Project
2D miR_HR1_12
Navigation
Downloads
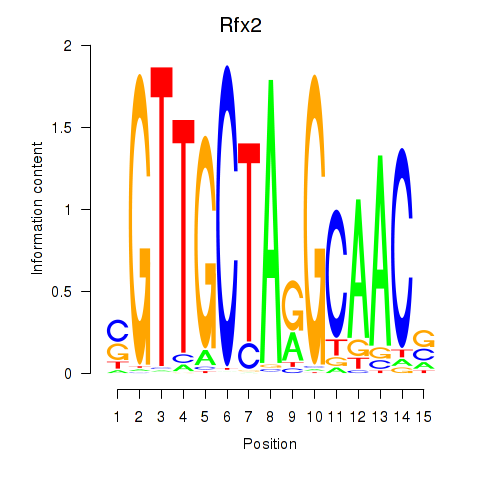
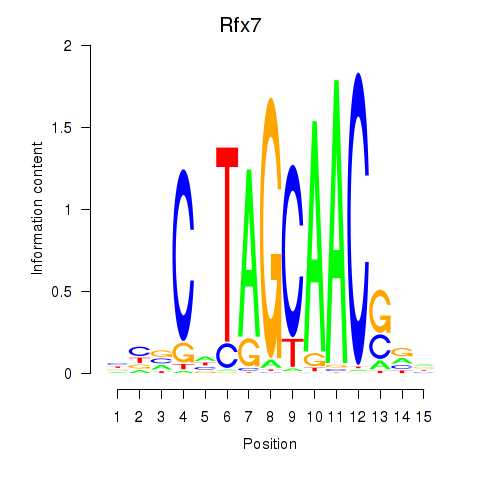
Results for Rfx2_Rfx7
Z-value: 1.79
Transcription factors associated with Rfx2_Rfx7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx2
|
ENSMUSG00000024206.8 | regulatory factor X, 2 (influences HLA class II expression) |
|
Rfx7
|
ENSMUSG00000037674.9 | regulatory factor X, 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx7 | mm10_v2_chr9_+_72532214_72532271 | -0.53 | 7.8e-02 | Click! |
| Rfx2 | mm10_v2_chr17_-_56830916_56831008 | 0.06 | 8.6e-01 | Click! |
Activity profile of Rfx2_Rfx7 motif
Sorted Z-values of Rfx2_Rfx7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_44468051 | 3.53 |
ENSMUST00000118493.1
|
Josd2
|
Josephin domain containing 2 |
| chr2_-_3419019 | 3.39 |
ENSMUST00000115084.1
ENSMUST00000115083.1 |
Meig1
|
meiosis expressed gene 1 |
| chr1_-_10009098 | 3.13 |
ENSMUST00000176398.1
ENSMUST00000027049.3 |
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr17_+_24696234 | 3.10 |
ENSMUST00000019464.7
|
Noxo1
|
NADPH oxidase organizer 1 |
| chr2_-_3419066 | 2.97 |
ENSMUST00000115082.3
|
Meig1
|
meiosis expressed gene 1 |
| chr2_-_28466266 | 2.81 |
ENSMUST00000127683.1
ENSMUST00000086370.4 |
1700007K13Rik
|
RIKEN cDNA 1700007K13 gene |
| chr2_-_152831665 | 2.77 |
ENSMUST00000156688.1
ENSMUST00000007803.5 |
Bcl2l1
|
BCL2-like 1 |
| chr11_+_114727384 | 2.72 |
ENSMUST00000069325.7
|
Dnaic2
|
dynein, axonemal, intermediate chain 2 |
| chr7_+_44896125 | 2.62 |
ENSMUST00000166552.1
ENSMUST00000168207.1 |
Fuz
|
fuzzy homolog (Drosophila) |
| chr5_+_30466044 | 2.52 |
ENSMUST00000031078.3
ENSMUST00000114743.1 |
1700001C02Rik
|
RIKEN cDNA 1700001C02 gene |
| chrX_+_52791179 | 2.51 |
ENSMUST00000101588.1
|
Ccdc160
|
coiled-coil domain containing 160 |
| chr10_+_75037066 | 2.45 |
ENSMUST00000147802.1
ENSMUST00000020391.5 |
Rab36
|
RAB36, member RAS oncogene family |
| chr8_+_116504973 | 2.40 |
ENSMUST00000078170.5
|
Dynlrb2
|
dynein light chain roadblock-type 2 |
| chr7_+_44468020 | 2.30 |
ENSMUST00000117324.1
ENSMUST00000120852.1 ENSMUST00000118628.1 |
Josd2
|
Josephin domain containing 2 |
| chr4_-_126325672 | 2.23 |
ENSMUST00000102616.1
|
Tekt2
|
tektin 2 |
| chr9_+_72958785 | 2.13 |
ENSMUST00000098567.2
ENSMUST00000034734.8 |
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 homolog (human) |
| chr2_-_152831112 | 2.12 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr7_+_44896077 | 1.98 |
ENSMUST00000071207.7
ENSMUST00000166849.1 ENSMUST00000168712.1 ENSMUST00000168389.1 |
Fuz
|
fuzzy homolog (Drosophila) |
| chr7_+_44467980 | 1.80 |
ENSMUST00000035844.4
|
Josd2
|
Josephin domain containing 2 |
| chr5_+_30281377 | 1.71 |
ENSMUST00000101448.3
|
Drc1
|
dynein regulatory complex subunit 1 |
| chr7_-_4546567 | 1.67 |
ENSMUST00000065957.5
|
Syt5
|
synaptotagmin V |
| chr11_+_100545607 | 1.58 |
ENSMUST00000092684.5
ENSMUST00000006976.7 |
Ttc25
|
tetratricopeptide repeat domain 25 |
| chr9_-_22002599 | 1.56 |
ENSMUST00000115336.2
ENSMUST00000044926.5 |
Ccdc151
|
coiled-coil domain containing 151 |
| chr11_+_119229092 | 1.53 |
ENSMUST00000053440.7
|
Ccdc40
|
coiled-coil domain containing 40 |
| chr12_-_119238794 | 1.50 |
ENSMUST00000026360.8
|
Itgb8
|
integrin beta 8 |
| chr4_+_138262189 | 1.48 |
ENSMUST00000030539.3
|
Kif17
|
kinesin family member 17 |
| chr12_+_112808914 | 1.40 |
ENSMUST00000037014.3
ENSMUST00000177808.1 |
BC022687
|
cDNA sequence BC022687 |
| chr18_-_3309858 | 1.37 |
ENSMUST00000144496.1
ENSMUST00000154715.1 |
Crem
|
cAMP responsive element modulator |
| chr11_+_102881204 | 1.32 |
ENSMUST00000021307.3
ENSMUST00000159834.1 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr4_-_12087912 | 1.32 |
ENSMUST00000050686.3
|
Tmem67
|
transmembrane protein 67 |
| chr7_+_141195047 | 1.30 |
ENSMUST00000047093.4
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr10_+_112163621 | 1.29 |
ENSMUST00000092176.1
|
Caps2
|
calcyphosphine 2 |
| chr12_-_110840905 | 1.28 |
ENSMUST00000177224.1
ENSMUST00000084974.4 ENSMUST00000070565.8 |
Stk30
|
serine/threonine kinase 30 |
| chr4_+_100478806 | 1.27 |
ENSMUST00000133493.2
ENSMUST00000092730.3 ENSMUST00000106979.3 |
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr17_+_25471564 | 1.26 |
ENSMUST00000025002.1
|
Tekt4
|
tektin 4 |
| chr1_+_46066738 | 1.26 |
ENSMUST00000069293.7
|
Dnah7b
|
dynein, axonemal, heavy chain 7B |
| chr4_-_126325641 | 1.24 |
ENSMUST00000131113.1
|
Tekt2
|
tektin 2 |
| chr2_-_32775584 | 1.24 |
ENSMUST00000161430.1
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr4_+_45012830 | 1.20 |
ENSMUST00000095105.1
|
1700055D18Rik
|
RIKEN cDNA 1700055D18 gene |
| chr2_-_32775330 | 1.20 |
ENSMUST00000161089.1
ENSMUST00000066478.2 ENSMUST00000161950.1 |
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr5_-_34187670 | 1.20 |
ENSMUST00000042701.6
ENSMUST00000119171.1 |
Mxd4
|
Max dimerization protein 4 |
| chr9_+_49102720 | 1.19 |
ENSMUST00000070390.5
ENSMUST00000167095.1 |
Tmprss5
|
transmembrane protease, serine 5 (spinesin) |
| chr5_-_5664196 | 1.16 |
ENSMUST00000061008.3
ENSMUST00000054865.6 |
A330021E22Rik
|
RIKEN cDNA A330021E22 gene |
| chr2_-_93462457 | 1.13 |
ENSMUST00000028644.4
|
Cd82
|
CD82 antigen |
| chr1_+_74601441 | 1.10 |
ENSMUST00000087183.4
ENSMUST00000148456.1 ENSMUST00000113694.1 |
Stk36
|
serine/threonine kinase 36 |
| chr11_-_97041395 | 1.08 |
ENSMUST00000021251.6
|
Lrrc46
|
leucine rich repeat containing 46 |
| chr6_+_49319274 | 1.05 |
ENSMUST00000055559.7
ENSMUST00000114491.1 |
Ccdc126
|
coiled-coil domain containing 126 |
| chr4_+_134864536 | 1.05 |
ENSMUST00000030627.7
|
Rhd
|
Rh blood group, D antigen |
| chr11_+_60353324 | 1.05 |
ENSMUST00000070805.6
ENSMUST00000094140.2 ENSMUST00000108723.2 ENSMUST00000108722.4 |
Lrrc48
|
leucine rich repeat containing 48 |
| chr18_-_3309723 | 1.03 |
ENSMUST00000136961.1
ENSMUST00000152108.1 |
Crem
|
cAMP responsive element modulator |
| chr4_+_117251951 | 1.02 |
ENSMUST00000062824.5
|
Tmem53
|
transmembrane protein 53 |
| chr3_-_146108047 | 1.02 |
ENSMUST00000160285.1
|
Wdr63
|
WD repeat domain 63 |
| chr9_+_110476985 | 1.01 |
ENSMUST00000084948.4
ENSMUST00000061155.6 ENSMUST00000140686.1 ENSMUST00000084952.5 |
Kif9
|
kinesin family member 9 |
| chr10_+_75037291 | 0.97 |
ENSMUST00000139384.1
|
Rab36
|
RAB36, member RAS oncogene family |
| chr6_+_8259327 | 0.97 |
ENSMUST00000159378.1
|
Gm16039
|
predicted gene 16039 |
| chr6_+_8259288 | 0.96 |
ENSMUST00000159335.1
|
Gm16039
|
predicted gene 16039 |
| chr7_+_75455534 | 0.96 |
ENSMUST00000147005.1
ENSMUST00000166315.1 |
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chrX_-_163761323 | 0.95 |
ENSMUST00000059320.2
|
Rnf138rt1
|
ring finger protein 138, retrogene 1 |
| chr4_+_117252010 | 0.92 |
ENSMUST00000125943.1
ENSMUST00000106434.1 |
Tmem53
|
transmembrane protein 53 |
| chr1_+_74601548 | 0.91 |
ENSMUST00000087186.4
|
Stk36
|
serine/threonine kinase 36 |
| chr2_-_168230575 | 0.91 |
ENSMUST00000109193.1
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr8_+_36993551 | 0.90 |
ENSMUST00000098825.3
|
AI429214
|
expressed sequence AI429214 |
| chr9_-_49486209 | 0.89 |
ENSMUST00000055096.4
|
Ttc12
|
tetratricopeptide repeat domain 12 |
| chr11_+_117115195 | 0.89 |
ENSMUST00000103026.3
ENSMUST00000090433.5 |
Sec14l1
|
SEC14-like 1 (S. cerevisiae) |
| chr4_-_132922547 | 0.87 |
ENSMUST00000030696.4
|
Fam76a
|
family with sequence similarity 76, member A |
| chr5_+_110879788 | 0.87 |
ENSMUST00000156290.2
ENSMUST00000040111.9 |
Ttc28
|
tetratricopeptide repeat domain 28 |
| chr7_+_4922251 | 0.87 |
ENSMUST00000047309.5
|
Nat14
|
N-acetyltransferase 14 |
| chr9_+_46012810 | 0.86 |
ENSMUST00000126865.1
|
Sik3
|
SIK family kinase 3 |
| chr2_-_168230353 | 0.86 |
ENSMUST00000154111.1
|
Dpm1
|
dolichol-phosphate (beta-D) mannosyltransferase 1 |
| chr11_-_60352869 | 0.85 |
ENSMUST00000095254.5
ENSMUST00000102683.4 ENSMUST00000093048.6 ENSMUST00000093046.6 ENSMUST00000064019.8 ENSMUST00000102682.4 |
Tom1l2
|
target of myb1-like 2 (chicken) |
| chr4_-_132922487 | 0.83 |
ENSMUST00000097856.3
|
Fam76a
|
family with sequence similarity 76, member A |
| chr6_+_8259379 | 0.82 |
ENSMUST00000162034.1
ENSMUST00000160705.1 ENSMUST00000159433.1 |
Gm16039
|
predicted gene 16039 |
| chr10_-_93081596 | 0.81 |
ENSMUST00000168617.1
ENSMUST00000168110.1 ENSMUST00000020200.7 |
Gm872
|
RIKEN cDNA 4930485B16 gene |
| chr8_-_22805596 | 0.81 |
ENSMUST00000163739.1
|
Ap3m2
|
adaptor-related protein complex 3, mu 2 subunit |
| chr8_-_120177420 | 0.79 |
ENSMUST00000135567.2
|
Fam92b
|
family with sequence similarity 92, member B |
| chr1_-_37865040 | 0.78 |
ENSMUST00000041815.8
|
Tsga10
|
testis specific 10 |
| chr2_-_69789568 | 0.77 |
ENSMUST00000094942.3
|
Ccdc173
|
coiled-coil domain containing 173 |
| chr7_-_99858872 | 0.75 |
ENSMUST00000036274.6
|
Spcs2
|
signal peptidase complex subunit 2 homolog (S. cerevisiae) |
| chr8_+_123212857 | 0.75 |
ENSMUST00000060133.6
|
Spata33
|
spermatogenesis associated 33 |
| chr17_+_80290206 | 0.74 |
ENSMUST00000061703.9
|
Morn2
|
MORN repeat containing 2 |
| chr9_-_53975246 | 0.74 |
ENSMUST00000048409.7
|
Elmod1
|
ELMO/CED-12 domain containing 1 |
| chr9_+_59578192 | 0.74 |
ENSMUST00000118549.1
ENSMUST00000034840.3 |
Celf6
|
CUGBP, Elav-like family member 6 |
| chr15_-_82338801 | 0.73 |
ENSMUST00000023088.7
|
Naga
|
N-acetyl galactosaminidase, alpha |
| chr2_+_69789621 | 0.73 |
ENSMUST00000151298.1
ENSMUST00000028494.2 |
Phospho2
|
phosphatase, orphan 2 |
| chr7_-_126976092 | 0.73 |
ENSMUST00000181859.1
|
D830044I16Rik
|
RIKEN cDNA D830044I16 gene |
| chr12_+_73286779 | 0.72 |
ENSMUST00000140523.1
|
Slc38a6
|
solute carrier family 38, member 6 |
| chr9_+_72985504 | 0.72 |
ENSMUST00000156879.1
|
Ccpg1
|
cell cycle progression 1 |
| chr18_+_75820174 | 0.71 |
ENSMUST00000058997.7
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr11_+_117115133 | 0.71 |
ENSMUST00000021177.8
|
Sec14l1
|
SEC14-like 1 (S. cerevisiae) |
| chr17_+_34644805 | 0.71 |
ENSMUST00000174796.1
|
Fkbpl
|
FK506 binding protein-like |
| chr9_+_53405280 | 0.70 |
ENSMUST00000005262.1
|
4930550C14Rik
|
RIKEN cDNA 4930550C14 gene |
| chr15_+_98708187 | 0.69 |
ENSMUST00000003444.4
|
Ccdc65
|
coiled-coil domain containing 65 |
| chr2_+_69789647 | 0.69 |
ENSMUST00000112266.1
|
Phospho2
|
phosphatase, orphan 2 |
| chr17_-_37280418 | 0.69 |
ENSMUST00000077585.2
|
Olfr99
|
olfactory receptor 99 |
| chr15_-_79834323 | 0.68 |
ENSMUST00000177316.2
ENSMUST00000175858.2 |
Nptxr
|
neuronal pentraxin receptor |
| chr17_-_6948283 | 0.68 |
ENSMUST00000024572.9
|
Rsph3b
|
radial spoke 3B homolog (Chlamydomonas) |
| chr7_+_16959714 | 0.68 |
ENSMUST00000038163.6
|
Pnmal1
|
PNMA-like 1 |
| chrX_-_17319316 | 0.67 |
ENSMUST00000026014.7
|
Efhc2
|
EF-hand domain (C-terminal) containing 2 |
| chr13_-_92131494 | 0.66 |
ENSMUST00000099326.3
ENSMUST00000146492.1 |
Rasgrf2
|
RAS protein-specific guanine nucleotide-releasing factor 2 |
| chr14_+_31641051 | 0.66 |
ENSMUST00000090147.6
|
Btd
|
biotinidase |
| chr10_+_69213084 | 0.64 |
ENSMUST00000163497.1
ENSMUST00000164212.1 ENSMUST00000067908.7 |
Rhobtb1
|
Rho-related BTB domain containing 1 |
| chr1_-_75210732 | 0.64 |
ENSMUST00000113623.1
|
Glb1l
|
galactosidase, beta 1-like |
| chr17_+_85090647 | 0.64 |
ENSMUST00000095188.5
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr1_-_130715734 | 0.63 |
ENSMUST00000066863.6
ENSMUST00000050406.4 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr7_-_68749170 | 0.63 |
ENSMUST00000118110.1
ENSMUST00000048068.7 |
Arrdc4
|
arrestin domain containing 4 |
| chr9_+_22003035 | 0.63 |
ENSMUST00000115331.2
ENSMUST00000003493.7 |
Prkcsh
|
protein kinase C substrate 80K-H |
| chr7_+_16944645 | 0.63 |
ENSMUST00000094807.5
|
Pnmal2
|
PNMA-like 2 |
| chr17_-_29078953 | 0.62 |
ENSMUST00000133221.1
|
Trp53cor1
|
tumor protein p53 pathway corepressor 1 |
| chr7_-_139978748 | 0.62 |
ENSMUST00000097970.2
|
6430531B16Rik
|
RIKEN cDNA 6430531B16 gene |
| chr8_+_105427634 | 0.61 |
ENSMUST00000067305.6
|
Lrrc36
|
leucine rich repeat containing 36 |
| chr7_-_141214080 | 0.61 |
ENSMUST00000026573.5
ENSMUST00000170841.1 |
1600016N20Rik
|
RIKEN cDNA 1600016N20 gene |
| chr9_+_72985410 | 0.60 |
ENSMUST00000037977.8
|
Ccpg1
|
cell cycle progression 1 |
| chr17_+_15396240 | 0.60 |
ENSMUST00000055352.6
|
Fam120b
|
family with sequence similarity 120, member B |
| chr10_+_89686365 | 0.60 |
ENSMUST00000181598.1
|
1500026H17Rik
|
RIKEN cDNA 1500026H17 gene |
| chr19_-_40612160 | 0.60 |
ENSMUST00000132452.1
ENSMUST00000135795.1 ENSMUST00000025981.8 |
Tctn3
|
tectonic family member 3 |
| chr14_+_31001414 | 0.60 |
ENSMUST00000022476.7
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr16_-_36874806 | 0.59 |
ENSMUST00000075946.5
|
Eaf2
|
ELL associated factor 2 |
| chr12_+_84114321 | 0.59 |
ENSMUST00000123491.1
ENSMUST00000046340.2 ENSMUST00000136159.1 |
Dnal1
|
dynein, axonemal, light chain 1 |
| chr8_-_95281590 | 0.59 |
ENSMUST00000120044.2
ENSMUST00000121162.2 |
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr11_+_102665566 | 0.58 |
ENSMUST00000100378.3
|
Gm1564
|
predicted gene 1564 |
| chr2_+_28840406 | 0.58 |
ENSMUST00000113853.2
|
Ddx31
|
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 31 |
| chr8_+_34154563 | 0.58 |
ENSMUST00000033933.5
|
Tmem66
|
transmembrane protein 66 |
| chr17_-_87265866 | 0.58 |
ENSMUST00000145895.1
ENSMUST00000129616.1 ENSMUST00000155904.1 ENSMUST00000151155.1 ENSMUST00000144236.1 ENSMUST00000024963.3 |
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr9_-_106685653 | 0.57 |
ENSMUST00000163441.1
|
Tex264
|
testis expressed gene 264 |
| chr17_-_35046726 | 0.57 |
ENSMUST00000097338.4
|
Msh5
|
mutS homolog 5 (E. coli) |
| chr14_-_31640878 | 0.56 |
ENSMUST00000167066.1
ENSMUST00000127204.2 ENSMUST00000022437.8 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr7_+_99859123 | 0.56 |
ENSMUST00000036155.8
|
Xrra1
|
X-ray radiation resistance associated 1 |
| chr9_+_72985568 | 0.56 |
ENSMUST00000150826.2
ENSMUST00000085350.4 ENSMUST00000140675.1 |
Ccpg1
|
cell cycle progression 1 |
| chr12_-_84148449 | 0.56 |
ENSMUST00000061425.2
|
Pnma1
|
paraneoplastic antigen MA1 |
| chrX_-_102906469 | 0.56 |
ENSMUST00000120808.1
ENSMUST00000121197.1 |
Dmrtc1a
|
DMRT-like family C1a |
| chrX_+_135839034 | 0.56 |
ENSMUST00000173804.1
ENSMUST00000113136.1 |
Gprasp2
|
G protein-coupled receptor associated sorting protein 2 |
| chr16_+_36875119 | 0.56 |
ENSMUST00000135406.1
ENSMUST00000114812.1 ENSMUST00000134616.1 ENSMUST00000023534.6 |
Golgb1
Golgb1
|
golgi autoantigen, golgin subfamily b, macrogolgin 1 golgi autoantigen, golgin subfamily b, macrogolgin 1 |
| chr2_-_32775625 | 0.56 |
ENSMUST00000161958.1
|
Ttc16
|
tetratricopeptide repeat domain 16 |
| chr7_+_28766747 | 0.56 |
ENSMUST00000170068.1
ENSMUST00000072965.4 |
Sirt2
|
sirtuin 2 |
| chr17_+_7945653 | 0.56 |
ENSMUST00000097423.2
|
Rsph3a
|
radial spoke 3A homolog (Chlamydomonas) |
| chr2_+_143915273 | 0.55 |
ENSMUST00000103172.3
|
Dstn
|
destrin |
| chr14_+_31001383 | 0.55 |
ENSMUST00000168584.1
|
Glt8d1
|
glycosyltransferase 8 domain containing 1 |
| chr15_+_85132080 | 0.54 |
ENSMUST00000023067.2
|
Ribc2
|
RIB43A domain with coiled-coils 2 |
| chr7_-_127588595 | 0.54 |
ENSMUST00000072155.3
|
Gm166
|
predicted gene 166 |
| chr1_+_85894281 | 0.53 |
ENSMUST00000027425.9
|
Itm2c
|
integral membrane protein 2C |
| chr2_+_102550012 | 0.53 |
ENSMUST00000028612.7
|
Pamr1
|
peptidase domain containing associated with muscle regeneration 1 |
| chr11_+_23008337 | 0.53 |
ENSMUST00000172602.1
|
Fam161a
|
family with sequence similarity 161, member A |
| chr9_-_123717576 | 0.52 |
ENSMUST00000026274.7
|
Lztfl1
|
leucine zipper transcription factor-like 1 |
| chr4_-_119415494 | 0.51 |
ENSMUST00000063642.2
|
Ccdc30
|
coiled-coil domain containing 30 |
| chr16_+_17561885 | 0.51 |
ENSMUST00000171002.1
ENSMUST00000023441.4 |
P2rx6
|
purinergic receptor P2X, ligand-gated ion channel, 6 |
| chr10_+_79669410 | 0.51 |
ENSMUST00000020552.5
|
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr14_+_57424054 | 0.50 |
ENSMUST00000122063.1
|
Ift88
|
intraflagellar transport 88 |
| chr9_-_108578657 | 0.50 |
ENSMUST00000068700.5
|
Wdr6
|
WD repeat domain 6 |
| chr2_+_156840966 | 0.50 |
ENSMUST00000109564.1
|
Tgif2
|
TGFB-induced factor homeobox 2 |
| chr9_-_106685892 | 0.49 |
ENSMUST00000169068.1
ENSMUST00000046735.4 |
Tex264
|
testis expressed gene 264 |
| chr7_+_90426312 | 0.49 |
ENSMUST00000061391.7
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr12_-_98259416 | 0.49 |
ENSMUST00000021390.7
|
Galc
|
galactosylceramidase |
| chr10_-_75032528 | 0.49 |
ENSMUST00000159994.1
ENSMUST00000179546.1 ENSMUST00000160450.1 ENSMUST00000160072.1 ENSMUST00000009214.3 ENSMUST00000166088.1 |
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr1_-_155146755 | 0.49 |
ENSMUST00000027744.8
|
Mr1
|
major histocompatibility complex, class I-related |
| chr7_-_119896285 | 0.49 |
ENSMUST00000106519.1
|
Dcun1d3
|
DCN1, defective in cullin neddylation 1, domain containing 3 (S. cerevisiae) |
| chr17_-_85090204 | 0.49 |
ENSMUST00000072406.3
ENSMUST00000171795.1 |
Prepl
|
prolyl endopeptidase-like |
| chr5_+_137030275 | 0.48 |
ENSMUST00000041543.8
|
Vgf
|
VGF nerve growth factor inducible |
| chr11_+_23008420 | 0.48 |
ENSMUST00000109557.2
|
Fam161a
|
family with sequence similarity 161, member A |
| chr3_-_89393294 | 0.48 |
ENSMUST00000142119.1
ENSMUST00000029677.8 ENSMUST00000148361.1 |
Zbtb7b
|
zinc finger and BTB domain containing 7B |
| chr7_-_45814277 | 0.48 |
ENSMUST00000107728.1
|
Cyth2
|
cytohesin 2 |
| chrX_-_122397351 | 0.47 |
ENSMUST00000079490.4
|
Nap1l3
|
nucleosome assembly protein 1-like 3 |
| chr9_-_72985344 | 0.47 |
ENSMUST00000124565.2
|
Gm5918
|
predicted gene 5918 |
| chr9_-_65908676 | 0.47 |
ENSMUST00000119245.1
ENSMUST00000134338.1 ENSMUST00000179395.1 |
Trip4
|
thyroid hormone receptor interactor 4 |
| chr2_-_144527341 | 0.47 |
ENSMUST00000163701.1
ENSMUST00000081982.5 |
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr4_+_12906838 | 0.46 |
ENSMUST00000143186.1
ENSMUST00000183345.1 |
Triqk
|
triple QxxK/R motif containing |
| chr3_-_107517321 | 0.46 |
ENSMUST00000166892.1
|
Slc6a17
|
solute carrier family 6 (neurotransmitter transporter), member 17 |
| chrX_+_37546975 | 0.46 |
ENSMUST00000081327.5
ENSMUST00000184210.1 ENSMUST00000184270.1 |
Rhox3e
|
reproductive homeobox 3E |
| chr7_-_73537621 | 0.46 |
ENSMUST00000172704.1
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr9_+_46012822 | 0.45 |
ENSMUST00000120463.2
ENSMUST00000120247.1 |
Sik3
|
SIK family kinase 3 |
| chr5_+_29195983 | 0.45 |
ENSMUST00000160888.1
ENSMUST00000159272.1 ENSMUST00000001247.5 ENSMUST00000161398.1 ENSMUST00000160246.1 |
Rnf32
|
ring finger protein 32 |
| chr10_+_5593718 | 0.44 |
ENSMUST00000051809.8
|
Myct1
|
myc target 1 |
| chr11_-_53423123 | 0.44 |
ENSMUST00000036045.5
|
Leap2
|
liver-expressed antimicrobial peptide 2 |
| chr16_+_52031549 | 0.44 |
ENSMUST00000114471.1
|
Cblb
|
Casitas B-lineage lymphoma b |
| chr18_+_63708689 | 0.44 |
ENSMUST00000072726.5
|
Wdr7
|
WD repeat domain 7 |
| chr7_-_139683797 | 0.44 |
ENSMUST00000129990.1
ENSMUST00000130453.1 |
9330101J02Rik
|
RIKEN cDNA 9330101J02 gene |
| chr7_-_45814302 | 0.44 |
ENSMUST00000107729.1
ENSMUST00000056820.6 |
Cyth2
|
cytohesin 2 |
| chr2_-_134644019 | 0.44 |
ENSMUST00000110120.1
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr12_+_33394854 | 0.43 |
ENSMUST00000020878.6
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr8_-_116978908 | 0.43 |
ENSMUST00000070577.5
|
1700030J22Rik
|
RIKEN cDNA 1700030J22 gene |
| chr8_-_120177440 | 0.43 |
ENSMUST00000048786.6
|
Fam92b
|
family with sequence similarity 92, member B |
| chr9_+_105642957 | 0.42 |
ENSMUST00000065778.6
|
Pik3r4
|
phosphatidylinositol 3 kinase, regulatory subunit, polypeptide 4, p150 |
| chr16_-_62847008 | 0.41 |
ENSMUST00000089289.5
|
Arl13b
|
ADP-ribosylation factor-like 13B |
| chr2_-_134644079 | 0.41 |
ENSMUST00000110119.1
|
Tmx4
|
thioredoxin-related transmembrane protein 4 |
| chr1_+_95313607 | 0.40 |
ENSMUST00000059975.6
|
Fam174a
|
family with sequence similarity 174, member A |
| chr11_+_62551167 | 0.40 |
ENSMUST00000019649.3
|
Ubb
|
ubiquitin B |
| chr5_-_116288978 | 0.40 |
ENSMUST00000050178.6
|
Ccdc60
|
coiled-coil domain containing 60 |
| chr4_+_126058557 | 0.40 |
ENSMUST00000035497.4
|
Oscp1
|
organic solute carrier partner 1 |
| chr7_-_126884678 | 0.39 |
ENSMUST00000071268.4
ENSMUST00000117394.1 |
Taok2
|
TAO kinase 2 |
| chr9_-_64341145 | 0.39 |
ENSMUST00000120760.1
ENSMUST00000168844.2 |
Dis3l
|
DIS3 mitotic control homolog (S. cerevisiae)-like |
| chr2_+_125859134 | 0.39 |
ENSMUST00000028636.6
ENSMUST00000125084.1 |
Galk2
|
galactokinase 2 |
| chr11_+_117523526 | 0.39 |
ENSMUST00000132261.1
|
Gm11734
|
predicted gene 11734 |
| chr3_-_155093346 | 0.39 |
ENSMUST00000066568.4
|
Fpgt
|
fucose-1-phosphate guanylyltransferase |
| chr4_+_41569775 | 0.38 |
ENSMUST00000102963.3
|
Dnaic1
|
dynein, axonemal, intermediate chain 1 |
| chr1_+_59119822 | 0.37 |
ENSMUST00000180570.1
|
G730003C15Rik
|
RIKEN cDNA G730003C15 gene |
| chr9_-_119937514 | 0.37 |
ENSMUST00000035099.7
|
Gorasp1
|
golgi reassembly stacking protein 1 |
| chrX_+_37255007 | 0.37 |
ENSMUST00000185028.1
ENSMUST00000185050.1 |
Rhox3a
|
reproductive homeobox 3A |
| chr4_+_43875524 | 0.37 |
ENSMUST00000030198.6
|
Reck
|
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr12_+_111166413 | 0.36 |
ENSMUST00000021706.4
|
Traf3
|
TNF receptor-associated factor 3 |
| chr13_+_21735055 | 0.36 |
ENSMUST00000087714.4
|
Hist1h4j
|
histone cluster 1, H4j |
| chr1_-_53706775 | 0.35 |
ENSMUST00000094964.5
|
Dnah7a
|
dynein, axonemal, heavy chain 7A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx2_Rfx7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.6 | GO:2000314 | negative regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000314) |
| 1.2 | 4.9 | GO:0046898 | response to cycloheximide(GO:0046898) |
| 0.7 | 10.1 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.5 | 6.9 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.5 | 2.0 | GO:0007228 | positive regulation of hh target transcription factor activity(GO:0007228) |
| 0.5 | 1.5 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.5 | 1.9 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.3 | 1.3 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.3 | 1.6 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.3 | 1.8 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.2 | 0.2 | GO:0018307 | enzyme active site formation(GO:0018307) |
| 0.2 | 0.7 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.2 | 0.5 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.2 | 1.2 | GO:0046477 | glycosylceramide catabolic process(GO:0046477) |
| 0.1 | 0.6 | GO:0060282 | positive regulation of oocyte development(GO:0060282) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) negative regulation of defense response to bacterium(GO:1900425) |
| 0.1 | 0.5 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.1 | 1.7 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.1 | 1.0 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.1 | GO:0007227 | signal transduction downstream of smoothened(GO:0007227) |
| 0.1 | 0.6 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.1 | 0.3 | GO:1903538 | meiotic sister chromatid cohesion, centromeric(GO:0051754) meiotic cell cycle process involved in oocyte maturation(GO:1903537) regulation of meiotic cell cycle process involved in oocyte maturation(GO:1903538) |
| 0.1 | 4.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.1 | 0.5 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.3 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.1 | 1.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.1 | 0.5 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.4 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.1 | 0.4 | GO:0007113 | endomitotic cell cycle(GO:0007113) thrombopoietin-mediated signaling pathway(GO:0038163) positive regulation of male germ cell proliferation(GO:2000256) |
| 0.1 | 0.4 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.1 | 0.5 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.1 | 0.3 | GO:1902953 | positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.1 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.5 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.1 | 0.3 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.1 | 1.3 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.1 | 0.7 | GO:0018095 | protein polyglutamylation(GO:0018095) |
| 0.1 | 1.5 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.1 | 0.2 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.3 | GO:0070829 | heterochromatin maintenance(GO:0070829) |
| 0.1 | 2.4 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.1 | 0.4 | GO:0021886 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.1 | 0.9 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.1 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.1 | 0.3 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.1 | 0.1 | GO:2000282 | regulation of cellular amino acid biosynthetic process(GO:2000282) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.3 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.1 | 0.8 | GO:0033133 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 7.9 | GO:0016579 | protein deubiquitination(GO:0016579) |
| 0.1 | 0.2 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.2 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.1 | 0.6 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.1 | 0.3 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.1 | 0.2 | GO:0060854 | patterning of lymph vessels(GO:0060854) |
| 0.1 | 0.4 | GO:0021943 | formation of radial glial scaffolds(GO:0021943) |
| 0.1 | 0.6 | GO:0051026 | chiasma assembly(GO:0051026) |
| 0.1 | 0.3 | GO:1904717 | regulation of AMPA glutamate receptor clustering(GO:1904717) |
| 0.0 | 0.2 | GO:0006049 | UDP-N-acetylglucosamine catabolic process(GO:0006049) |
| 0.0 | 0.7 | GO:2000310 | regulation of N-methyl-D-aspartate selective glutamate receptor activity(GO:2000310) |
| 0.0 | 0.3 | GO:2001170 | negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 1.7 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0006616 | SRP-dependent cotranslational protein targeting to membrane, translocation(GO:0006616) |
| 0.0 | 0.1 | GO:0046341 | CDP-diacylglycerol metabolic process(GO:0046341) |
| 0.0 | 0.1 | GO:1903334 | positive regulation of protein folding(GO:1903334) |
| 0.0 | 0.3 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.3 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.4 | GO:0090503 | RNA phosphodiester bond hydrolysis, exonucleolytic(GO:0090503) |
| 0.0 | 0.8 | GO:0099514 | anterograde synaptic vesicle transport(GO:0048490) synaptic vesicle cytoskeletal transport(GO:0099514) synaptic vesicle transport along microtubule(GO:0099517) |
| 0.0 | 0.5 | GO:1900113 | negative regulation of histone H3-K9 trimethylation(GO:1900113) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 1.3 | GO:2000171 | negative regulation of dendrite development(GO:2000171) |
| 0.0 | 0.2 | GO:0060316 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) |
| 0.0 | 0.2 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 1.3 | GO:0060351 | cartilage development involved in endochondral bone morphogenesis(GO:0060351) |
| 0.0 | 0.2 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.0 | 0.2 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.0 | 1.0 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.4 | GO:0006012 | galactose metabolic process(GO:0006012) |
| 0.0 | 0.2 | GO:2001168 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.3 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.5 | GO:0043084 | penile erection(GO:0043084) |
| 0.0 | 0.2 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.0 | 0.1 | GO:0001830 | trophectodermal cell fate commitment(GO:0001830) |
| 0.0 | 1.7 | GO:0016574 | histone ubiquitination(GO:0016574) |
| 0.0 | 0.1 | GO:1903061 | positive regulation of protein lipidation(GO:1903061) negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.7 | GO:0050907 | detection of chemical stimulus involved in sensory perception(GO:0050907) |
| 0.0 | 0.2 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.3 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.6 | GO:0035357 | peroxisome proliferator activated receptor signaling pathway(GO:0035357) |
| 0.0 | 0.5 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:1990592 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 3.1 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.5 | GO:2000300 | regulation of synaptic vesicle exocytosis(GO:2000300) |
| 0.0 | 1.8 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.3 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.5 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.3 | GO:0048280 | vesicle fusion with Golgi apparatus(GO:0048280) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.6 | GO:0006767 | water-soluble vitamin metabolic process(GO:0006767) |
| 0.0 | 0.2 | GO:2000601 | positive regulation of Arp2/3 complex-mediated actin nucleation(GO:2000601) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.1 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.5 | GO:0032620 | interleukin-17 production(GO:0032620) |
| 0.0 | 0.1 | GO:1901525 | negative regulation of macromitophagy(GO:1901525) |
| 0.0 | 0.1 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.8 | GO:0002437 | inflammatory response to antigenic stimulus(GO:0002437) |
| 0.0 | 0.1 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.4 | GO:0010165 | response to X-ray(GO:0010165) |
| 0.0 | 0.1 | GO:0007182 | common-partner SMAD protein phosphorylation(GO:0007182) |
| 0.0 | 0.2 | GO:1902176 | negative regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902176) |
| 0.0 | 0.2 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) |
| 0.0 | 0.1 | GO:0046929 | negative regulation of neurotransmitter secretion(GO:0046929) |
| 0.0 | 0.5 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.5 | GO:0061512 | protein localization to cilium(GO:0061512) |
| 0.0 | 0.8 | GO:0045454 | cell redox homeostasis(GO:0045454) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.3 | GO:0071625 | vocalization behavior(GO:0071625) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.3 | GO:0060765 | regulation of androgen receptor signaling pathway(GO:0060765) |
| 0.0 | 0.4 | GO:0000186 | activation of MAPKK activity(GO:0000186) |
| 0.0 | 0.3 | GO:0097120 | receptor localization to synapse(GO:0097120) |
| 0.0 | 0.3 | GO:1905145 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 6.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.4 | 4.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.4 | 1.8 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.3 | 1.7 | GO:1990769 | proximal neuron projection(GO:1990769) |
| 0.3 | 3.3 | GO:0002177 | manchette(GO:0002177) |
| 0.3 | 3.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.2 | 1.5 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.2 | 1.4 | GO:0033503 | HULC complex(GO:0033503) |
| 0.2 | 0.6 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.2 | 1.6 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.2 | 1.3 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.4 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.5 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.1 | 1.4 | GO:0036038 | MKS complex(GO:0036038) |
| 0.1 | 0.2 | GO:1990429 | Pex17p-Pex14p docking complex(GO:1990415) peroxisomal importomer complex(GO:1990429) |
| 0.1 | 0.5 | GO:0030991 | intraciliary transport particle A(GO:0030991) |
| 0.1 | 0.3 | GO:0031021 | interphase microtubule organizing center(GO:0031021) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.8 | GO:1904115 | axon cytoplasm(GO:1904115) |
| 0.0 | 0.5 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.8 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.7 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 1.5 | GO:0008305 | integrin complex(GO:0008305) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.9 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.4 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.5 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.2 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 1.0 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.2 | GO:0005786 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) |
| 0.0 | 0.3 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 1.0 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.2 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.3 | GO:0097539 | ciliary transition fiber(GO:0097539) |
| 0.0 | 0.2 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.3 | GO:0031011 | Ino80 complex(GO:0031011) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.1 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.4 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.8 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 1.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.3 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 1.5 | GO:0031514 | motile cilium(GO:0031514) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.3 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.2 | GO:0031209 | SCAR complex(GO:0031209) |
| 0.0 | 0.6 | GO:0031201 | SNARE complex(GO:0031201) |
| 0.0 | 0.2 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.9 | GO:0005930 | axoneme(GO:0005930) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 4.9 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.4 | 1.8 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.3 | 3.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.2 | 4.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.2 | 1.5 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.9 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.2 | 1.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.2 | 1.4 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.2 | 0.7 | GO:0033883 | pyridoxal phosphatase activity(GO:0033883) |
| 0.2 | 0.7 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 1.3 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.6 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.4 | GO:0033699 | DNA 5'-adenosine monophosphate hydrolase activity(GO:0033699) |
| 0.1 | 0.4 | GO:0004335 | galactokinase activity(GO:0004335) |
| 0.1 | 1.5 | GO:0008574 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) |
| 0.1 | 1.0 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 3.0 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.1 | 0.3 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 7.9 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.1 | 2.9 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.1 | 0.4 | GO:0070568 | guanylyltransferase activity(GO:0070568) |
| 0.1 | 0.5 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.1 | 0.2 | GO:0001132 | RNA polymerase II transcription factor activity, TBP-class protein binding, involved in preinitiation complex assembly(GO:0001129) RNA polymerase II transcription factor activity, TBP-class protein binding(GO:0001132) |
| 0.1 | 0.4 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.1 | 0.6 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.3 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.3 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.9 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.1 | 0.3 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.1 | 0.6 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 0.2 | GO:0019002 | GMP binding(GO:0019002) |
| 0.1 | 0.6 | GO:0030983 | mismatched DNA binding(GO:0030983) |
| 0.1 | 1.3 | GO:0003756 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.1 | 1.3 | GO:0031005 | filamin binding(GO:0031005) |
| 0.1 | 0.4 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.1 | 0.2 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0003827 | alpha-1,3-mannosylglycoprotein 2-beta-N-acetylglucosaminyltransferase activity(GO:0003827) |
| 0.0 | 0.3 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 1.1 | GO:0008519 | ammonium transmembrane transporter activity(GO:0008519) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.2 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.2 | GO:0004342 | glucosamine-6-phosphate deaminase activity(GO:0004342) |
| 0.0 | 2.9 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.0 | 0.3 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.5 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.3 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 2.1 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.3 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.3 | GO:0043262 | adenosine-diphosphatase activity(GO:0043262) |
| 0.0 | 0.5 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.2 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.2 | GO:0008312 | signal recognition particle binding(GO:0005047) 7S RNA binding(GO:0008312) |
| 0.0 | 1.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.3 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.5 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.5 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.3 | GO:0043855 | cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.3 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 2.9 | GO:0001046 | core promoter sequence-specific DNA binding(GO:0001046) |
| 0.0 | 1.0 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.7 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.4 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.7 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.6 | GO:0016278 | lysine N-methyltransferase activity(GO:0016278) protein-lysine N-methyltransferase activity(GO:0016279) |
| 0.0 | 0.2 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.4 | GO:0000175 | 3'-5'-exoribonuclease activity(GO:0000175) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0004579 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 5.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.1 | 1.5 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 2.4 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 2.5 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 2.1 | PID HEDGEHOG GLI PATHWAY | Hedgehog signaling events mediated by Gli proteins |
| 0.0 | 0.7 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 1.0 | PID THROMBIN PAR1 PATHWAY | PAR1-mediated thrombin signaling events |
| 0.0 | 0.3 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.8 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 1.0 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.6 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 4.9 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.1 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 1.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.7 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.6 | REACTOME MEIOSIS | Genes involved in Meiosis |
| 0.0 | 0.8 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.5 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.6 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.7 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 1.2 | REACTOME REGULATION OF INSULIN SECRETION | Genes involved in Regulation of Insulin Secretion |
| 0.0 | 1.2 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.4 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |