | NADP metabolic process(GO:0006739) |
| 0.0 | 0.4 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 1.1 | GO:0071357 | type I interferon signaling pathway(GO:0060337) cellular response to type I interferon(GO:0071357) |
| 0.0 | 0.4 | GO:0090205 | positive regulation of cholesterol biosynthetic process(GO:0045542) positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.6 | GO:0046069 | cGMP catabolic process(GO:0046069) |
| 0.0 | 0.3 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.4 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.2 | GO:0046549 | retinal cone cell development(GO:0046549) |
| 0.0 | 1.3 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.7 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 1.8 | GO:0006805 | xenobiotic metabolic process(GO:0006805) |
| 0.0 | 0.7 | GO:0035094 | response to nicotine(GO:0035094) |
| 0.0 | 0.2 | GO:1900122 | positive regulation of receptor binding(GO:1900122) |
| 0.0 | 0.9 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.5 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.2 | GO:0042359 | vitamin D metabolic process(GO:0042359) |
| 0.0 | 1.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.5 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.4 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.2 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 1.3 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.4 | GO:0030194 | positive regulation of blood coagulation(GO:0030194) positive regulation of hemostasis(GO:1900048) |
| 0.0 | 0.4 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:1901907 | diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.3 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.4 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 1.1 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.8 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.1 | GO:1902477 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 1.1 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 1.0 | GO:0006040 | amino sugar metabolic process(GO:0006040) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 1.5 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 1.4 | GO:0032024 | positive regulation of insulin secretion(GO:0032024) |
| 0.0 | 2.7 | GO:0071805 | potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
| 0.0 | 0.5 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.1 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0060767 | epithelial cell proliferation involved in prostate gland development(GO:0060767) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.3 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.4 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.8 | GO:0051453 | regulation of intracellular pH(GO:0051453) |
| 0.0 | 0.3 | GO:0010718 | positive regulation of epithelial to mesenchymal transition(GO:0010718) |
| 0.0 | 0.4 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 1.0 | GO:0048704 | embryonic skeletal system morphogenesis(GO:0048704) |
| 0.0 | 1.7 | GO:0009755 | hormone-mediated signaling pathway(GO:0009755) |
| 0.0 | 0.1 | GO:1901897 | regulation of relaxation of cardiac muscle(GO:1901897) |
| 0.0 | 0.1 | GO:0071320 | cellular response to cAMP(GO:0071320) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.4 | 1.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.4 | 4.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.4 | 1.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.3 | 1.0 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.3 | 5.9 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.3 | 1.9 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 1.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.2 | 0.7 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.2 | 1.5 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 1.3 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.2 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 3.7 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.7 | GO:0044322 | endoplasmic reticulum quality control compartment(GO:0044322) |
| 0.1 | 1.9 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.1 | 1.3 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.1 | 1.2 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.9 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 2.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 1.0 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.1 | 0.4 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.1 | 1.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.1 | 0.8 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.1 | 0.5 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 1.4 | GO:0005767 | secondary lysosome(GO:0005767) |
| 0.1 | 0.4 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.1 | 1.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.1 | 0.7 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.3 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.8 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.5 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.3 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.7 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.8 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 3.1 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.8 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 2.5 | GO:0008076 | voltage-gated potassium channel complex(GO:0008076) |
| 0.0 | 0.9 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 1.2 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.4 | GO:0005782 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.6 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.0 | 1.6 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.9 | GO:0005793 | endoplasmic reticulum-Golgi intermediate compartment(GO:0005793) |
| 0.0 | 0.9 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
| 0.0 | 0.5 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.9 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 1.5 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.1 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 22.3 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 2.0 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.4 | GO:0005769 | early endosome(GO:0005769) |
| 0.0 | 0.4 | GO:0000407 | pre-autophagosomal structure(GO:0000407) |
| 0.0 | 1.1 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.3 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 1.5 | GO:0031234 | extrinsic component of cytoplasmic side of plasma membrane(GO:0031234) |
| 0.0 | 0.2 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 2.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.8 | 3.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.7 | 2.0 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.6 | 1.9 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.5 | 1.6 | GO:0070737 | protein-glycine ligase activity, elongating(GO:0070737) |
| 0.5 | 1.9 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.5 | 2.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.4 | 6.1 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.4 | 1.3 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.4 | 1.5 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.3 | 2.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.3 | 1.3 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.3 | 1.0 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.3 | 1.0 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.3 | 1.3 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.3 | 4.5 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) |
| 0.3 | 0.9 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.3 | 1.4 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.3 | 1.7 | GO:0043546 | molybdenum ion binding(GO:0030151) molybdopterin cofactor binding(GO:0043546) |
| 0.3 | 1.5 | GO:0004028 | 3-chloroallyl aldehyde dehydrogenase activity(GO:0004028) |
| 0.2 | 0.7 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 0.7 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.2 | 1.7 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.2 | 5.9 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.2 | 1.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.2 | 0.8 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.2 | 2.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.2 | 1.0 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.2 | 1.4 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.2 | 1.7 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) |
| 0.2 | 1.4 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 1.4 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.2 | 1.5 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.4 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 2.4 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.7 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.1 | 0.8 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.1 | 0.8 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.4 | GO:0019778 | Atg12 activating enzyme activity(GO:0019778) Atg8 activating enzyme activity(GO:0019779) |
| 0.1 | 4.0 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.1 | 0.7 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.6 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.1 | 1.3 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.1 | 1.5 | GO:0015250 | water channel activity(GO:0015250) |
| 0.1 | 1.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0047936 | glucose 1-dehydrogenase [NAD(P)] activity(GO:0047936) |
| 0.1 | 1.7 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.1 | 1.5 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.1 | 0.4 | GO:0004096 | catalase activity(GO:0004096) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 15.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.5 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 1.3 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.1 | 1.0 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.1 | 0.3 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.8 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.1 | 0.4 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.6 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 2.9 | GO:0005272 | sodium channel activity(GO:0005272) |
| 0.1 | 0.8 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.1 | 2.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 1.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 3.6 | GO:0005160 | transforming growth factor beta receptor binding(GO:0005160) |
| 0.1 | 1.2 | GO:0046933 | proton-transporting ATP synthase activity, rotational mechanism(GO:0046933) |
| 0.1 | 0.6 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.1 | 0.5 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.5 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.1 | 1.3 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.1 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 0.1 | GO:0098809 | nitrite reductase activity(GO:0098809) |
| 0.1 | 1.2 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.1 | 1.4 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.1 | 1.7 | GO:0032183 | SUMO binding(GO:0032183) |
| 0.1 | 0.5 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.1 | 1.9 | GO:0005112 | Notch binding(GO:0005112) |
| 0.1 | 1.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 1.0 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.5 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.9 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.2 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.0 | 2.1 | GO:0017046 | peptide hormone binding(GO:0017046) |
| 0.0 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 4.1 | GO:0005249 | voltage-gated potassium channel activity(GO:0005249) |
| 0.0 | 0.5 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.9 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.8 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.5 | GO:0008329 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.5 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 1.6 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.9 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.9 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.7 | GO:0008395 | steroid hydroxylase activity(GO:0008395) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.5 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.3 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 1.0 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0034431 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) |
| 0.0 | 0.9 | GO:0016836 | hydro-lyase activity(GO:0016836) |
| 0.0 | 0.1 | GO:0055131 | C3HC4-type RING finger domain binding(GO:0055131) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 2.0 | GO:0019905 | syntaxin binding(GO:0019905) |
| 0.0 | 0.8 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.6 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.3 | GO:0034987 | immunoglobulin receptor binding(GO:0034987) |
| 0.0 | 1.1 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.4 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.2 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 1.8 | GO:0002020 | protease binding(GO:0002020) |
| 0.0 | 0.5 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 2.5 | GO:0052689 | carboxylic ester hydrolase activity(GO:0052689) |
| 0.0 | 0.4 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 1.7 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.7 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
| 0.0 | 1.0 | GO:0061733 | peptide-lysine-N-acetyltransferase activity(GO:0061733) |
| 0.0 | 0.1 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 2.5 | GO:0051015 | actin filament binding(GO:0051015) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.1 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.2 | 1.4 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.1 | 1.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.1 | 2.5 | PID S1P META PATHWAY | Sphingosine 1-phosphate (S1P) pathway |
| 0.1 | 4.4 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.1 | 2.8 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.1 | 1.0 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.1 | 1.3 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.1 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.1 | 0.8 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.1 | 1.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.1 | 3.2 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.1 | 2.7 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.1 | 2.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.6 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.1 | 1.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.8 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.8 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 1.2 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 2.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.6 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.1 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.8 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.0 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 4.6 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.2 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.5 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.9 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 2.2 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.4 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.8 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 1.3 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 1.7 | PID BETA CATENIN NUC PATHWAY | Regulation of nuclear beta catenin signaling and target gene transcription |
| 0.0 | 0.5 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.5 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.8 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.7 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 4.5 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 3.3 | NABA MATRISOME ASSOCIATED | Ensemble of genes encoding ECM-associated proteins including ECM-affilaited proteins, ECM regulators and secreted factors |
| 0.0 | 0.4 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.8 | PID P53 DOWNSTREAM PATHWAY | Direct p53 effectors |
| 0.0 | 0.1 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.2 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.6 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.1 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.2 | 2.4 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.2 | 2.0 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 5.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.5 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.9 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.9 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.1 | 2.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.5 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 2.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.1 | 4.5 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.1 | 2.7 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.1 | 0.7 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 0.9 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 0.9 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.1 | 2.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.1 | 1.1 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.1 | 0.6 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 2.8 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.1 | 1.2 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 1.7 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.1 | 2.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 0.5 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.9 | REACTOME PHASE1 FUNCTIONALIZATION OF COMPOUNDS | Genes involved in Phase 1 - Functionalization of compounds |
| 0.0 | 1.3 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.3 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.7 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.6 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.4 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.0 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.8 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.7 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.4 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 4.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.5 | REACTOME PROCESSING OF INTRONLESS PRE MRNAS | Genes involved in Processing of Intronless Pre-mRNAs |
| 0.0 | 1.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.2 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 5.7 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.0 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.8 | REACTOME SPHINGOLIPID METABOLISM | Genes involved in Sphingolipid metabolism |
| 0.0 | 1.3 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.4 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 1.4 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.9 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |