Project
2D miR_HR1_12
Navigation
Downloads
Results for Hnf1b
Z-value: 1.23
Transcription factors associated with Hnf1b
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Hnf1b
|
ENSMUSG00000020679.5 | HNF1 homeobox B |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Hnf1b | mm10_v2_chr11_+_83852135_83852242 | -0.35 | 2.7e-01 | Click! |
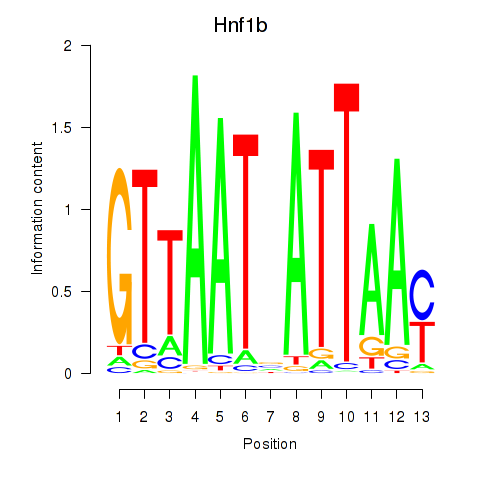
Activity profile of Hnf1b motif
Sorted Z-values of Hnf1b motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_158814469 | 5.09 |
ENSMUST00000161589.2
|
Pappa2
|
pappalysin 2 |
| chr7_+_30493622 | 2.04 |
ENSMUST00000058280.6
ENSMUST00000133318.1 ENSMUST00000142575.1 ENSMUST00000131040.1 |
Prodh2
|
proline dehydrogenase (oxidase) 2 |
| chr5_+_90518932 | 1.77 |
ENSMUST00000113179.2
ENSMUST00000128740.1 |
Afm
|
afamin |
| chr16_-_45492962 | 1.72 |
ENSMUST00000114585.2
|
Gm609
|
predicted gene 609 |
| chr3_+_115080965 | 1.56 |
ENSMUST00000051309.8
|
Olfm3
|
olfactomedin 3 |
| chrX_+_52791179 | 1.49 |
ENSMUST00000101588.1
|
Ccdc160
|
coiled-coil domain containing 160 |
| chrX_+_164140447 | 1.49 |
ENSMUST00000073973.4
|
Ace2
|
angiotensin I converting enzyme (peptidyl-dipeptidase A) 2 |
| chr6_-_52183828 | 1.46 |
ENSMUST00000134831.1
|
Hoxa3
|
homeobox A3 |
| chr13_-_53286052 | 1.45 |
ENSMUST00000021918.8
|
Ror2
|
receptor tyrosine kinase-like orphan receptor 2 |
| chr13_+_89540636 | 1.43 |
ENSMUST00000022108.7
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr3_-_72967854 | 1.39 |
ENSMUST00000167334.1
|
Sis
|
sucrase isomaltase (alpha-glucosidase) |
| chr7_-_101870711 | 1.39 |
ENSMUST00000151706.1
|
Folr1
|
folate receptor 1 (adult) |
| chr2_-_62412219 | 1.37 |
ENSMUST00000047812.7
|
Dpp4
|
dipeptidylpeptidase 4 |
| chr6_-_147243794 | 1.37 |
ENSMUST00000153786.1
|
Gm15767
|
predicted gene 15767 |
| chr1_-_20617992 | 1.32 |
ENSMUST00000088448.5
|
Pkhd1
|
polycystic kidney and hepatic disease 1 |
| chr17_+_23660477 | 1.22 |
ENSMUST00000062967.8
|
Ccdc64b
|
coiled-coil domain containing 64B |
| chr12_+_58211772 | 1.20 |
ENSMUST00000110671.2
ENSMUST00000044299.2 |
Sstr1
|
somatostatin receptor 1 |
| chr1_+_88211956 | 1.17 |
ENSMUST00000073049.6
|
Ugt1a1
|
UDP glucuronosyltransferase 1 family, polypeptide A1 |
| chr4_-_53262547 | 1.15 |
ENSMUST00000098075.2
|
AI427809
|
expressed sequence AI427809 |
| chr2_-_35100677 | 1.14 |
ENSMUST00000045776.4
ENSMUST00000113050.3 |
AI182371
|
expressed sequence AI182371 |
| chr5_+_102845007 | 1.12 |
ENSMUST00000070000.4
|
Arhgap24
|
Rho GTPase activating protein 24 |
| chr18_+_33464163 | 1.08 |
ENSMUST00000097634.3
|
Gm10549
|
predicted gene 10549 |
| chr16_+_96361654 | 1.08 |
ENSMUST00000113794.1
|
Igsf5
|
immunoglobulin superfamily, member 5 |
| chr9_+_72958785 | 1.08 |
ENSMUST00000098567.2
ENSMUST00000034734.8 |
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 homolog (human) |
| chr1_+_88138364 | 1.06 |
ENSMUST00000014263.4
|
Ugt1a6a
|
UDP glucuronosyltransferase 1 family, polypeptide A6A |
| chr1_+_125676969 | 1.03 |
ENSMUST00000027581.6
|
Gpr39
|
G protein-coupled receptor 39 |
| chr1_+_88070765 | 1.01 |
ENSMUST00000073772.4
|
Ugt1a9
|
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chrX_+_140907602 | 0.98 |
ENSMUST00000033806.4
|
Vsig1
|
V-set and immunoglobulin domain containing 1 |
| chr4_-_140617062 | 0.96 |
ENSMUST00000154979.1
|
Arhgef10l
|
Rho guanine nucleotide exchange factor (GEF) 10-like |
| chr12_+_59095653 | 0.94 |
ENSMUST00000021384.4
|
Mia2
|
melanoma inhibitory activity 2 |
| chr5_-_87569023 | 0.94 |
ENSMUST00000113314.2
|
Sult1d1
|
sulfotransferase family 1D, member 1 |
| chr1_+_88166004 | 0.93 |
ENSMUST00000097659.4
|
Ugt1a5
|
UDP glucuronosyltransferase 1 family, polypeptide A5 |
| chr16_+_96361749 | 0.92 |
ENSMUST00000000163.6
ENSMUST00000081093.3 ENSMUST00000113795.1 |
Igsf5
|
immunoglobulin superfamily, member 5 |
| chrX_-_139085230 | 0.90 |
ENSMUST00000152457.1
|
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chr15_+_10215955 | 0.89 |
ENSMUST00000130720.1
|
Prlr
|
prolactin receptor |
| chr8_-_107065632 | 0.89 |
ENSMUST00000034393.5
|
Tmed6
|
transmembrane emp24 protein transport domain containing 6 |
| chr11_+_78037959 | 0.89 |
ENSMUST00000073660.6
|
Flot2
|
flotillin 2 |
| chr5_-_87535113 | 0.88 |
ENSMUST00000120150.1
|
Sult1b1
|
sulfotransferase family 1B, member 1 |
| chr11_+_78037931 | 0.86 |
ENSMUST00000072289.5
ENSMUST00000100784.2 |
Flot2
|
flotillin 2 |
| chr3_-_116712696 | 0.86 |
ENSMUST00000169530.1
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr19_+_42045792 | 0.86 |
ENSMUST00000172244.1
ENSMUST00000081714.4 |
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr6_-_137571007 | 0.84 |
ENSMUST00000100841.2
|
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr9_+_21835506 | 0.83 |
ENSMUST00000058777.6
|
Gm6484
|
predicted gene 6484 |
| chr19_+_43782181 | 0.83 |
ENSMUST00000026208.4
|
Abcc2
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr19_-_46672883 | 0.82 |
ENSMUST00000026012.7
|
Cyp17a1
|
cytochrome P450, family 17, subfamily a, polypeptide 1 |
| chr11_+_101367542 | 0.82 |
ENSMUST00000019469.2
|
G6pc
|
glucose-6-phosphatase, catalytic |
| chr1_+_88055377 | 0.82 |
ENSMUST00000138182.1
ENSMUST00000113142.3 |
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chrX_-_139085211 | 0.80 |
ENSMUST00000033626.8
ENSMUST00000060824.3 |
Serpina7
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 |
| chrX_+_101376359 | 0.79 |
ENSMUST00000119080.1
|
Gjb1
|
gap junction protein, beta 1 |
| chr10_-_30842765 | 0.78 |
ENSMUST00000019924.8
|
Hey2
|
hairy/enhancer-of-split related with YRPW motif 2 |
| chr11_+_3983636 | 0.77 |
ENSMUST00000078757.1
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr11_+_78465697 | 0.77 |
ENSMUST00000001126.3
|
Slc46a1
|
solute carrier family 46, member 1 |
| chr5_+_150952607 | 0.74 |
ENSMUST00000078856.6
|
Kl
|
klotho |
| chr13_-_23914998 | 0.74 |
ENSMUST00000021769.8
ENSMUST00000110407.2 |
Slc17a4
|
solute carrier family 17 (sodium phosphate), member 4 |
| chr11_+_3983704 | 0.71 |
ENSMUST00000063004.7
|
Gal3st1
|
galactose-3-O-sulfotransferase 1 |
| chr6_-_24168083 | 0.71 |
ENSMUST00000031713.8
|
Slc13a1
|
solute carrier family 13 (sodium/sulfate symporters), member 1 |
| chr12_+_76533540 | 0.64 |
ENSMUST00000075249.4
|
Plekhg3
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr3_-_113291449 | 0.64 |
ENSMUST00000179568.1
|
Amy2a4
|
amylase 2a4 |
| chr8_-_93535707 | 0.63 |
ENSMUST00000077816.6
|
Ces5a
|
carboxylesterase 5A |
| chr6_-_116716888 | 0.63 |
ENSMUST00000056623.6
|
Tmem72
|
transmembrane protein 72 |
| chr5_+_107497718 | 0.62 |
ENSMUST00000112671.2
|
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr19_+_56287911 | 0.61 |
ENSMUST00000095948.4
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr14_+_118787894 | 0.60 |
ENSMUST00000047761.6
ENSMUST00000071546.7 |
Cldn10
|
claudin 10 |
| chr5_-_87337165 | 0.60 |
ENSMUST00000031195.2
|
Ugt2a3
|
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr7_+_90348699 | 0.59 |
ENSMUST00000107211.1
ENSMUST00000107210.1 |
Sytl2
|
synaptotagmin-like 2 |
| chr4_+_138967112 | 0.58 |
ENSMUST00000116094.2
|
Rnf186
|
ring finger protein 186 |
| chr13_-_21453714 | 0.58 |
ENSMUST00000032820.7
ENSMUST00000110485.1 |
Zscan26
|
zinc finger and SCAN domain containing 26 |
| chr12_-_12940600 | 0.58 |
ENSMUST00000130990.1
|
Mycn
|
v-myc myelocytomatosis viral related oncogene, neuroblastoma derived (avian) |
| chr16_+_24448082 | 0.57 |
ENSMUST00000078988.2
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr11_+_7197780 | 0.55 |
ENSMUST00000020704.7
|
Igfbp1
|
insulin-like growth factor binding protein 1 |
| chr1_+_88055467 | 0.54 |
ENSMUST00000173325.1
|
Ugt1a10
|
UDP glycosyltransferase 1 family, polypeptide A10 |
| chr17_+_57062231 | 0.53 |
ENSMUST00000097299.3
ENSMUST00000169543.1 ENSMUST00000163763.1 |
Crb3
|
crumbs homolog 3 (Drosophila) |
| chr17_+_57062486 | 0.53 |
ENSMUST00000163628.1
|
Crb3
|
crumbs homolog 3 (Drosophila) |
| chr4_-_139968026 | 0.52 |
ENSMUST00000105031.2
|
Klhdc7a
|
kelch domain containing 7A |
| chr18_-_43737186 | 0.51 |
ENSMUST00000025381.2
|
Spink3
|
serine peptidase inhibitor, Kazal type 3 |
| chr4_+_11758147 | 0.51 |
ENSMUST00000029871.5
ENSMUST00000108303.1 |
Cdh17
|
cadherin 17 |
| chr9_-_77045788 | 0.49 |
ENSMUST00000034911.6
|
Tinag
|
tubulointerstitial nephritis antigen |
| chr10_-_10472314 | 0.49 |
ENSMUST00000179956.1
ENSMUST00000172530.1 ENSMUST00000132573.1 |
Adgb
|
androglobin |
| chr11_+_44518959 | 0.49 |
ENSMUST00000019333.3
|
Rnf145
|
ring finger protein 145 |
| chr7_+_131032061 | 0.49 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr13_+_54701457 | 0.48 |
ENSMUST00000037145.7
|
Cdhr2
|
cadherin-related family member 2 |
| chr4_+_150853919 | 0.48 |
ENSMUST00000073600.2
|
Errfi1
|
ERBB receptor feedback inhibitor 1 |
| chr4_-_111898695 | 0.47 |
ENSMUST00000102720.1
|
Slc5a9
|
solute carrier family 5 (sodium/glucose cotransporter), member 9 |
| chr14_+_26894557 | 0.46 |
ENSMUST00000090337.4
ENSMUST00000165929.2 |
Asb14
|
ankyrin repeat and SOCS box-containing 14 |
| chr13_+_4191163 | 0.45 |
ENSMUST00000021634.2
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr9_+_54538984 | 0.45 |
ENSMUST00000060242.5
ENSMUST00000118413.1 |
Sh2d7
|
SH2 domain containing 7 |
| chr11_-_6230127 | 0.45 |
ENSMUST00000004505.2
|
Npc1l1
|
NPC1-like 1 |
| chr13_+_55152640 | 0.44 |
ENSMUST00000005452.5
|
Fgfr4
|
fibroblast growth factor receptor 4 |
| chr6_+_38918969 | 0.43 |
ENSMUST00000003017.6
|
Tbxas1
|
thromboxane A synthase 1, platelet |
| chr19_-_57197556 | 0.42 |
ENSMUST00000099294.2
|
Ablim1
|
actin-binding LIM protein 1 |
| chr7_-_119895446 | 0.42 |
ENSMUST00000098080.2
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr5_+_90490714 | 0.41 |
ENSMUST00000042755.3
|
Afp
|
alpha fetoprotein |
| chr15_+_99393574 | 0.41 |
ENSMUST00000162624.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr1_+_40439627 | 0.41 |
ENSMUST00000097772.3
|
Il1rl1
|
interleukin 1 receptor-like 1 |
| chr11_+_115834314 | 0.40 |
ENSMUST00000173289.1
ENSMUST00000137900.1 |
Llgl2
|
lethal giant larvae homolog 2 (Drosophila) |
| chr8_+_70724064 | 0.40 |
ENSMUST00000034307.7
ENSMUST00000110095.2 |
Pde4c
|
phosphodiesterase 4C, cAMP specific |
| chr19_-_39740999 | 0.40 |
ENSMUST00000099472.3
|
Cyp2c68
|
cytochrome P450, family 2, subfamily c, polypeptide 68 |
| chr10_-_89533550 | 0.40 |
ENSMUST00000105297.1
|
Nr1h4
|
nuclear receptor subfamily 1, group H, member 4 |
| chr7_-_45062393 | 0.40 |
ENSMUST00000129101.1
|
Prrg2
|
proline-rich Gla (G-carboxyglutamic acid) polypeptide 2 |
| chr3_+_90537306 | 0.39 |
ENSMUST00000107335.1
|
S100a16
|
S100 calcium binding protein A16 |
| chr11_+_85832551 | 0.38 |
ENSMUST00000000095.6
|
Tbx2
|
T-box 2 |
| chr11_-_75937726 | 0.38 |
ENSMUST00000121287.1
|
Rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr1_+_130731963 | 0.37 |
ENSMUST00000039323.6
|
AA986860
|
expressed sequence AA986860 |
| chr8_-_122476036 | 0.37 |
ENSMUST00000014614.3
|
Rnf166
|
ring finger protein 166 |
| chr11_-_76027726 | 0.37 |
ENSMUST00000021207.6
|
Fam101b
|
family with sequence similarity 101, member B |
| chr5_-_108675569 | 0.37 |
ENSMUST00000051757.7
|
Slc26a1
|
solute carrier family 26 (sulfate transporter), member 1 |
| chrX_+_107792583 | 0.36 |
ENSMUST00000101292.2
|
Fam46d
|
family with sequence similarity 46, member D |
| chr15_+_99393610 | 0.36 |
ENSMUST00000159531.1
|
Tmbim6
|
transmembrane BAX inhibitor motif containing 6 |
| chr16_-_79091078 | 0.36 |
ENSMUST00000023566.4
ENSMUST00000060402.5 |
Tmprss15
|
transmembrane protease, serine 15 |
| chr6_-_129100903 | 0.36 |
ENSMUST00000032258.6
|
Clec2e
|
C-type lectin domain family 2, member e |
| chr19_+_60889749 | 0.35 |
ENSMUST00000003313.8
|
Grk5
|
G protein-coupled receptor kinase 5 |
| chr3_-_144819494 | 0.35 |
ENSMUST00000029929.7
|
Clca2
|
chloride channel calcium activated 2 |
| chr7_-_144751968 | 0.35 |
ENSMUST00000155175.1
|
Ano1
|
anoctamin 1, calcium activated chloride channel |
| chr11_+_110968016 | 0.34 |
ENSMUST00000106636.1
ENSMUST00000180023.1 |
Kcnj16
|
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr5_-_114444036 | 0.34 |
ENSMUST00000031560.7
|
Mmab
|
methylmalonic aciduria (cobalamin deficiency) type B homolog (human) |
| chr4_+_19280850 | 0.32 |
ENSMUST00000102999.1
|
Cngb3
|
cyclic nucleotide gated channel beta 3 |
| chr7_-_119479249 | 0.32 |
ENSMUST00000033263.4
|
Umod
|
uromodulin |
| chrX_+_101449078 | 0.32 |
ENSMUST00000033674.5
|
Itgb1bp2
|
integrin beta 1 binding protein 2 |
| chr2_+_91265252 | 0.31 |
ENSMUST00000028691.6
|
Arfgap2
|
ADP-ribosylation factor GTPase activating protein 2 |
| chr1_+_181051232 | 0.31 |
ENSMUST00000036819.6
|
9130409I23Rik
|
RIKEN cDNA 9130409I23 gene |
| chr5_-_106926245 | 0.31 |
ENSMUST00000117588.1
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog (S. cerevisiae) |
| chr16_+_33251415 | 0.31 |
ENSMUST00000023502.4
|
Snx4
|
sorting nexin 4 |
| chr7_+_141746736 | 0.30 |
ENSMUST00000026590.8
|
Muc2
|
mucin 2 |
| chrX_+_164090187 | 0.30 |
ENSMUST00000015545.3
|
Tmem27
|
transmembrane protein 27 |
| chr6_-_129876659 | 0.29 |
ENSMUST00000014687.4
ENSMUST00000122219.1 |
Klra17
|
killer cell lectin-like receptor, subfamily A, member 17 |
| chr19_+_56287943 | 0.28 |
ENSMUST00000166049.1
|
Habp2
|
hyaluronic acid binding protein 2 |
| chr4_-_99654983 | 0.28 |
ENSMUST00000136525.1
|
Gm12688
|
predicted gene 12688 |
| chr1_+_58795371 | 0.28 |
ENSMUST00000027189.8
|
Casp8
|
caspase 8 |
| chr3_+_90537242 | 0.28 |
ENSMUST00000098911.3
|
S100a16
|
S100 calcium binding protein A16 |
| chr15_-_36879816 | 0.27 |
ENSMUST00000100713.2
|
Gm10384
|
predicted gene 10384 |
| chr5_+_107497762 | 0.27 |
ENSMUST00000152474.1
ENSMUST00000060553.7 |
A830010M20Rik
|
RIKEN cDNA A830010M20 gene |
| chr19_-_11856001 | 0.26 |
ENSMUST00000079875.3
|
Olfr1418
|
olfactory receptor 1418 |
| chr5_-_100373484 | 0.25 |
ENSMUST00000182433.1
|
Sec31a
|
Sec31 homolog A (S. cerevisiae) |
| chr1_-_187215421 | 0.24 |
ENSMUST00000110945.3
ENSMUST00000183931.1 ENSMUST00000027908.6 |
Spata17
|
spermatogenesis associated 17 |
| chr19_+_5878622 | 0.24 |
ENSMUST00000136833.1
ENSMUST00000141362.1 |
Slc25a45
|
solute carrier family 25, member 45 |
| chr6_-_38124568 | 0.24 |
ENSMUST00000040259.4
|
Atp6v0a4
|
ATPase, H+ transporting, lysosomal V0 subunit A4 |
| chr4_-_134767940 | 0.23 |
ENSMUST00000037828.6
|
Ldlrap1
|
low density lipoprotein receptor adaptor protein 1 |
| chr7_-_28962265 | 0.23 |
ENSMUST00000068045.7
|
Actn4
|
actinin alpha 4 |
| chr6_+_134920401 | 0.23 |
ENSMUST00000067327.4
ENSMUST00000003115.6 |
Cdkn1b
|
cyclin-dependent kinase inhibitor 1B |
| chr11_+_83852135 | 0.23 |
ENSMUST00000146786.1
|
Hnf1b
|
HNF1 homeobox B |
| chr4_+_44943727 | 0.22 |
ENSMUST00000154177.1
|
Gm12678
|
predicted gene 12678 |
| chr17_+_74395607 | 0.22 |
ENSMUST00000179074.1
ENSMUST00000024870.6 |
Slc30a6
|
solute carrier family 30 (zinc transporter), member 6 |
| chr2_+_70039114 | 0.22 |
ENSMUST00000060208.4
|
Myo3b
|
myosin IIIB |
| chr3_+_69222412 | 0.22 |
ENSMUST00000183126.1
|
Arl14
|
ADP-ribosylation factor-like 14 |
| chr9_+_54980880 | 0.22 |
ENSMUST00000093844.3
|
Chrna5
|
cholinergic receptor, nicotinic, alpha polypeptide 5 |
| chr7_+_45062429 | 0.21 |
ENSMUST00000107830.1
ENSMUST00000003513.3 ENSMUST00000107829.1 ENSMUST00000150609.1 |
Nosip
|
nitric oxide synthase interacting protein |
| chr1_+_133309778 | 0.21 |
ENSMUST00000094557.4
ENSMUST00000183457.1 ENSMUST00000183738.1 ENSMUST00000185157.1 ENSMUST00000184603.1 |
Golt1a
Kiss1
GOLT1A
|
golgi transport 1 homolog A (S. cerevisiae) KiSS-1 metastasis-suppressor KISS1 isoform e |
| chr8_-_67974567 | 0.20 |
ENSMUST00000098696.3
ENSMUST00000038959.9 ENSMUST00000093469.4 |
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_90460889 | 0.20 |
ENSMUST00000031314.8
|
Alb
|
albumin |
| chr12_-_103631404 | 0.20 |
ENSMUST00000121625.1
ENSMUST00000044231.5 |
Serpina10
|
serine (or cysteine) peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr19_+_39113898 | 0.19 |
ENSMUST00000087234.2
|
Cyp2c66
|
cytochrome P450, family 2, subfamily c, polypeptide 66 |
| chr4_-_108144986 | 0.19 |
ENSMUST00000130776.1
|
Scp2
|
sterol carrier protein 2, liver |
| chr5_-_38491948 | 0.19 |
ENSMUST00000129099.1
|
Slc2a9
|
solute carrier family 2 (facilitated glucose transporter), member 9 |
| chr6_-_129622685 | 0.18 |
ENSMUST00000032252.5
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr14_+_61309753 | 0.18 |
ENSMUST00000055159.7
|
Arl11
|
ADP-ribosylation factor-like 11 |
| chr17_-_33718591 | 0.17 |
ENSMUST00000174040.1
ENSMUST00000173015.1 ENSMUST00000066121.6 ENSMUST00000172767.1 ENSMUST00000173329.1 |
March2
|
membrane-associated ring finger (C3HC4) 2 |
| chr18_-_66022580 | 0.17 |
ENSMUST00000143990.1
|
Lman1
|
lectin, mannose-binding, 1 |
| chr13_-_73678005 | 0.17 |
ENSMUST00000022105.7
ENSMUST00000109680.2 ENSMUST00000109679.2 |
Slc6a18
|
solute carrier family 6 (neurotransmitter transporter), member 18 |
| chr11_-_75937677 | 0.16 |
ENSMUST00000108420.2
|
Rph3al
|
rabphilin 3A-like (without C2 domains) |
| chr7_-_6011010 | 0.16 |
ENSMUST00000086338.1
|
Vmn1r65
|
vomeronasal 1 receptor 65 |
| chr10_-_89621253 | 0.16 |
ENSMUST00000020102.7
|
Slc17a8
|
solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 8 |
| chrX_+_68761839 | 0.16 |
ENSMUST00000069731.5
|
Fmr1nb
|
fragile X mental retardation 1 neighbor |
| chr12_+_4769375 | 0.15 |
ENSMUST00000178879.1
|
Pfn4
|
profilin family, member 4 |
| chr17_+_47737030 | 0.15 |
ENSMUST00000086932.3
|
Tfeb
|
transcription factor EB |
| chr10_+_102159000 | 0.15 |
ENSMUST00000020039.6
|
Mgat4c
|
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme C (putative) |
| chr3_-_116712644 | 0.15 |
ENSMUST00000029569.2
|
Slc35a3
|
solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member 3 |
| chr4_+_48279794 | 0.15 |
ENSMUST00000030029.3
|
Invs
|
inversin |
| chr4_+_99030946 | 0.15 |
ENSMUST00000030280.6
|
Angptl3
|
angiopoietin-like 3 |
| chr9_-_44767792 | 0.14 |
ENSMUST00000034607.9
|
Arcn1
|
archain 1 |
| chr1_-_36273425 | 0.14 |
ENSMUST00000056946.6
|
Neurl3
|
neuralized homolog 3 homolog (Drosophila) |
| chr13_+_23782572 | 0.14 |
ENSMUST00000074067.2
|
Trim38
|
tripartite motif-containing 38 |
| chr4_-_103026709 | 0.13 |
ENSMUST00000084382.5
ENSMUST00000106869.2 |
Insl5
|
insulin-like 5 |
| chr7_+_24897381 | 0.13 |
ENSMUST00000003469.7
|
Cd79a
|
CD79A antigen (immunoglobulin-associated alpha) |
| chr9_+_67840386 | 0.13 |
ENSMUST00000077879.5
|
Vps13c
|
vacuolar protein sorting 13C (yeast) |
| chr3_-_83049797 | 0.13 |
ENSMUST00000048246.3
|
Fgb
|
fibrinogen beta chain |
| chr14_+_46882854 | 0.12 |
ENSMUST00000022386.8
ENSMUST00000100672.3 |
Samd4
|
sterile alpha motif domain containing 4 |
| chr7_-_29505447 | 0.12 |
ENSMUST00000183096.1
ENSMUST00000085809.4 |
Sipa1l3
|
signal-induced proliferation-associated 1 like 3 |
| chr7_-_28962223 | 0.12 |
ENSMUST00000127210.1
|
Actn4
|
actinin alpha 4 |
| chr14_+_34085978 | 0.12 |
ENSMUST00000022519.8
|
Anxa8
|
annexin A8 |
| chr5_+_65348386 | 0.11 |
ENSMUST00000031096.7
|
Klb
|
klotho beta |
| chr14_+_48446340 | 0.11 |
ENSMUST00000111735.2
|
Tmem260
|
transmembrane protein 260 |
| chr7_+_27731398 | 0.10 |
ENSMUST00000130997.1
|
Zfp60
|
zinc finger protein 60 |
| chr10_-_107719978 | 0.10 |
ENSMUST00000050702.7
|
Ptprq
|
protein tyrosine phosphatase, receptor type, Q |
| chr14_+_34086008 | 0.10 |
ENSMUST00000120077.1
|
Anxa8
|
annexin A8 |
| chr11_-_84167466 | 0.09 |
ENSMUST00000050771.7
|
Gm11437
|
predicted gene 11437 |
| chr1_+_172126750 | 0.09 |
ENSMUST00000075895.2
ENSMUST00000111252.3 |
Pex19
|
peroxisomal biogenesis factor 19 |
| chr18_+_35599219 | 0.09 |
ENSMUST00000115734.1
|
Paip2
|
polyadenylate-binding protein-interacting protein 2 |
| chr1_+_133246092 | 0.08 |
ENSMUST00000038295.8
ENSMUST00000105082.2 |
Plekha6
|
pleckstrin homology domain containing, family A member 6 |
| chr9_+_77046680 | 0.08 |
ENSMUST00000184488.1
|
RP23-282G22.2
|
RP23-282G22.2 |
| chr19_+_27217011 | 0.08 |
ENSMUST00000164746.1
ENSMUST00000172302.1 |
Vldlr
|
very low density lipoprotein receptor |
| chr14_+_79515618 | 0.08 |
ENSMUST00000110835.1
|
Elf1
|
E74-like factor 1 |
| chr3_-_69859875 | 0.07 |
ENSMUST00000051239.7
ENSMUST00000171529.1 |
Sptssb
|
serine palmitoyltransferase, small subunit B |
| chr9_-_71163224 | 0.07 |
ENSMUST00000074465.2
|
Aqp9
|
aquaporin 9 |
| chr2_+_162987502 | 0.07 |
ENSMUST00000117123.1
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr8_+_68276567 | 0.07 |
ENSMUST00000066594.3
|
Sh2d4a
|
SH2 domain containing 4A |
| chr6_+_52177498 | 0.07 |
ENSMUST00000070587.3
|
5730596B20Rik
|
RIKEN cDNA 5730596B20 gene |
| chr7_+_141215852 | 0.06 |
ENSMUST00000046890.5
ENSMUST00000133763.1 |
Rassf7
|
Ras association (RalGDS/AF-6) domain family (N-terminal) member 7 |
| chr10_+_128411616 | 0.06 |
ENSMUST00000096386.5
ENSMUST00000171342.1 |
Rnf41
|
ring finger protein 41 |
| chr11_+_115912001 | 0.06 |
ENSMUST00000132961.1
|
Smim6
|
small integral membrane protein 6 |
| chr1_-_187215454 | 0.06 |
ENSMUST00000183819.1
|
Spata17
|
spermatogenesis associated 17 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Hnf1b
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.6 | 1.7 | GO:1903903 | regulation of establishment of T cell polarity(GO:1903903) |
| 0.6 | 1.7 | GO:0033189 | response to vitamin A(GO:0033189) |
| 0.5 | 5.5 | GO:0052697 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.5 | 1.4 | GO:0010716 | negative regulation of extracellular matrix disassembly(GO:0010716) |
| 0.4 | 1.5 | GO:0015827 | aromatic amino acid transport(GO:0015801) tryptophan transport(GO:0015827) |
| 0.3 | 1.0 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.3 | 0.6 | GO:0052695 | cellular glucuronidation(GO:0052695) |
| 0.3 | 0.9 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.3 | 1.5 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.3 | 0.9 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.2 | 1.5 | GO:0021615 | specification of organ position(GO:0010159) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.2 | 0.8 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.2 | 1.0 | GO:0015788 | UDP-N-acetylglucosamine transport(GO:0015788) |
| 0.2 | 0.8 | GO:0035905 | ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.2 | 0.8 | GO:0015886 | heme transport(GO:0015886) |
| 0.2 | 0.9 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.2 | 0.5 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.2 | 1.5 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.2 | 1.3 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.2 | 0.8 | GO:0031438 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.2 | 1.4 | GO:0071231 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) cellular response to folic acid(GO:0071231) |
| 0.1 | 0.9 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.1 | 0.4 | GO:0061144 | alveolar secondary septum development(GO:0061144) |
| 0.1 | 0.8 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.4 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.1 | 0.4 | GO:0060596 | muscle cell fate determination(GO:0007521) mammary placode formation(GO:0060596) |
| 0.1 | 1.1 | GO:0035021 | negative regulation of Rac protein signal transduction(GO:0035021) |
| 0.1 | 0.9 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.1 | 0.2 | GO:0090118 | receptor-mediated endocytosis of low-density lipoprotein particle involved in cholesterol transport(GO:0090118) |
| 0.1 | 1.1 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.1 | 0.4 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.1 | 0.3 | GO:0072236 | metanephric loop of Henle development(GO:0072236) |
| 0.1 | 0.4 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.1 | 0.5 | GO:0042536 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) |
| 0.1 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.1 | 1.4 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.1 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.1 | 0.6 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 1.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.1 | 0.8 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.1 | 1.1 | GO:0036158 | outer dynein arm assembly(GO:0036158) inner dynein arm assembly(GO:0036159) |
| 0.1 | 0.5 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.2 | GO:0051659 | maintenance of mitochondrion location(GO:0051659) |
| 0.1 | 0.4 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.1 | 0.2 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.1 | 0.7 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.1 | 0.4 | GO:0002826 | negative regulation of T-helper 1 type immune response(GO:0002826) |
| 0.1 | 0.2 | GO:1901146 | regulation of pronephros size(GO:0035565) mesonephros morphogenesis(GO:0061206) mesonephric nephron development(GO:0061215) mesonephric nephron morphogenesis(GO:0061228) mesenchymal stem cell maintenance involved in mesonephric nephron morphogenesis(GO:0061235) regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061295) negative regulation of mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:0061296) mesenchymal cell apoptotic process involved in mesonephric nephron morphogenesis(GO:1901146) |
| 0.1 | 0.2 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) |
| 0.1 | 0.4 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.6 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0021691 | cerebellar Purkinje cell layer maturation(GO:0021691) |
| 0.0 | 0.3 | GO:0002349 | histamine production involved in inflammatory response(GO:0002349) histamine secretion involved in inflammatory response(GO:0002441) histamine secretion by mast cell(GO:0002553) |
| 0.0 | 0.3 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 5.1 | GO:0060349 | bone morphogenesis(GO:0060349) |
| 0.0 | 1.0 | GO:0032933 | response to sterol depletion(GO:0006991) SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.6 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:1902477 | defense response to bacterium, incompatible interaction(GO:0009816) regulation of defense response to bacterium, incompatible interaction(GO:1902477) |
| 0.0 | 0.4 | GO:0030299 | intestinal cholesterol absorption(GO:0030299) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 1.3 | GO:0010824 | regulation of centrosome duplication(GO:0010824) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.4 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 0.2 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.5 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 1.7 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 1.2 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.5 | GO:0043567 | regulation of insulin-like growth factor receptor signaling pathway(GO:0043567) |
| 0.0 | 0.3 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) |
| 0.0 | 0.2 | GO:0070072 | vacuolar proton-transporting V-type ATPase complex assembly(GO:0070072) |
| 0.0 | 0.2 | GO:0061088 | sequestering of zinc ion(GO:0032119) regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 1.2 | GO:0007218 | neuropeptide signaling pathway(GO:0007218) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.0 | GO:0034442 | regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.0 | 0.3 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.2 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0015851 | nucleobase transport(GO:0015851) pyrimidine nucleobase transport(GO:0015855) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0046598 | positive regulation of viral entry into host cell(GO:0046598) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0044830 | modulation by host of viral RNA genome replication(GO:0044830) |
| 0.0 | 0.2 | GO:0060088 | auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 1.7 | GO:0001931 | uropod(GO:0001931) flotillin complex(GO:0016600) cell trailing edge(GO:0031254) |
| 0.1 | 0.3 | GO:0030690 | Noc1p-Noc2p complex(GO:0030690) |
| 0.1 | 1.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.1 | 0.5 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.1 | 0.8 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 1.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.1 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.1 | 0.8 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.5 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.0 | 1.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.8 | GO:0016580 | Sin3 complex(GO:0016580) |
| 0.0 | 1.6 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 3.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
| 0.0 | 0.4 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.6 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 1.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 1.0 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.1 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0030126 | COPI vesicle coat(GO:0030126) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.8 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.4 | 1.5 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.4 | 1.8 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.3 | 1.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.3 | 1.2 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.3 | 0.8 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.3 | 0.8 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.3 | 1.8 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.2 | 1.2 | GO:0004994 | somatostatin receptor activity(GO:0004994) |
| 0.2 | 0.9 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.2 | 0.8 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.2 | 0.9 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.2 | 5.8 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.2 | 0.5 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.1 | 0.8 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.1 | 1.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 1.4 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.1 | 0.6 | GO:0004556 | alpha-amylase activity(GO:0004556) |
| 0.1 | 0.4 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.1 | 0.4 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.3 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |
| 0.1 | 0.3 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.1 | 0.8 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.1 | 2.0 | GO:0071949 | FAD binding(GO:0071949) |
| 0.1 | 1.1 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.1 | 0.3 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.1 | 0.3 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.1 | 0.2 | GO:0034875 | testosterone 16-alpha-hydroxylase activity(GO:0008390) oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.3 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.1 | 1.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.4 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.1 | 0.8 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0050632 | propanoyl-CoA C-acyltransferase activity(GO:0033814) propionyl-CoA C2-trimethyltridecanoyltransferase activity(GO:0050632) phosphatidylethanolamine transporter activity(GO:1904121) |
| 0.0 | 1.5 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.5 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 4.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 1.0 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 1.5 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.6 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.4 | GO:0008392 | arachidonic acid epoxygenase activity(GO:0008392) |
| 0.0 | 1.7 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.5 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 1.9 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.5 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 2.4 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.9 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.8 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.2 | GO:0042166 | acetylcholine-gated cation channel activity(GO:0022848) acetylcholine binding(GO:0042166) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0015205 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 1.6 | GO:0005089 | Rho guanyl-nucleotide exchange factor activity(GO:0005089) |
| 0.0 | 0.8 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 8.0 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 1.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 3.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.8 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.9 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.6 | PID FGF PATHWAY | FGF signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.1 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.3 | 5.6 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.1 | 0.8 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.9 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 1.3 | REACTOME FGFR LIGAND BINDING AND ACTIVATION | Genes involved in FGFR ligand binding and activation |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.9 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 1.5 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.4 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.9 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 1.0 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.4 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 7ALPHA HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |