Project
2D miR_HR1_12
Navigation
Downloads
Results for Ezh2_Atf2_Ikzf1
Z-value: 1.69
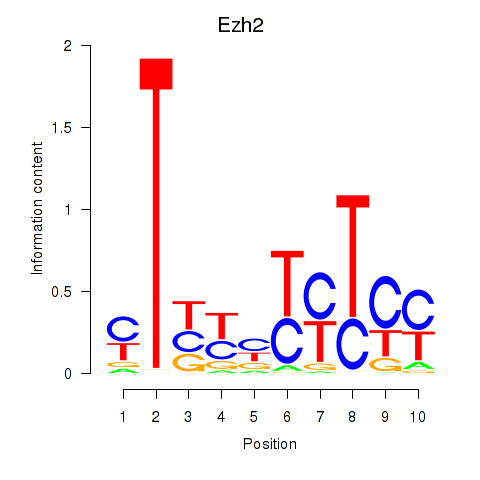
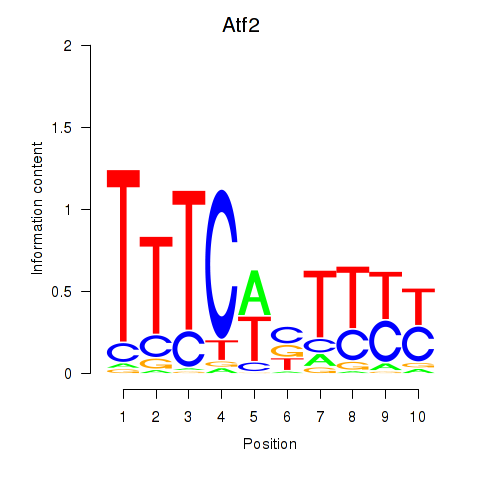
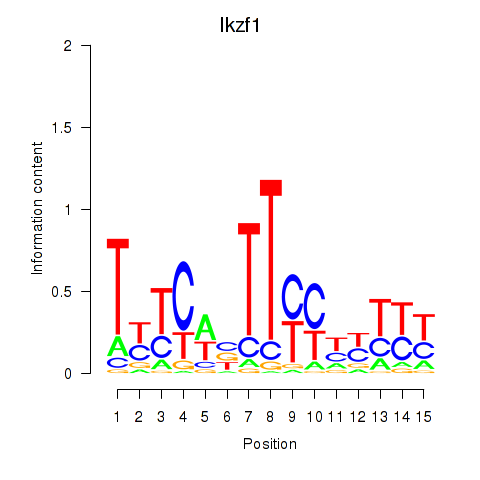
Transcription factors associated with Ezh2_Atf2_Ikzf1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ezh2
|
ENSMUSG00000029687.10 | enhancer of zeste 2 polycomb repressive complex 2 subunit |
|
Atf2
|
ENSMUSG00000027104.12 | activating transcription factor 2 |
|
Ikzf1
|
ENSMUSG00000018654.11 | IKAROS family zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ezh2 | mm10_v2_chr6_-_47594967_47595047 | -0.87 | 2.1e-04 | Click! |
| Ikzf1 | mm10_v2_chr11_+_11684967_11685027 | -0.71 | 1.0e-02 | Click! |
| Atf2 | mm10_v2_chr2_-_73892530_73892555 | -0.55 | 6.6e-02 | Click! |
Activity profile of Ezh2_Atf2_Ikzf1 motif
Sorted Z-values of Ezh2_Atf2_Ikzf1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_76521902 | 4.82 |
ENSMUST00000164703.1
ENSMUST00000096365.3 |
Scrt1
|
scratch homolog 1, zinc finger protein (Drosophila) |
| chr5_+_21372642 | 3.78 |
ENSMUST00000035799.5
|
Fgl2
|
fibrinogen-like protein 2 |
| chr9_-_117252450 | 3.73 |
ENSMUST00000111773.3
ENSMUST00000068962.7 ENSMUST00000044901.7 |
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr11_+_23020464 | 2.98 |
ENSMUST00000094363.3
ENSMUST00000151877.1 |
Fam161a
|
family with sequence similarity 161, member A |
| chr11_+_87760533 | 2.75 |
ENSMUST00000039627.5
ENSMUST00000100644.3 |
Bzrap1
|
benzodiazepine receptor associated protein 1 |
| chr6_-_55175019 | 2.57 |
ENSMUST00000003569.5
|
Inmt
|
indolethylamine N-methyltransferase |
| chr18_+_37489465 | 2.33 |
ENSMUST00000055949.2
|
Pcdhb18
|
protocadherin beta 18 |
| chr2_-_181314500 | 2.32 |
ENSMUST00000103045.3
|
Stmn3
|
stathmin-like 3 |
| chr14_-_124677089 | 2.21 |
ENSMUST00000095529.3
|
Fgf14
|
fibroblast growth factor 14 |
| chr3_+_96219858 | 1.96 |
ENSMUST00000073115.4
|
Hist2h2ab
|
histone cluster 2, H2ab |
| chr2_+_152081529 | 1.81 |
ENSMUST00000064061.3
|
Scrt2
|
scratch homolog 2, zinc finger protein (Drosophila) |
| chr6_+_96113146 | 1.76 |
ENSMUST00000122120.1
|
Fam19a1
|
family with sequence similarity 19, member A1 |
| chr1_-_174921813 | 1.75 |
ENSMUST00000055294.3
|
Grem2
|
gremlin 2 homolog, cysteine knot superfamily (Xenopus laevis) |
| chr4_+_41762309 | 1.74 |
ENSMUST00000108042.2
|
Il11ra1
|
interleukin 11 receptor, alpha chain 1 |
| chr7_+_104244449 | 1.71 |
ENSMUST00000106849.2
ENSMUST00000060315.5 |
Trim34a
|
tripartite motif-containing 34A |
| chr2_-_164857542 | 1.70 |
ENSMUST00000109316.1
ENSMUST00000156255.1 ENSMUST00000128110.1 ENSMUST00000109317.3 |
Pltp
|
phospholipid transfer protein |
| chrX_+_164140447 | 1.63 |
ENSMUST00000073973.4
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr2_-_129699833 | 1.62 |
ENSMUST00000028883.5
|
Pdyn
|
prodynorphin |
| chr10_-_44004846 | 1.62 |
ENSMUST00000020017.8
|
Aim1
|
absent in melanoma 1 |
| chr12_-_78980758 | 1.61 |
ENSMUST00000174072.1
|
Tmem229b
|
transmembrane protein 229B |
| chr4_-_110290884 | 1.59 |
ENSMUST00000142722.1
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) |
| chr8_+_23411490 | 1.59 |
ENSMUST00000033952.7
|
Sfrp1
|
secreted frizzled-related protein 1 |
| chr14_+_103513328 | 1.57 |
ENSMUST00000095576.3
|
Scel
|
sciellin |
| chr16_-_42340595 | 1.53 |
ENSMUST00000102817.4
|
Gap43
|
growth associated protein 43 |
| chr11_-_97699634 | 1.51 |
ENSMUST00000103148.1
ENSMUST00000169807.1 |
Pcgf2
|
polycomb group ring finger 2 |
| chr16_+_32735886 | 1.51 |
ENSMUST00000132475.1
ENSMUST00000096106.3 |
Muc4
|
mucin 4 |
| chr5_-_92328068 | 1.48 |
ENSMUST00000113093.3
|
Cxcl9
|
chemokine (C-X-C motif) ligand 9 |
| chr9_-_117252126 | 1.48 |
ENSMUST00000174868.1
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chrX_-_73869804 | 1.48 |
ENSMUST00000066576.5
ENSMUST00000114430.1 |
L1cam
|
L1 cell adhesion molecule |
| chr11_+_69964758 | 1.43 |
ENSMUST00000108597.1
ENSMUST00000060651.5 ENSMUST00000108596.1 |
Cldn7
|
claudin 7 |
| chr14_+_54936456 | 1.43 |
ENSMUST00000037814.6
|
Cmtm5
|
CKLF-like MARVEL transmembrane domain containing 5 |
| chrX_+_41401304 | 1.42 |
ENSMUST00000076349.5
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3) |
| chr7_-_31054815 | 1.41 |
ENSMUST00000071697.4
ENSMUST00000108110.3 |
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr11_+_61022560 | 1.40 |
ENSMUST00000089184.4
|
Kcnj12
|
potassium inwardly-rectifying channel, subfamily J, member 12 |
| chr1_-_155146755 | 1.40 |
ENSMUST00000027744.8
|
Mr1
|
major histocompatibility complex, class I-related |
| chr10_+_34483400 | 1.39 |
ENSMUST00000019913.7
ENSMUST00000170771.1 |
Frk
|
fyn-related kinase |
| chr10_+_57784859 | 1.38 |
ENSMUST00000020024.5
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr4_-_49549523 | 1.38 |
ENSMUST00000029987.9
|
Aldob
|
aldolase B, fructose-bisphosphate |
| chrX_+_8271642 | 1.37 |
ENSMUST00000115590.1
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr4_+_54947976 | 1.34 |
ENSMUST00000098070.3
|
Zfp462
|
zinc finger protein 462 |
| chr3_+_8509477 | 1.34 |
ENSMUST00000029002.7
|
Stmn2
|
stathmin-like 2 |
| chr5_-_103211251 | 1.33 |
ENSMUST00000060871.5
ENSMUST00000112846.1 ENSMUST00000170792.1 ENSMUST00000112847.2 |
Mapk10
|
mitogen-activated protein kinase 10 |
| chr14_-_31577318 | 1.33 |
ENSMUST00000112027.2
|
Colq
|
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr4_-_148500449 | 1.31 |
ENSMUST00000030840.3
|
Angptl7
|
angiopoietin-like 7 |
| chr15_-_5063741 | 1.30 |
ENSMUST00000110689.3
|
C7
|
complement component 7 |
| chr11_-_78697370 | 1.30 |
ENSMUST00000142739.1
|
Nlk
|
nemo like kinase |
| chr6_-_123289862 | 1.28 |
ENSMUST00000032239.4
ENSMUST00000177367.1 |
Clec4e
|
C-type lectin domain family 4, member e |
| chr16_-_33056174 | 1.28 |
ENSMUST00000115100.1
ENSMUST00000040309.8 |
Iqcg
|
IQ motif containing G |
| chr4_+_102421518 | 1.26 |
ENSMUST00000106904.2
|
Pde4b
|
phosphodiesterase 4B, cAMP specific |
| chr11_-_69605829 | 1.25 |
ENSMUST00000047889.6
|
Atp1b2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide |
| chr2_+_84734050 | 1.24 |
ENSMUST00000090729.2
|
Ypel4
|
yippee-like 4 (Drosophila) |
| chr10_+_97565436 | 1.24 |
ENSMUST00000038160.4
|
Lum
|
lumican |
| chr1_-_158958367 | 1.19 |
ENSMUST00000159861.2
|
Pappa2
|
pappalysin 2 |
| chr5_+_52363925 | 1.19 |
ENSMUST00000101208.4
|
Sod3
|
superoxide dismutase 3, extracellular |
| chr2_-_24048857 | 1.18 |
ENSMUST00000114497.1
|
Hnmt
|
histamine N-methyltransferase |
| chr17_+_6673610 | 1.18 |
ENSMUST00000097430.2
|
Sytl3
|
synaptotagmin-like 3 |
| chr9_-_117252111 | 1.18 |
ENSMUST00000111772.3
|
Rbms3
|
RNA binding motif, single stranded interacting protein |
| chr5_+_30913398 | 1.17 |
ENSMUST00000031055.5
|
Emilin1
|
elastin microfibril interfacer 1 |
| chr3_+_137341067 | 1.16 |
ENSMUST00000122064.1
|
Emcn
|
endomucin |
| chr15_+_9436028 | 1.15 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chr3_-_113574242 | 1.15 |
ENSMUST00000142505.2
|
Amy1
|
amylase 1, salivary |
| chr1_+_131599239 | 1.12 |
ENSMUST00000027690.6
|
Avpr1b
|
arginine vasopressin receptor 1B |
| chr7_-_126585775 | 1.10 |
ENSMUST00000084589.4
|
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr7_+_104244465 | 1.09 |
ENSMUST00000106848.1
|
Trim34a
|
tripartite motif-containing 34A |
| chr1_-_51478390 | 1.09 |
ENSMUST00000027279.5
|
Nabp1
|
nucleic acid binding protein 1 |
| chrX_+_93286499 | 1.09 |
ENSMUST00000046565.7
ENSMUST00000113947.2 |
Arx
|
aristaless related homeobox |
| chr3_+_127791374 | 1.08 |
ENSMUST00000171621.1
|
Tifa
|
TRAF-interacting protein with forkhead-associated domain |
| chr3_+_89421619 | 1.07 |
ENSMUST00000094378.3
ENSMUST00000137793.1 |
Shc1
|
src homology 2 domain-containing transforming protein C1 |
| chr3_+_137341103 | 1.07 |
ENSMUST00000119475.1
|
Emcn
|
endomucin |
| chr6_+_41392356 | 1.07 |
ENSMUST00000049079.7
|
Gm5771
|
predicted gene 5771 |
| chr10_-_128176568 | 1.07 |
ENSMUST00000092033.2
|
Rbms2
|
RNA binding motif, single stranded interacting protein 2 |
| chr18_+_37484955 | 1.07 |
ENSMUST00000053856.4
|
Pcdhb17
|
protocadherin beta 17 |
| chr14_-_66868572 | 1.06 |
ENSMUST00000022629.8
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr6_+_125321409 | 1.05 |
ENSMUST00000176442.1
ENSMUST00000177329.1 |
Scnn1a
|
sodium channel, nonvoltage-gated 1 alpha |
| chr13_+_40917626 | 1.04 |
ENSMUST00000067778.6
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr3_+_97628804 | 1.04 |
ENSMUST00000107050.1
ENSMUST00000029729.8 ENSMUST00000107049.1 |
Fmo5
|
flavin containing monooxygenase 5 |
| chr10_-_127041513 | 1.03 |
ENSMUST00000116231.2
|
Mettl21b
|
methyltransferase like 21B |
| chr3_-_106406090 | 1.01 |
ENSMUST00000029510.7
|
BC051070
|
cDNA sequence BC051070 |
| chr4_-_53159885 | 0.98 |
ENSMUST00000030010.3
|
Abca1
|
ATP-binding cassette, sub-family A (ABC1), member 1 |
| chr2_+_70563435 | 0.98 |
ENSMUST00000123330.1
|
Gad1
|
glutamate decarboxylase 1 |
| chr6_+_78370877 | 0.98 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr1_-_20617992 | 0.97 |
ENSMUST00000088448.5
|
Pkhd1
|
polycystic kidney and hepatic disease 1 |
| chr11_+_67025144 | 0.97 |
ENSMUST00000079077.5
ENSMUST00000061786.5 |
Tmem220
|
transmembrane protein 220 |
| chr7_-_114636299 | 0.96 |
ENSMUST00000032906.4
ENSMUST00000032907.7 |
Calca
|
calcitonin/calcitonin-related polypeptide, alpha |
| chr5_+_57718021 | 0.96 |
ENSMUST00000094783.3
ENSMUST00000068110.7 |
Pcdh7
|
protocadherin 7 |
| chr16_-_23890805 | 0.95 |
ENSMUST00000004480.3
|
Sst
|
somatostatin |
| chr19_+_55741810 | 0.92 |
ENSMUST00000111657.3
ENSMUST00000061496.9 ENSMUST00000041717.7 ENSMUST00000111662.4 |
Tcf7l2
|
transcription factor 7 like 2, T cell specific, HMG box |
| chrX_+_164162167 | 0.92 |
ENSMUST00000131543.1
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr3_+_96576984 | 0.90 |
ENSMUST00000148290.1
|
Gm16253
|
predicted gene 16253 |
| chr10_+_128267997 | 0.90 |
ENSMUST00000050901.2
|
Apof
|
apolipoprotein F |
| chr11_+_29692937 | 0.90 |
ENSMUST00000102843.3
ENSMUST00000102842.3 ENSMUST00000078830.4 ENSMUST00000170731.1 |
Rtn4
|
reticulon 4 |
| chr6_-_137169710 | 0.90 |
ENSMUST00000117919.1
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr10_+_97479470 | 0.90 |
ENSMUST00000105287.3
|
Dcn
|
decorin |
| chr2_-_29253001 | 0.89 |
ENSMUST00000071201.4
|
Ntng2
|
netrin G2 |
| chr12_+_73286779 | 0.89 |
ENSMUST00000140523.1
|
Slc38a6
|
solute carrier family 38, member 6 |
| chr6_+_41521782 | 0.88 |
ENSMUST00000070380.4
|
Prss2
|
protease, serine, 2 |
| chr11_-_109298121 | 0.88 |
ENSMUST00000020920.3
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr5_+_66968416 | 0.86 |
ENSMUST00000038188.7
|
Limch1
|
LIM and calponin homology domains 1 |
| chr12_+_109545390 | 0.86 |
ENSMUST00000146701.1
|
Meg3
|
maternally expressed 3 |
| chr13_-_34919992 | 0.86 |
ENSMUST00000021851.6
|
Fam217a
|
family with sequence similarity 217, member A |
| chrX_-_70365052 | 0.86 |
ENSMUST00000101509.2
|
Ids
|
iduronate 2-sulfatase |
| chr3_+_115080965 | 0.85 |
ENSMUST00000051309.8
|
Olfm3
|
olfactomedin 3 |
| chr15_+_10215955 | 0.85 |
ENSMUST00000130720.1
|
Prlr
|
prolactin receptor |
| chr6_-_124769548 | 0.85 |
ENSMUST00000149652.1
ENSMUST00000112476.1 ENSMUST00000004378.8 |
Eno2
|
enolase 2, gamma neuronal |
| chr10_-_109010955 | 0.84 |
ENSMUST00000105276.1
ENSMUST00000064054.7 |
Syt1
|
synaptotagmin I |
| chr8_-_25101734 | 0.83 |
ENSMUST00000098866.4
|
Plekha2
|
pleckstrin homology domain-containing, family A (phosphoinositide binding specific) member 2 |
| chr1_-_169747634 | 0.83 |
ENSMUST00000027991.5
ENSMUST00000111357.1 |
Rgs4
|
regulator of G-protein signaling 4 |
| chr12_-_119238794 | 0.83 |
ENSMUST00000026360.8
|
Itgb8
|
integrin beta 8 |
| chr18_+_37447641 | 0.82 |
ENSMUST00000052387.3
|
Pcdhb14
|
protocadherin beta 14 |
| chr18_-_31317043 | 0.81 |
ENSMUST00000139924.1
ENSMUST00000153060.1 |
Rit2
|
Ras-like without CAAX 2 |
| chr5_+_137030275 | 0.81 |
ENSMUST00000041543.8
|
Vgf
|
VGF nerve growth factor inducible |
| chr14_-_46390501 | 0.81 |
ENSMUST00000100676.2
|
Bmp4
|
bone morphogenetic protein 4 |
| chr1_-_136230289 | 0.81 |
ENSMUST00000150163.1
ENSMUST00000144464.1 |
5730559C18Rik
|
RIKEN cDNA 5730559C18 gene |
| chr7_-_30861470 | 0.80 |
ENSMUST00000052700.3
|
Ffar1
|
free fatty acid receptor 1 |
| chr10_-_34207551 | 0.79 |
ENSMUST00000048010.7
|
Dse
|
dermatan sulfate epimerase |
| chr2_-_66568312 | 0.79 |
ENSMUST00000112354.1
|
Scn9a
|
sodium channel, voltage-gated, type IX, alpha |
| chr18_-_3309858 | 0.79 |
ENSMUST00000144496.1
ENSMUST00000154715.1 |
Crem
|
cAMP responsive element modulator |
| chr6_+_80018877 | 0.79 |
ENSMUST00000147663.1
ENSMUST00000128718.1 ENSMUST00000126005.1 ENSMUST00000133918.1 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chr15_+_98571004 | 0.78 |
ENSMUST00000023728.6
|
4930415O20Rik
|
RIKEN cDNA 4930415O20 gene |
| chr14_-_46390576 | 0.77 |
ENSMUST00000074077.5
|
Bmp4
|
bone morphogenetic protein 4 |
| chr13_+_42709482 | 0.77 |
ENSMUST00000066928.5
ENSMUST00000148891.1 |
Phactr1
|
phosphatase and actin regulator 1 |
| chr10_+_69151427 | 0.77 |
ENSMUST00000167286.1
|
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr6_-_41314700 | 0.77 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr15_-_95528228 | 0.77 |
ENSMUST00000075275.2
|
Nell2
|
NEL-like 2 |
| chr17_+_33919332 | 0.76 |
ENSMUST00000025161.7
|
Tapbp
|
TAP binding protein |
| chr18_+_33464163 | 0.76 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chr4_-_111898695 | 0.76 |
ENSMUST00000102720.1
|
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr11_+_101246405 | 0.76 |
ENSMUST00000122006.1
ENSMUST00000151830.1 |
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chrX_+_8271381 | 0.75 |
ENSMUST00000033512.4
|
Slc38a5
|
solute carrier family 38, member 5 |
| chr7_-_127930066 | 0.75 |
ENSMUST00000032988.8
|
Prss8
|
protease, serine, 8 (prostasin) |
| chr10_+_127866457 | 0.74 |
ENSMUST00000092058.3
|
BC089597
|
cDNA sequence BC089597 |
| chr17_+_85028347 | 0.74 |
ENSMUST00000024944.7
|
Slc3a1
|
solute carrier family 3, member 1 |
| chr11_-_109298066 | 0.74 |
ENSMUST00000106706.1
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr9_-_39604124 | 0.73 |
ENSMUST00000042485.4
ENSMUST00000141370.1 |
AW551984
|
expressed sequence AW551984 |
| chr6_+_41302265 | 0.73 |
ENSMUST00000031913.4
|
Try4
|
trypsin 4 |
| chr11_-_119086221 | 0.73 |
ENSMUST00000026665.7
|
Cbx4
|
chromobox 4 |
| chr7_+_104244496 | 0.73 |
ENSMUST00000106854.1
ENSMUST00000143414.1 |
Trim34a
|
tripartite motif-containing 34A |
| chr3_+_19985612 | 0.72 |
ENSMUST00000172860.1
|
Cp
|
ceruloplasmin |
| chr11_-_115367667 | 0.72 |
ENSMUST00000123428.1
ENSMUST00000044152.6 ENSMUST00000106542.2 |
Hid1
|
HID1 domain containing |
| chr5_+_90460889 | 0.72 |
ENSMUST00000031314.8
|
Alb
|
albumin |
| chr19_-_58455161 | 0.72 |
ENSMUST00000135730.1
ENSMUST00000152507.1 |
Gfra1
|
glial cell line derived neurotrophic factor family receptor alpha 1 |
| chr9_+_99470440 | 0.71 |
ENSMUST00000056103.4
|
1600029I14Rik
|
RIKEN cDNA 1600029I14 gene |
| chr12_+_35992900 | 0.71 |
ENSMUST00000020898.5
|
Agr2
|
anterior gradient 2 |
| chr14_-_13914478 | 0.70 |
ENSMUST00000144914.1
|
Gm281
|
predicted gene 281 |
| chr8_+_62951361 | 0.70 |
ENSMUST00000119068.1
|
Spock3
|
sparc/osteonectin, cwcv and kazal-like domains proteoglycan 3 |
| chr7_+_126950687 | 0.70 |
ENSMUST00000106333.1
|
Sez6l2
|
seizure related 6 homolog like 2 |
| chr4_+_43641262 | 0.70 |
ENSMUST00000123351.1
ENSMUST00000128549.1 |
Npr2
|
natriuretic peptide receptor 2 |
| chr3_+_146121655 | 0.70 |
ENSMUST00000039450.4
|
Mcoln3
|
mucolipin 3 |
| chr18_-_3281712 | 0.69 |
ENSMUST00000182204.1
ENSMUST00000154705.1 ENSMUST00000182833.1 ENSMUST00000151084.1 |
Crem
|
cAMP responsive element modulator |
| chr8_+_105427634 | 0.69 |
ENSMUST00000067305.6
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr17_-_90455872 | 0.69 |
ENSMUST00000174337.1
ENSMUST00000172466.1 |
Nrxn1
|
neurexin I |
| chr14_+_32991379 | 0.69 |
ENSMUST00000038956.4
|
Lrrc18
|
leucine rich repeat containing 18 |
| chr5_-_53707532 | 0.68 |
ENSMUST00000031093.3
|
Cckar
|
cholecystokinin A receptor |
| chr7_-_4522427 | 0.67 |
ENSMUST00000098859.3
|
Tnni3
|
troponin I, cardiac 3 |
| chr6_-_129507107 | 0.67 |
ENSMUST00000183258.1
ENSMUST00000182784.1 ENSMUST00000032265.6 ENSMUST00000162815.1 |
Olr1
|
oxidized low density lipoprotein (lectin-like) receptor 1 |
| chr18_+_37518341 | 0.66 |
ENSMUST00000097609.1
|
Pcdhb22
|
protocadherin beta 22 |
| chr15_-_84447037 | 0.66 |
ENSMUST00000080751.2
|
1810041L15Rik
|
RIKEN cDNA 1810041L15 gene |
| chr5_-_100719675 | 0.66 |
ENSMUST00000112908.1
ENSMUST00000045617.8 |
Hpse
|
heparanase |
| chrX_-_95658416 | 0.66 |
ENSMUST00000044382.6
|
Zc4h2
|
zinc finger, C4H2 domain containing |
| chr1_-_9298499 | 0.65 |
ENSMUST00000132064.1
|
Sntg1
|
syntrophin, gamma 1 |
| chr7_+_28071230 | 0.65 |
ENSMUST00000138392.1
ENSMUST00000076648.7 |
Fcgbp
|
Fc fragment of IgG binding protein |
| chr5_+_136967859 | 0.64 |
ENSMUST00000001790.5
|
Cldn15
|
claudin 15 |
| chr8_+_82863351 | 0.64 |
ENSMUST00000078525.5
|
Rnf150
|
ring finger protein 150 |
| chr11_+_29718563 | 0.64 |
ENSMUST00000060992.5
|
Rtn4
|
reticulon 4 |
| chr6_+_125145235 | 0.64 |
ENSMUST00000119527.1
ENSMUST00000088276.6 ENSMUST00000051171.7 ENSMUST00000117675.1 |
Iffo1
|
intermediate filament family orphan 1 |
| chr1_-_88205674 | 0.63 |
ENSMUST00000119972.2
|
Dnajb3
|
DnaJ (Hsp40) homolog, subfamily B, member 3 |
| chr1_-_130729249 | 0.62 |
ENSMUST00000171479.1
|
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr18_-_36726730 | 0.62 |
ENSMUST00000061829.6
|
Cd14
|
CD14 antigen |
| chr8_+_116504973 | 0.62 |
ENSMUST00000078170.5
|
Dynlrb2
|
dynein light chain roadblock-type 2 |
| chr5_-_101665195 | 0.62 |
ENSMUST00000044125.8
|
Nkx6-1
|
NK6 homeobox 1 |
| chr5_+_145204523 | 0.62 |
ENSMUST00000085671.3
ENSMUST00000031601.7 |
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr13_-_41358990 | 0.62 |
ENSMUST00000163623.1
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated gene 9 |
| chr8_-_33641940 | 0.61 |
ENSMUST00000095349.4
|
Ubxn8
|
UBX domain protein 8 |
| chr11_+_32000452 | 0.61 |
ENSMUST00000020537.2
ENSMUST00000109409.1 |
Nsg2
|
neuron specific gene family member 2 |
| chr14_+_54464141 | 0.61 |
ENSMUST00000022782.8
|
Lrp10
|
low-density lipoprotein receptor-related protein 10 |
| chr4_-_110287479 | 0.61 |
ENSMUST00000106598.1
ENSMUST00000102723.4 ENSMUST00000153906.1 |
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D) |
| chr11_+_98664341 | 0.60 |
ENSMUST00000017348.2
|
Gsdma
|
gasdermin A |
| chr5_+_19907502 | 0.60 |
ENSMUST00000101558.3
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr14_-_70635946 | 0.59 |
ENSMUST00000022695.9
|
Dmtn
|
dematin actin binding protein |
| chr9_-_121792478 | 0.59 |
ENSMUST00000035110.4
|
Hhatl
|
hedgehog acyltransferase-like |
| chr6_-_137169678 | 0.59 |
ENSMUST00000119610.1
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr3_-_113574758 | 0.59 |
ENSMUST00000106540.1
|
Amy1
|
amylase 1, salivary |
| chr11_+_101627942 | 0.59 |
ENSMUST00000010506.3
|
Rdm1
|
RAD52 motif 1 |
| chr1_-_45503282 | 0.58 |
ENSMUST00000086430.4
|
Col5a2
|
collagen, type V, alpha 2 |
| chr11_+_93099284 | 0.58 |
ENSMUST00000092780.3
ENSMUST00000107863.2 |
Car10
|
carbonic anhydrase 10 |
| chr5_+_90518932 | 0.58 |
ENSMUST00000113179.2
ENSMUST00000128740.1 |
Afm
|
afamin |
| chr7_+_90380646 | 0.57 |
ENSMUST00000098310.2
|
Sytl2
|
synaptotagmin-like 2 |
| chr10_+_57784914 | 0.57 |
ENSMUST00000165013.1
|
Fabp7
|
fatty acid binding protein 7, brain |
| chr11_-_99493112 | 0.57 |
ENSMUST00000006969.7
|
Krt23
|
keratin 23 |
| chrX_+_101376359 | 0.57 |
ENSMUST00000119080.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr13_-_9878998 | 0.57 |
ENSMUST00000063093.9
|
Chrm3
|
cholinergic receptor, muscarinic 3, cardiac |
| chr14_+_55561060 | 0.57 |
ENSMUST00000117701.1
|
Dcaf11
|
DDB1 and CUL4 associated factor 11 |
| chrX_-_7574120 | 0.57 |
ENSMUST00000045924.7
ENSMUST00000115742.2 ENSMUST00000150787.1 |
Ppp1r3f
|
protein phosphatase 1, regulatory (inhibitor) subunit 3F |
| chr8_+_105413614 | 0.57 |
ENSMUST00000109355.2
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr10_+_115817247 | 0.56 |
ENSMUST00000035563.7
ENSMUST00000080630.3 ENSMUST00000179196.1 |
Tspan8
|
tetraspanin 8 |
| chr14_-_54409469 | 0.56 |
ENSMUST00000000984.4
|
Slc7a7
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 |
| chr11_+_101245996 | 0.55 |
ENSMUST00000129680.1
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr16_-_22161450 | 0.55 |
ENSMUST00000115379.1
|
Igf2bp2
|
insulin-like growth factor 2 mRNA binding protein 2 |
| chr7_+_100006404 | 0.55 |
ENSMUST00000032977.4
|
Chrdl2
|
chordin-like 2 |
| chr12_-_90738438 | 0.55 |
ENSMUST00000082432.3
|
Dio2
|
deiodinase, iodothyronine, type II |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ezh2_Atf2_Ikzf1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:0060300 | regulation of cytokine activity(GO:0060300) |
| 0.6 | 2.6 | GO:0015801 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.6 | 1.7 | GO:0072098 | intermediate mesoderm development(GO:0048389) pattern specification involved in mesonephros development(GO:0061227) anterior/posterior pattern specification involved in kidney development(GO:0072098) |
| 0.6 | 1.7 | GO:2000041 | regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.4 | 2.7 | GO:1903278 | positive regulation of sodium ion export(GO:1903275) positive regulation of sodium ion export from cell(GO:1903278) |
| 0.4 | 2.2 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.4 | 1.2 | GO:0021759 | globus pallidus development(GO:0021759) |
| 0.4 | 2.0 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.4 | 1.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.4 | 1.1 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) |
| 0.3 | 1.4 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.3 | 1.4 | GO:0006001 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) glycolytic process through fructose-1-phosphate(GO:0061625) |
| 0.3 | 2.0 | GO:0002835 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.3 | 1.0 | GO:0001983 | baroreceptor response to increased systemic arterial blood pressure(GO:0001983) |
| 0.3 | 1.8 | GO:0032489 | regulation of Cdc42 protein signal transduction(GO:0032489) |
| 0.3 | 1.4 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.3 | 1.7 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.3 | 1.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.3 | 0.8 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.3 | 1.3 | GO:0008582 | regulation of synaptic growth at neuromuscular junction(GO:0008582) |
| 0.3 | 2.3 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.3 | 1.5 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.2 | 1.2 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.2 | 0.7 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.2 | 0.7 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.2 | 1.2 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.2 | 0.7 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.2 | 0.7 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.2 | 0.4 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.2 | 1.3 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.2 | 0.7 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.2 | 1.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 0.6 | GO:0071725 | response to triacyl bacterial lipopeptide(GO:0071725) cellular response to triacyl bacterial lipopeptide(GO:0071727) |
| 0.2 | 1.6 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.2 | 1.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.2 | 1.0 | GO:1990839 | response to endothelin(GO:1990839) |
| 0.2 | 0.6 | GO:0035585 | calcium-mediated signaling using extracellular calcium source(GO:0035585) |
| 0.2 | 1.2 | GO:0050748 | negative regulation of lipoprotein metabolic process(GO:0050748) |
| 0.2 | 1.9 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.2 | 0.4 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.2 | 0.5 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.2 | 0.5 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.2 | 0.7 | GO:1903896 | positive regulation of IRE1-mediated unfolded protein response(GO:1903896) |
| 0.2 | 0.5 | GO:0002591 | positive regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002591) peptide antigen transport(GO:0046968) |
| 0.2 | 0.3 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.2 | 0.7 | GO:0001980 | regulation of systemic arterial blood pressure by ischemic conditions(GO:0001980) |
| 0.2 | 0.5 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 1.0 | GO:0036476 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.2 | 0.5 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.2 | 0.3 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.2 | 0.3 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.2 | 0.5 | GO:1900248 | cytoplasmic translational elongation(GO:0002182) regulation of cytoplasmic translational elongation(GO:1900247) negative regulation of cytoplasmic translational elongation(GO:1900248) |
| 0.2 | 0.3 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.2 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.1 | GO:2000481 | positive regulation of cAMP-dependent protein kinase activity(GO:2000481) |
| 0.1 | 0.3 | GO:0072199 | mesenchymal cell proliferation involved in ureter development(GO:0072198) regulation of mesenchymal cell proliferation involved in ureter development(GO:0072199) |
| 0.1 | 0.3 | GO:0002589 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) |
| 0.1 | 0.1 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) |
| 0.1 | 0.6 | GO:0002408 | myeloid dendritic cell chemotaxis(GO:0002408) |
| 0.1 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.3 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.4 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.4 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.1 | 1.7 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.1 | 1.0 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.1 | 0.7 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 2.2 | GO:0015816 | glycine transport(GO:0015816) |
| 0.1 | 1.2 | GO:0048625 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) embryonic hindgut morphogenesis(GO:0048619) myoblast fate commitment(GO:0048625) |
| 0.1 | 0.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.7 | GO:0097118 | neuroligin clustering involved in postsynaptic membrane assembly(GO:0097118) |
| 0.1 | 4.1 | GO:0046688 | response to copper ion(GO:0046688) |
| 0.1 | 0.8 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.1 | 1.8 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.1 | 0.8 | GO:1904075 | regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.1 | 0.3 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 0.1 | 0.5 | GO:1900736 | regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900736) |
| 0.1 | 0.3 | GO:0033092 | positive regulation of immature T cell proliferation in thymus(GO:0033092) |
| 0.1 | 0.5 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.1 | 0.4 | GO:0010958 | regulation of amino acid import(GO:0010958) |
| 0.1 | 0.5 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.1 | 0.9 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.1 | 0.7 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.1 | GO:1902488 | cholangiocyte apoptotic process(GO:1902488) regulation of cholangiocyte apoptotic process(GO:1904192) negative regulation of cholangiocyte apoptotic process(GO:1904193) |
| 0.1 | 1.2 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.1 | 0.5 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 6.1 | GO:2001222 | regulation of neuron migration(GO:2001222) |
| 0.1 | 0.5 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.2 | GO:0032741 | positive regulation of interleukin-18 production(GO:0032741) positive regulation of superoxide dismutase activity(GO:1901671) positive regulation of removal of superoxide radicals(GO:1904833) |
| 0.1 | 0.5 | GO:0008594 | photoreceptor cell morphogenesis(GO:0008594) |
| 0.1 | 0.6 | GO:0021984 | adenohypophysis development(GO:0021984) |
| 0.1 | 0.6 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.4 | GO:0060376 | positive regulation of mast cell differentiation(GO:0060376) |
| 0.1 | 4.1 | GO:0019835 | cytolysis(GO:0019835) |
| 0.1 | 0.1 | GO:0036022 | limb joint morphogenesis(GO:0036022) embryonic skeletal limb joint morphogenesis(GO:0036023) |
| 0.1 | 0.1 | GO:0009744 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.1 | 0.5 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.1 | 0.5 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.1 | 0.4 | GO:2000525 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.1 | 0.4 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.1 | 0.4 | GO:0070459 | prolactin secretion(GO:0070459) |
| 0.1 | 0.4 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.1 | 0.4 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.4 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.1 | 1.0 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.1 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.1 | 0.3 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.1 | 2.5 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.1 | 0.1 | GO:0072190 | ureter urothelium development(GO:0072190) |
| 0.1 | 1.4 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 2.0 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.1 | 0.4 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.7 | GO:0060923 | cardiac muscle cell fate commitment(GO:0060923) |
| 0.1 | 1.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.1 | 0.4 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.1 | 0.8 | GO:0043084 | penile erection(GO:0043084) |
| 0.1 | 0.5 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.1 | 0.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.3 | GO:0010814 | substance P catabolic process(GO:0010814) calcitonin catabolic process(GO:0010816) endothelin maturation(GO:0034959) |
| 0.1 | 0.1 | GO:0060197 | cloacal septation(GO:0060197) |
| 0.1 | 3.5 | GO:0007212 | dopamine receptor signaling pathway(GO:0007212) |
| 0.1 | 0.3 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0006068 | ethanol catabolic process(GO:0006068) primary alcohol catabolic process(GO:0034310) |
| 0.1 | 0.6 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 1.4 | GO:0032463 | negative regulation of protein homooligomerization(GO:0032463) |
| 0.1 | 0.8 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.1 | 0.2 | GO:0006114 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.1 | 0.2 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.1 | 0.2 | GO:1900019 | regulation of protein kinase C activity(GO:1900019) positive regulation of protein kinase C activity(GO:1900020) |
| 0.1 | 0.2 | GO:0042660 | positive regulation of cell fate specification(GO:0042660) |
| 0.1 | 0.3 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.7 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.8 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.1 | 0.2 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.1 | 0.4 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.1 | 0.8 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.1 | 1.5 | GO:0046697 | decidualization(GO:0046697) |
| 0.1 | 0.2 | GO:0045062 | extrathymic T cell selection(GO:0045062) |
| 0.1 | 0.4 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.1 | 0.4 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.1 | 0.3 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.1 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.1 | 0.1 | GO:0043380 | regulation of memory T cell differentiation(GO:0043380) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.4 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) |
| 0.1 | 0.4 | GO:0039019 | pronephric nephron development(GO:0039019) |
| 0.1 | 0.1 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.1 | 0.3 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.1 | 0.5 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.1 | 0.2 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 0.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 0.7 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.1 | 0.6 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.8 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.1 | 0.2 | GO:0021934 | hindbrain tangential cell migration(GO:0021934) |
| 0.1 | 0.3 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.1 | 0.2 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.1 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.1 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.3 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.1 | 0.3 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) |
| 0.1 | 0.8 | GO:1901629 | regulation of presynaptic membrane organization(GO:1901629) |
| 0.1 | 0.5 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.1 | 0.2 | GO:0019255 | UDP-glucose metabolic process(GO:0006011) glucose 1-phosphate metabolic process(GO:0019255) |
| 0.1 | 0.3 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.1 | 1.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.1 | 0.3 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.1 | 0.3 | GO:1902608 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.1 | 0.3 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 1.4 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.5 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.1 | 0.2 | GO:0010936 | detection of oxygen(GO:0003032) negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.7 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.1 | 0.2 | GO:0019859 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.1 | 0.2 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.1 | 0.5 | GO:0021612 | facial nerve structural organization(GO:0021612) |
| 0.1 | 0.5 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.2 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.3 | GO:0061734 | parkin-mediated mitophagy in response to mitochondrial depolarization(GO:0061734) |
| 0.1 | 0.3 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.1 | 0.2 | GO:1905077 | negative regulation of interleukin-17 secretion(GO:1905077) |
| 0.1 | 0.4 | GO:0051660 | establishment of centrosome localization(GO:0051660) |
| 0.1 | 0.2 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.1 | 0.2 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.3 | GO:0015888 | thiamine transport(GO:0015888) |
| 0.1 | 0.2 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.1 | 0.2 | GO:0034241 | positive regulation of macrophage fusion(GO:0034241) |
| 0.1 | 0.3 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 1.0 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.1 | 0.7 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.1 | 0.4 | GO:0031547 | brain-derived neurotrophic factor receptor signaling pathway(GO:0031547) |
| 0.1 | 0.7 | GO:0070633 | transepithelial transport(GO:0070633) |
| 0.1 | 0.2 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.1 | 0.3 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.3 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.1 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.1 | 0.2 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.1 | 0.6 | GO:2000465 | regulation of glycogen (starch) synthase activity(GO:2000465) |
| 0.1 | 0.3 | GO:1901678 | iron coordination entity transport(GO:1901678) |
| 0.1 | 0.2 | GO:2000538 | signal complex assembly(GO:0007172) regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.3 | GO:0055095 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) |
| 0.1 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.3 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) |
| 0.0 | 0.4 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 1.9 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.5 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.0 | 0.6 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.2 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.1 | GO:1903817 | negative regulation of delayed rectifier potassium channel activity(GO:1902260) negative regulation of voltage-gated potassium channel activity(GO:1903817) |
| 0.0 | 1.2 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0002220 | innate immune response activating cell surface receptor signaling pathway(GO:0002220) |
| 0.0 | 0.1 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.2 | GO:0010519 | negative regulation of phospholipase activity(GO:0010519) |
| 0.0 | 0.2 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.4 | GO:0071763 | nuclear membrane organization(GO:0071763) |
| 0.0 | 0.3 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.7 | GO:0042759 | long-chain fatty acid biosynthetic process(GO:0042759) |
| 0.0 | 0.0 | GO:0036301 | macrophage colony-stimulating factor production(GO:0036301) granulocyte colony-stimulating factor production(GO:0071611) regulation of granulocyte colony-stimulating factor production(GO:0071655) regulation of macrophage colony-stimulating factor production(GO:1901256) |
| 0.0 | 0.3 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.1 | GO:0060741 | prostate gland stromal morphogenesis(GO:0060741) |
| 0.0 | 0.2 | GO:0033578 | protein glycosylation in Golgi(GO:0033578) |
| 0.0 | 0.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.3 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0006707 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0002461 | tolerance induction dependent upon immune response(GO:0002461) |
| 0.0 | 0.1 | GO:1901492 | positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.2 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.1 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 1.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.5 | GO:0009143 | nucleoside triphosphate catabolic process(GO:0009143) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) |
| 0.0 | 0.4 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.5 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.5 | GO:0048194 | Golgi vesicle budding(GO:0048194) |
| 0.0 | 0.2 | GO:1904453 | regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904451) positive regulation of hydrogen:potassium-exchanging ATPase activity(GO:1904453) |
| 0.0 | 0.2 | GO:0010796 | regulation of multivesicular body size(GO:0010796) |
| 0.0 | 0.1 | GO:0033693 | peripheral nervous system axon regeneration(GO:0014012) neurofilament bundle assembly(GO:0033693) response to sodium arsenite(GO:1903935) |
| 0.0 | 0.5 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.2 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.2 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 1.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.4 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.3 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.4 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.0 | 0.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.0 | 0.3 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0010986 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0031022 | modulation of microtubule cytoskeleton involved in cerebral cortex radial glia guided migration(GO:0021815) nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.2 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 1.9 | GO:0051591 | response to cAMP(GO:0051591) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.3 | GO:0098909 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) |
| 0.0 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.2 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.7 | GO:0010801 | negative regulation of peptidyl-threonine phosphorylation(GO:0010801) |
| 0.0 | 0.1 | GO:0042539 | hypotonic salinity response(GO:0042539) cellular hypotonic salinity response(GO:0071477) |
| 0.0 | 5.3 | GO:0007416 | synapse assembly(GO:0007416) |
| 0.0 | 0.3 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.0 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.1 | GO:0090425 | hepatocyte cell migration(GO:0002194) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.0 | 0.2 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.1 | GO:0040030 | regulation of molecular function, epigenetic(GO:0040030) |
| 0.0 | 0.7 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.1 | GO:0072553 | terminal button organization(GO:0072553) |
| 0.0 | 0.4 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.3 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.4 | GO:0097205 | renal filtration(GO:0097205) |
| 0.0 | 0.6 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.2 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.1 | GO:1904528 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) positive regulation of microtubule binding(GO:1904528) |
| 0.0 | 0.3 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.0 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.1 | GO:0010626 | negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.9 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.5 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.1 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.3 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) |
| 0.0 | 0.1 | GO:0042758 | long-chain fatty acid catabolic process(GO:0042758) |
| 0.0 | 0.2 | GO:0003406 | retinal pigment epithelium development(GO:0003406) |
| 0.0 | 0.2 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.0 | 0.1 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.0 | 0.1 | GO:0021993 | initiation of neural tube closure(GO:0021993) |
| 0.0 | 0.2 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.2 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.2 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0006013 | mannose metabolic process(GO:0006013) |
| 0.0 | 0.2 | GO:1902748 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0006699 | bile acid biosynthetic process(GO:0006699) |
| 0.0 | 0.1 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.4 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.3 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 0.0 | 0.1 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.2 | GO:0010575 | positive regulation of vascular endothelial growth factor production(GO:0010575) |
| 0.0 | 0.2 | GO:0070072 | proton-transporting V-type ATPase complex assembly(GO:0070070) vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.1 | GO:0060718 | chorionic trophoblast cell differentiation(GO:0060718) |
| 0.0 | 0.2 | GO:0051572 | negative regulation of histone H3-K4 methylation(GO:0051572) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.8 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.3 | GO:0006614 | SRP-dependent cotranslational protein targeting to membrane(GO:0006614) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:2000659 | regulation of interleukin-1-mediated signaling pathway(GO:2000659) |
| 0.0 | 0.3 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.1 | GO:0045988 | negative regulation of striated muscle contraction(GO:0045988) |
| 0.0 | 1.0 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.2 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.5 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.3 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0009950 | dorsal/ventral axis specification(GO:0009950) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of gonadotropin secretion(GO:0032277) negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.3 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.1 | GO:0048104 | establishment of body hair or bristle planar orientation(GO:0048104) establishment of body hair planar orientation(GO:0048105) |
| 0.0 | 0.1 | GO:1904457 | positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 2.5 | GO:0016052 | carbohydrate catabolic process(GO:0016052) |
| 0.0 | 0.2 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.6 | GO:0001919 | regulation of receptor recycling(GO:0001919) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.6 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell mediated immunity(GO:0002448) mast cell degranulation(GO:0043303) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.0 | 0.8 | GO:0071514 | genetic imprinting(GO:0071514) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) |
| 0.0 | 0.6 | GO:0030514 | negative regulation of BMP signaling pathway(GO:0030514) |
| 0.0 | 0.1 | GO:0045822 | negative regulation of heart contraction(GO:0045822) |
| 0.0 | 0.1 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.0 | 0.5 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.5 | GO:0097352 | autophagosome maturation(GO:0097352) |
| 0.0 | 0.6 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.0 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.0 | 0.1 | GO:0070294 | renal sodium ion transport(GO:0003096) renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.2 | GO:0033344 | cholesterol efflux(GO:0033344) |
| 0.0 | 0.2 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:1902477 | defense response, incompatible interaction(GO:0009814) defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.1 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.0 | 0.2 | GO:0002755 | MyD88-dependent toll-like receptor signaling pathway(GO:0002755) |
| 0.0 | 0.2 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 1.2 | GO:0038083 | peptidyl-tyrosine autophosphorylation(GO:0038083) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.3 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.2 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.4 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.2 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.1 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.0 | 0.5 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) ATP hydrolysis coupled transmembrane transport(GO:0090662) |
| 0.0 | 0.2 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 0.6 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.9 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.3 | GO:0006691 | leukotriene metabolic process(GO:0006691) |
| 0.0 | 0.2 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.2 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.5 | GO:0006891 | intra-Golgi vesicle-mediated transport(GO:0006891) |
| 0.0 | 0.2 | GO:0001783 | B cell apoptotic process(GO:0001783) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.0 | GO:0021553 | olfactory nerve development(GO:0021553) |
| 0.0 | 0.1 | GO:0086042 | cardiac muscle cell-cardiac muscle cell adhesion(GO:0086042) bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.0 | GO:0035729 | cellular response to hepatocyte growth factor stimulus(GO:0035729) |
| 0.0 | 0.0 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.0 | 0.2 | GO:0051156 | glucose 6-phosphate metabolic process(GO:0051156) |
| 0.0 | 0.2 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.0 | GO:0002759 | regulation of antimicrobial humoral response(GO:0002759) |
| 0.0 | 1.4 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.2 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 1.2 | GO:0007586 | digestion(GO:0007586) |
| 0.0 | 0.1 | GO:0001964 | startle response(GO:0001964) |
| 0.0 | 0.8 | GO:0046889 | positive regulation of lipid biosynthetic process(GO:0046889) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.3 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.3 | GO:0010971 | positive regulation of G2/M transition of mitotic cell cycle(GO:0010971) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.0 | GO:0045404 | positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.4 | GO:0042491 | auditory receptor cell differentiation(GO:0042491) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.0 | GO:2000553 | regulation of T-helper 2 cell cytokine production(GO:2000551) positive regulation of T-helper 2 cell cytokine production(GO:2000553) |
| 0.0 | 0.1 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.0 | 0.3 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0045217 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.5 | GO:0070542 | response to fatty acid(GO:0070542) |
| 0.0 | 0.3 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.1 | GO:0032596 | protein transport into membrane raft(GO:0032596) |
| 0.0 | 0.4 | GO:1903078 | positive regulation of protein localization to plasma membrane(GO:1903078) |
| 0.0 | 0.6 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.5 | GO:0045744 | negative regulation of G-protein coupled receptor protein signaling pathway(GO:0045744) |
| 0.0 | 1.1 | GO:0009308 | amine metabolic process(GO:0009308) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0010883 | regulation of lipid storage(GO:0010883) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0050926 | regulation of positive chemotaxis(GO:0050926) |
| 0.0 | 0.2 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.4 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) |
| 0.0 | 0.1 | GO:0055090 | acylglycerol homeostasis(GO:0055090) triglyceride homeostasis(GO:0070328) |
| 0.0 | 0.9 | GO:0030038 | contractile actin filament bundle assembly(GO:0030038) stress fiber assembly(GO:0043149) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.3 | GO:0009948 | anterior/posterior axis specification(GO:0009948) |
| 0.0 | 0.3 | GO:0097320 | membrane tubulation(GO:0097320) |
| 0.0 | 0.2 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.7 | GO:0006888 | ER to Golgi vesicle-mediated transport(GO:0006888) |
| 0.0 | 0.1 | GO:0033234 | negative regulation of protein sumoylation(GO:0033234) |
| 0.0 | 0.1 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.1 | GO:0099612 | protein localization to axon(GO:0099612) |
| 0.0 | 0.4 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 1.0 | GO:0045727 | positive regulation of translation(GO:0045727) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0002053 | positive regulation of mesenchymal cell proliferation(GO:0002053) |
| 0.0 | 0.2 | GO:0006353 | DNA-templated transcription, termination(GO:0006353) |
| 0.0 | 0.0 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.0 | 0.2 | GO:0016973 | poly(A)+ mRNA export from nucleus(GO:0016973) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.4 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.3 | 1.3 | GO:0090498 | extrinsic component of Golgi membrane(GO:0090498) |
| 0.2 | 1.3 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.2 | 0.8 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.2 | 0.6 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 2.2 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.2 | 1.8 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.2 | 0.9 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.2 | 1.7 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.2 | 1.7 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.2 | 0.7 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.2 | 0.5 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.2 | 1.3 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.2 | 1.4 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.2 | 1.3 | GO:0042825 | TAP complex(GO:0042825) |
| 0.2 | 2.7 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 0.6 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.2 | 2.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.1 | 0.6 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 1.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.1 | 0.5 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.1 | 0.9 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.1 | 2.6 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.1 | 1.2 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 0.5 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.1 | 0.3 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.1 | 0.3 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.4 | GO:0097487 | multivesicular body, internal vesicle(GO:0097487) |
| 0.1 | 0.8 | GO:0098644 | complex of collagen trimers(GO:0098644) |
| 0.1 | 0.7 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.6 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 0.4 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.1 | 0.1 | GO:0016939 | kinesin II complex(GO:0016939) |
| 0.1 | 0.2 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 1.0 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.4 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.1 | 0.4 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.1 | 0.2 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 0.7 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.1 | 2.1 | GO:0034364 | high-density lipoprotein particle(GO:0034364) |
| 0.1 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 3.4 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.1 | 0.8 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.1 | 0.3 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.1 | 1.8 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.1 | 1.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.5 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.4 | GO:0044308 | axonal spine(GO:0044308) |
| 0.1 | 3.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.1 | 1.5 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 1.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.1 | 0.6 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.1 | 2.2 | GO:0008305 | integrin complex(GO:0008305) |
| 0.1 | 0.4 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.1 | 1.4 | GO:0071782 | endoplasmic reticulum tubular network(GO:0071782) |
| 0.1 | 1.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.5 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:1990257 | piccolo-bassoon transport vesicle(GO:1990257) |
| 0.0 | 0.4 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.3 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.5 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0032983 | kainate selective glutamate receptor complex(GO:0032983) |
| 0.0 | 0.4 | GO:0031983 | vesicle lumen(GO:0031983) |
| 0.0 | 0.3 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.3 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.2 | GO:1990794 | lateral part of cell(GO:0097574) basolateral part of cell(GO:1990794) rod bipolar cell terminal bouton(GO:1990795) photoreceptor cell terminal bouton(GO:1990796) |
| 0.0 | 2.1 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.7 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) |
| 0.0 | 0.3 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.7 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.7 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 2.3 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 2.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.2 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.5 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0099522 | region of cytosol(GO:0099522) postsynaptic cytosol(GO:0099524) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.4 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 29.1 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.0 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 2.6 | GO:0019897 | extrinsic component of plasma membrane(GO:0019897) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 1.3 | GO:0000786 | nucleosome(GO:0000786) |
| 0.0 | 0.2 | GO:0070578 | RISC-loading complex(GO:0070578) |
| 0.0 | 0.5 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.0 | GO:0020016 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.7 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 1.0 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 1.7 | GO:0070160 | bicellular tight junction(GO:0005923) occluding junction(GO:0070160) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.0 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0044853 | plasma membrane raft(GO:0044853) |
| 0.0 | 0.5 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.1 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 2.6 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.5 | 2.0 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.4 | 1.7 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.4 | 1.5 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.4 | 2.6 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.4 | 1.5 | GO:0048408 | epidermal growth factor binding(GO:0048408) |
| 0.3 | 1.7 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.3 | 1.3 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.3 | 1.0 | GO:0030226 | apolipoprotein receptor activity(GO:0030226) apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.3 | 1.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 1.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.3 | 1.2 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.2 | 1.7 | GO:0031404 | alpha-amylase activity(GO:0004556) chloride ion binding(GO:0031404) |
| 0.2 | 0.7 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.2 | 1.9 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.2 | 1.9 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.2 | 1.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.2 | 1.6 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.2 | 1.8 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.2 | 1.3 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.2 | 1.5 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
| 0.2 | 0.6 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.2 | 1.4 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 1.6 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.6 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.2 | 0.8 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.2 | 0.6 | GO:2001069 | glycogen binding(GO:2001069) |
| 0.2 | 2.2 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.2 | 1.3 | GO:0046979 | TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.2 | 0.5 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.2 | 1.5 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.2 | 2.0 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.2 | 0.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.2 | 1.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.2 | 0.3 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) |
| 0.2 | 1.4 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.2 | 1.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.2 | 0.5 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.2 | 0.5 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.1 | 0.4 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.1 | 0.4 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 0.1 | 1.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.1 | 1.5 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.4 | GO:0070401 | NADP+ binding(GO:0070401) |
| 0.1 | 0.7 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) procollagen-proline dioxygenase activity(GO:0019798) |
| 0.1 | 0.5 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.5 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.1 | 0.4 | GO:0050253 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.1 | 0.4 | GO:0030617 | transforming growth factor beta receptor, inhibitory cytoplasmic mediator activity(GO:0030617) |
| 0.1 | 0.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.1 | 1.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.7 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.1 | 1.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.1 | 2.6 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.3 | GO:0008390 | testosterone 16-alpha-hydroxylase activity(GO:0008390) oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.8 | GO:0050694 | galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.1 | 0.3 | GO:0071820 | N-box binding(GO:0071820) |
| 0.1 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.1 | 0.8 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.1 | 0.5 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.6 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.8 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 0.3 | GO:0030116 | glial cell-derived neurotrophic factor receptor binding(GO:0030116) |
| 0.1 | 0.4 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 3.6 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.1 | 2.9 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.1 | 1.9 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 0.3 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.1 | 0.3 | GO:0045127 | N-acetylglucosamine kinase activity(GO:0045127) |
| 0.1 | 1.3 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.1 | 0.3 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.1 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 0.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.1 | GO:0016501 | prostacyclin receptor activity(GO:0016501) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.9 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.1 | 0.4 | GO:0004850 | uridine phosphorylase activity(GO:0004850) |
| 0.1 | 0.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 5.8 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.1 | 0.4 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.1 | 0.5 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 0.3 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.1 | 0.4 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 2.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.6 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.2 | GO:0050479 | glyceryl-ether monooxygenase activity(GO:0050479) |
| 0.1 | 0.7 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0030942 | endoplasmic reticulum signal peptide binding(GO:0030942) |
| 0.1 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.1 | 0.6 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.2 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.2 | GO:0071568 | UFM1 transferase activity(GO:0071568) |
| 0.1 | 0.5 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.1 | 0.4 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.4 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 2.9 | GO:0005504 | fatty acid binding(GO:0005504) |
| 0.1 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.4 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.1 | 0.4 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.1 | 2.9 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.1 | 0.6 | GO:0008430 | selenium binding(GO:0008430) |
| 0.1 | 0.2 | GO:0015185 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.1 | 1.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.1 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.2 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.1 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.1 | 0.9 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.2 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.1 | 0.7 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.1 | 0.8 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.1 | 1.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.1 | 0.1 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.3 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.3 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.2 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.2 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.8 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.4 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0035717 | chemokine (C-C motif) ligand 7 binding(GO:0035717) |
| 0.0 | 0.7 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 1.9 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.3 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.9 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.4 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 1.4 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.3 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.4 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.4 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) |
| 0.0 | 0.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 2.0 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.2 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.4 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.2 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.2 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0070991 | medium-chain-acyl-CoA dehydrogenase activity(GO:0070991) |
| 0.0 | 0.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 1.0 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.0 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 0.1 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 4.1 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.3 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0030549 | acetylcholine receptor activator activity(GO:0030549) |
| 0.0 | 0.2 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0016634 | oxidoreductase activity, acting on the CH-CH group of donors, oxygen as acceptor(GO:0016634) |
| 0.0 | 0.6 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.3 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.4 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.2 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.3 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 1.9 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0035827 | rubidium ion transmembrane transporter activity(GO:0035827) |
| 0.0 | 1.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.2 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.3 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.8 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.5 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.5 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.2 | GO:0070290 | phospholipase D activity(GO:0004630) N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.0 | 0.2 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) |
| 0.0 | 0.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.8 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.3 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0001962 | alpha-1,3-galactosyltransferase activity(GO:0001962) |
| 0.0 | 1.1 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.7 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.3 | GO:0097153 | cysteine-type endopeptidase activity involved in apoptotic process(GO:0097153) |
| 0.0 | 0.3 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.2 | GO:0032557 | pyrimidine ribonucleotide binding(GO:0032557) |
| 0.0 | 0.3 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.1 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) inositol hexakisphosphate 1-kinase activity(GO:0052723) inositol hexakisphosphate 3-kinase activity(GO:0052724) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.3 | GO:0016709 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, NAD(P)H as one donor, and incorporation of one atom of oxygen(GO:0016709) |
| 0.0 | 0.7 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 1.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.7 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.3 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.1 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.6 | GO:0004712 | protein serine/threonine/tyrosine kinase activity(GO:0004712) |
| 0.0 | 0.4 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.1 | GO:0005093 | Rab GDP-dissociation inhibitor activity(GO:0005093) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.4 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.5 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.2 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.2 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.1 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.3 | GO:0016504 | peptidase activator activity(GO:0016504) |
| 0.0 | 0.9 | GO:0016765 | transferase activity, transferring alkyl or aryl (other than methyl) groups(GO:0016765) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.0 | GO:0005333 | norepinephrine transmembrane transporter activity(GO:0005333) |
| 0.0 | 0.0 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.2 | GO:0001134 | transcription factor activity, transcription factor recruiting(GO:0001134) |
| 0.0 | 1.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) acylglycerol O-acyltransferase activity(GO:0016411) lysophosphatidic acid acyltransferase activity(GO:0042171) |
| 0.0 | 0.0 | GO:0019807 | aspartoacylase activity(GO:0019807) |
| 0.0 | 0.4 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.7 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.1 | 2.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 4.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.1 | 1.6 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.1 | 0.9 | ST INTEGRIN SIGNALING PATHWAY | Integrin Signaling Pathway |
| 0.1 | 2.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 1.9 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 2.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 1.6 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.4 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 1.7 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 6.7 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 1.7 | PID P75 NTR PATHWAY | p75(NTR)-mediated signaling |
| 0.0 | 0.2 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 1.6 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.4 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.4 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.9 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 1.6 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.7 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.9 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.5 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.0 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.6 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.8 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 2.0 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.7 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 1.0 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 3.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.3 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.5 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.2 | 1.8 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 0.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 3.5 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 2.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.1 | 2.3 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 3.3 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.1 | 0.9 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.1 | 1.8 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.1 | 1.9 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 3.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.1 | 1.7 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.1 | 2.5 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.1 | 2.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.1 | 0.6 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.1 | 0.3 | REACTOME ER PHAGOSOME PATHWAY | Genes involved in ER-Phagosome pathway |
| 0.1 | 0.6 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.1 | 1.6 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.1 | 2.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 0.7 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 1.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.1 | 0.5 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.1 | 0.5 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.1 | 0.7 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 1.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.1 | 0.6 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.1 | 1.1 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.7 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.6 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 2.4 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 1.7 | REACTOME G PROTEIN ACTIVATION | Genes involved in G-protein activation |
| 0.0 | 1.1 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 6.0 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.6 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 1.9 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 3.0 | REACTOME G ALPHA S SIGNALLING EVENTS | Genes involved in G alpha (s) signalling events |
| 0.0 | 0.1 | REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | Genes involved in Downstream signal transduction |
| 0.0 | 1.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.6 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.3 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.4 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.2 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.6 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.6 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.2 | REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | Genes involved in Activation of Kainate Receptors upon glutamate binding |
| 0.0 | 0.9 | REACTOME PHASE II CONJUGATION | Genes involved in Phase II conjugation |
| 0.0 | 0.5 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.0 | REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | Genes involved in Incretin Synthesis, Secretion, and Inactivation |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PA | Genes involved in Synthesis of PA |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.6 | REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | Genes involved in Cell surface interactions at the vascular wall |
| 0.0 | 0.1 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.6 | REACTOME METABOLISM OF VITAMINS AND COFACTORS | Genes involved in Metabolism of vitamins and cofactors |
| 0.0 | 0.2 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 1.3 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 1.3 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |