Project
2D miR_HR1_12
Navigation
Downloads
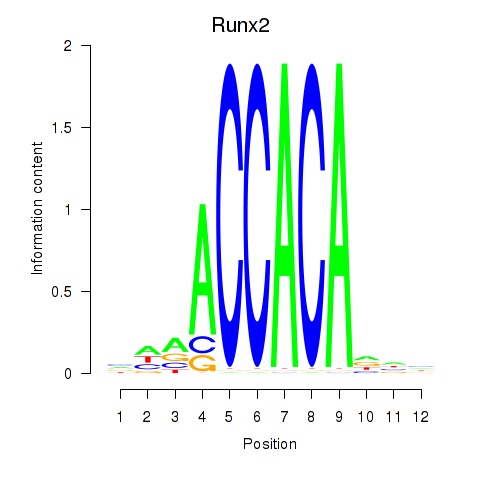
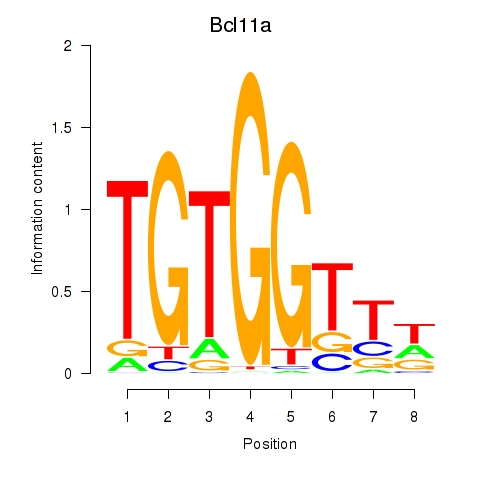
Results for Runx2_Bcl11a
Z-value: 1.88
Transcription factors associated with Runx2_Bcl11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx2
|
ENSMUSG00000039153.10 | runt related transcription factor 2 |
|
Bcl11a
|
ENSMUSG00000000861.9 | B cell CLL/lymphoma 11A (zinc finger protein) |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Bcl11a | mm10_v2_chr11_+_24078173_24078219 | -0.76 | 4.1e-03 | Click! |
| Runx2 | mm10_v2_chr17_-_44814581_44814595 | -0.49 | 1.0e-01 | Click! |
Activity profile of Runx2_Bcl11a motif
Sorted Z-values of Runx2_Bcl11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_109579056 | 4.84 |
ENSMUST00000074898.6
|
Hp
|
haptoglobin |
| chr10_-_81291227 | 3.01 |
ENSMUST00000045744.6
|
Tjp3
|
tight junction protein 3 |
| chr16_-_17838173 | 2.96 |
ENSMUST00000118960.1
|
Car15
|
carbonic anhydrase 15 |
| chr7_-_126676357 | 2.78 |
ENSMUST00000106371.1
ENSMUST00000106372.3 ENSMUST00000155419.1 |
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr1_-_164935522 | 2.42 |
ENSMUST00000027860.7
|
Xcl1
|
chemokine (C motif) ligand 1 |
| chr15_+_9436028 | 2.33 |
ENSMUST00000042360.3
|
Capsl
|
calcyphosine-like |
| chr7_-_126676428 | 2.28 |
ENSMUST00000106373.1
|
Sult1a1
|
sulfotransferase family 1A, phenol-preferring, member 1 |
| chr13_+_89540636 | 2.17 |
ENSMUST00000022108.7
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chrX_-_101734125 | 2.10 |
ENSMUST00000056614.6
|
Cxcr3
|
chemokine (C-X-C motif) receptor 3 |
| chr7_+_80246375 | 2.05 |
ENSMUST00000058266.6
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr14_+_55853997 | 2.02 |
ENSMUST00000100529.3
|
Nynrin
|
NYN domain and retroviral integrase containing |
| chr15_-_75567176 | 2.00 |
ENSMUST00000156032.1
ENSMUST00000127095.1 |
Ly6h
|
lymphocyte antigen 6 complex, locus H |
| chr2_+_70562007 | 1.90 |
ENSMUST00000094934.4
|
Gad1
|
glutamate decarboxylase 1 |
| chr11_+_96849863 | 1.82 |
ENSMUST00000018816.7
|
Copz2
|
coatomer protein complex, subunit zeta 2 |
| chr11_-_101967005 | 1.80 |
ENSMUST00000001534.6
|
Sost
|
sclerostin |
| chr6_+_123123423 | 1.79 |
ENSMUST00000032248.7
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr7_+_121865070 | 1.77 |
ENSMUST00000033161.5
|
Scnn1b
|
sodium channel, nonvoltage-gated 1 beta |
| chr7_+_80246529 | 1.69 |
ENSMUST00000107381.1
|
Ttll13
|
tubulin tyrosine ligase-like family, member 13 |
| chr10_+_127866457 | 1.69 |
ENSMUST00000092058.3
|
BC089597
|
cDNA sequence BC089597 |
| chr6_+_123123313 | 1.64 |
ENSMUST00000041779.6
|
Clec4a2
|
C-type lectin domain family 4, member a2 |
| chr6_-_83121385 | 1.63 |
ENSMUST00000146328.1
ENSMUST00000113936.3 ENSMUST00000032111.4 |
Wbp1
|
WW domain binding protein 1 |
| chr3_+_96181151 | 1.57 |
ENSMUST00000035371.8
|
Sv2a
|
synaptic vesicle glycoprotein 2 a |
| chr16_-_94370994 | 1.52 |
ENSMUST00000113914.1
ENSMUST00000113905.1 |
Pigp
|
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr9_+_53405280 | 1.49 |
ENSMUST00000005262.1
|
4930550C14Rik
|
RIKEN cDNA 4930550C14 gene |
| chr2_-_94264745 | 1.47 |
ENSMUST00000134563.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chr3_-_90514250 | 1.43 |
ENSMUST00000107340.1
ENSMUST00000060738.8 |
S100a1
|
S100 calcium binding protein A1 |
| chrX_-_57338598 | 1.42 |
ENSMUST00000033468.4
ENSMUST00000114736.1 |
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 |
| chr11_+_87663087 | 1.39 |
ENSMUST00000165679.1
|
Rnf43
|
ring finger protein 43 |
| chr3_-_89093358 | 1.39 |
ENSMUST00000090929.5
ENSMUST00000052539.6 |
Rusc1
|
RUN and SH3 domain containing 1 |
| chr2_+_21367532 | 1.37 |
ENSMUST00000055946.7
|
Gpr158
|
G protein-coupled receptor 158 |
| chr6_+_129350237 | 1.35 |
ENSMUST00000065289.4
|
Clec12a
|
C-type lectin domain family 12, member a |
| chr2_+_70562147 | 1.35 |
ENSMUST00000148210.1
|
Gad1
|
glutamate decarboxylase 1 |
| chr14_-_56262233 | 1.32 |
ENSMUST00000015581.4
|
Gzmb
|
granzyme B |
| chr11_+_82045705 | 1.31 |
ENSMUST00000021011.2
|
Ccl7
|
chemokine (C-C motif) ligand 7 |
| chr8_+_124576105 | 1.28 |
ENSMUST00000093033.5
ENSMUST00000133086.1 |
Capn9
|
calpain 9 |
| chr11_-_100414829 | 1.27 |
ENSMUST00000066489.6
|
Leprel4
|
leprecan-like 4 |
| chr15_-_76660108 | 1.27 |
ENSMUST00000066677.8
ENSMUST00000177359.1 |
Cyhr1
|
cysteine and histidine rich 1 |
| chr4_-_43040279 | 1.26 |
ENSMUST00000107958.1
ENSMUST00000107959.1 ENSMUST00000152846.1 |
Fam214b
|
family with sequence similarity 214, member B |
| chr4_-_143299498 | 1.24 |
ENSMUST00000030317.7
|
Pdpn
|
podoplanin |
| chr15_-_100584075 | 1.24 |
ENSMUST00000184908.1
|
POU6F1
|
POU domain, class 6, transcription factor 1 (Pou6f1), mRNA |
| chr4_-_137430517 | 1.23 |
ENSMUST00000102522.4
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr2_-_94264713 | 1.22 |
ENSMUST00000129661.1
|
E530001K10Rik
|
RIKEN cDNA E530001K10 gene |
| chr4_-_114908892 | 1.22 |
ENSMUST00000068654.3
|
Foxd2
|
forkhead box D2 |
| chr11_+_78322965 | 1.22 |
ENSMUST00000017534.8
|
Aldoc
|
aldolase C, fructose-bisphosphate |
| chr1_-_10009098 | 1.21 |
ENSMUST00000176398.1
ENSMUST00000027049.3 |
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr7_+_30459713 | 1.17 |
ENSMUST00000006825.8
|
Nphs1
|
nephrosis 1, nephrin |
| chr11_-_79523760 | 1.16 |
ENSMUST00000179322.1
|
Evi2b
|
ecotropic viral integration site 2b |
| chr11_-_106388066 | 1.14 |
ENSMUST00000106813.2
ENSMUST00000141146.1 |
Icam2
|
intercellular adhesion molecule 2 |
| chr10_-_40025253 | 1.14 |
ENSMUST00000163705.2
|
AI317395
|
expressed sequence AI317395 |
| chr2_-_144332146 | 1.12 |
ENSMUST00000037423.3
|
Ovol2
|
ovo-like 2 (Drosophila) |
| chr15_-_54278420 | 1.10 |
ENSMUST00000079772.3
|
Tnfrsf11b
|
tumor necrosis factor receptor superfamily, member 11b (osteoprotegerin) |
| chr17_+_21690766 | 1.10 |
ENSMUST00000097384.1
|
Gm10509
|
predicted gene 10509 |
| chr7_+_49974864 | 1.09 |
ENSMUST00000081872.5
ENSMUST00000151721.1 |
Nell1
|
NEL-like 1 |
| chr3_+_90514435 | 1.09 |
ENSMUST00000048138.6
ENSMUST00000181271.1 |
S100a13
|
S100 calcium binding protein A13 |
| chr10_+_76562270 | 1.08 |
ENSMUST00000009259.4
ENSMUST00000105414.1 |
Spatc1l
|
spermatogenesis and centriole associated 1 like |
| chr18_-_3299452 | 1.08 |
ENSMUST00000126578.1
|
Crem
|
cAMP responsive element modulator |
| chrX_-_8090442 | 1.07 |
ENSMUST00000033505.6
|
Was
|
Wiskott-Aldrich syndrome homolog (human) |
| chr3_+_3634145 | 1.05 |
ENSMUST00000108394.1
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr4_-_143299463 | 1.05 |
ENSMUST00000119654.1
|
Pdpn
|
podoplanin |
| chr8_-_111691002 | 1.05 |
ENSMUST00000034435.5
|
Ctrb1
|
chymotrypsinogen B1 |
| chr10_-_24101951 | 1.04 |
ENSMUST00000170267.1
|
Taar8c
|
trace amine-associated receptor 8C |
| chr4_+_144892813 | 1.03 |
ENSMUST00000105744.1
ENSMUST00000171001.1 |
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr17_+_35821675 | 1.03 |
ENSMUST00000003635.6
|
Ier3
|
immediate early response 3 |
| chr7_+_44572370 | 1.03 |
ENSMUST00000002274.8
|
Napsa
|
napsin A aspartic peptidase |
| chr4_+_128058962 | 1.02 |
ENSMUST00000184063.1
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr4_-_57916283 | 1.02 |
ENSMUST00000063816.5
|
D630039A03Rik
|
RIKEN cDNA D630039A03 gene |
| chr11_+_116030304 | 0.99 |
ENSMUST00000021116.5
ENSMUST00000106452.1 |
Unk
|
unkempt homolog (Drosophila) |
| chr11_-_83649349 | 0.98 |
ENSMUST00000001008.5
|
Ccl3
|
chemokine (C-C motif) ligand 3 |
| chr10_-_62379852 | 0.98 |
ENSMUST00000143236.1
ENSMUST00000133429.1 ENSMUST00000132926.1 ENSMUST00000116238.2 |
Hk1
|
hexokinase 1 |
| chr5_+_104435112 | 0.98 |
ENSMUST00000031243.8
ENSMUST00000086833.6 ENSMUST00000112748.1 ENSMUST00000112746.1 ENSMUST00000145084.1 ENSMUST00000132457.1 |
Spp1
|
secreted phosphoprotein 1 |
| chr13_-_37050237 | 0.96 |
ENSMUST00000164727.1
|
F13a1
|
coagulation factor XIII, A1 subunit |
| chr13_+_49608030 | 0.96 |
ENSMUST00000021822.5
|
Ogn
|
osteoglycin |
| chr13_+_54789377 | 0.96 |
ENSMUST00000026993.7
ENSMUST00000131692.2 ENSMUST00000163796.1 |
Tspan17
|
tetraspanin 17 |
| chr13_+_54789500 | 0.96 |
ENSMUST00000163915.1
ENSMUST00000099503.3 ENSMUST00000171859.1 |
Tspan17
|
tetraspanin 17 |
| chr18_-_62179948 | 0.95 |
ENSMUST00000053640.3
|
Adrb2
|
adrenergic receptor, beta 2 |
| chr4_+_118527229 | 0.94 |
ENSMUST00000030261.5
|
2610528J11Rik
|
RIKEN cDNA 2610528J11 gene |
| chr5_-_137858034 | 0.94 |
ENSMUST00000110978.2
|
Pilrb1
|
paired immunoglobin-like type 2 receptor beta 1 |
| chr14_+_31641051 | 0.93 |
ENSMUST00000090147.6
|
Btd
|
biotinidase |
| chr19_+_37436707 | 0.92 |
ENSMUST00000128184.1
|
Hhex
|
hematopoietically expressed homeobox |
| chr7_+_143005677 | 0.92 |
ENSMUST00000082008.5
ENSMUST00000105925.1 ENSMUST00000105924.1 |
Tspan32
|
tetraspanin 32 |
| chr9_-_70657121 | 0.91 |
ENSMUST00000049031.5
|
Fam63b
|
family with sequence similarity 63, member B |
| chr12_+_87514315 | 0.91 |
ENSMUST00000110152.2
|
Gm8300
|
predicted gene 8300 |
| chr18_-_11051479 | 0.90 |
ENSMUST00000180789.1
|
1010001N08Rik
|
RIKEN cDNA 1010001N08 gene |
| chr17_-_48167187 | 0.90 |
ENSMUST00000053612.6
ENSMUST00000027764.8 |
A530064D06Rik
|
RIKEN cDNA A530064D06 gene |
| chr7_-_43533171 | 0.90 |
ENSMUST00000004728.5
ENSMUST00000039861.5 |
Cd33
|
CD33 antigen |
| chr9_-_53706211 | 0.90 |
ENSMUST00000068449.3
|
Rab39
|
RAB39, member RAS oncogene family |
| chr4_-_137409777 | 0.89 |
ENSMUST00000024200.6
|
Gm13011
|
predicted gene 13011 |
| chr10_-_62507737 | 0.89 |
ENSMUST00000020271.6
|
Srgn
|
serglycin |
| chr8_-_11008458 | 0.89 |
ENSMUST00000040514.6
|
Irs2
|
insulin receptor substrate 2 |
| chr14_-_31640878 | 0.88 |
ENSMUST00000167066.1
ENSMUST00000127204.2 ENSMUST00000022437.8 |
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr18_-_3299537 | 0.88 |
ENSMUST00000129435.1
ENSMUST00000122958.1 |
Crem
|
cAMP responsive element modulator |
| chr7_+_28540863 | 0.87 |
ENSMUST00000119180.2
|
Sycn
|
syncollin |
| chr4_+_40920047 | 0.87 |
ENSMUST00000030122.4
|
Spink4
|
serine peptidase inhibitor, Kazal type 4 |
| chr10_+_43579161 | 0.87 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chrX_+_48623737 | 0.87 |
ENSMUST00000114936.1
|
Slc25a14
|
solute carrier family 25 (mitochondrial carrier, brain), member 14 |
| chr19_-_36736653 | 0.87 |
ENSMUST00000087321.2
|
Ppp1r3c
|
protein phosphatase 1, regulatory (inhibitor) subunit 3C |
| chr1_-_135167606 | 0.85 |
ENSMUST00000027682.8
|
Gpr37l1
|
G protein-coupled receptor 37-like 1 |
| chr6_+_49036518 | 0.84 |
ENSMUST00000031840.7
|
Gpnmb
|
glycoprotein (transmembrane) nmb |
| chr5_+_90772435 | 0.84 |
ENSMUST00000031320.6
|
Pf4
|
platelet factor 4 |
| chr15_+_31568791 | 0.84 |
ENSMUST00000162532.1
|
Cmbl
|
carboxymethylenebutenolidase-like (Pseudomonas) |
| chr16_-_20730544 | 0.83 |
ENSMUST00000076422.5
|
Thpo
|
thrombopoietin |
| chr7_-_126584220 | 0.83 |
ENSMUST00000128970.1
ENSMUST00000116269.2 |
Cln3
|
ceroid lipofuscinosis, neuronal 3, juvenile (Batten, Spielmeyer-Vogt disease) |
| chr16_+_29210108 | 0.83 |
ENSMUST00000162747.1
|
Hrasls
|
HRAS-like suppressor |
| chr19_-_20390944 | 0.83 |
ENSMUST00000025561.7
|
Anxa1
|
annexin A1 |
| chr6_-_38637220 | 0.83 |
ENSMUST00000096030.3
|
Klrg2
|
killer cell lectin-like receptor subfamily G, member 2 |
| chr16_-_88563166 | 0.82 |
ENSMUST00000049697.4
|
Cldn8
|
claudin 8 |
| chr5_-_53707532 | 0.82 |
ENSMUST00000031093.3
|
Cckar
|
cholecystokinin A receptor |
| chr14_+_33923582 | 0.81 |
ENSMUST00000168727.1
|
Gdf10
|
growth differentiation factor 10 |
| chr11_-_3931960 | 0.80 |
ENSMUST00000109990.1
ENSMUST00000020710.4 ENSMUST00000109989.3 ENSMUST00000109991.1 ENSMUST00000109993.2 |
Tcn2
|
transcobalamin 2 |
| chr2_-_168741752 | 0.80 |
ENSMUST00000029060.4
|
Atp9a
|
ATPase, class II, type 9A |
| chr4_-_133263042 | 0.80 |
ENSMUST00000105908.3
ENSMUST00000030674.7 |
Sytl1
|
synaptotagmin-like 1 |
| chr18_+_37496997 | 0.80 |
ENSMUST00000059571.5
|
Pcdhb19
|
protocadherin beta 19 |
| chr19_+_8850785 | 0.79 |
ENSMUST00000096257.2
|
Lrrn4cl
|
LRRN4 C-terminal like |
| chr12_+_36314160 | 0.79 |
ENSMUST00000041407.5
|
Sostdc1
|
sclerostin domain containing 1 |
| chr2_-_152831112 | 0.79 |
ENSMUST00000128172.1
|
Bcl2l1
|
BCL2-like 1 |
| chr7_+_141078188 | 0.79 |
ENSMUST00000106039.2
|
Pkp3
|
plakophilin 3 |
| chr5_+_114146525 | 0.78 |
ENSMUST00000102582.1
|
Acacb
|
acetyl-Coenzyme A carboxylase beta |
| chr7_+_16891755 | 0.78 |
ENSMUST00000078182.4
|
Gng8
|
guanine nucleotide binding protein (G protein), gamma 8 |
| chr2_-_58160495 | 0.78 |
ENSMUST00000028175.6
|
Cytip
|
cytohesin 1 interacting protein |
| chr4_-_131672133 | 0.78 |
ENSMUST00000144212.1
|
Gm12962
|
predicted gene 12962 |
| chr3_+_96219858 | 0.78 |
ENSMUST00000073115.4
|
Hist2h2ab
|
histone cluster 2, H2ab |
| chr3_+_28697901 | 0.77 |
ENSMUST00000029240.7
|
Slc2a2
|
solute carrier family 2 (facilitated glucose transporter), member 2 |
| chr4_+_144893127 | 0.77 |
ENSMUST00000142808.1
|
Dhrs3
|
dehydrogenase/reductase (SDR family) member 3 |
| chr4_+_135120640 | 0.77 |
ENSMUST00000056977.7
|
Runx3
|
runt related transcription factor 3 |
| chr7_-_44306903 | 0.77 |
ENSMUST00000004587.9
|
Clec11a
|
C-type lectin domain family 11, member a |
| chr6_-_87533219 | 0.76 |
ENSMUST00000113637.2
ENSMUST00000071024.6 |
Arhgap25
|
Rho GTPase activating protein 25 |
| chr9_+_54764748 | 0.75 |
ENSMUST00000034830.8
|
Crabp1
|
cellular retinoic acid binding protein I |
| chr2_+_126034967 | 0.75 |
ENSMUST00000110442.1
|
Fgf7
|
fibroblast growth factor 7 |
| chr14_-_64949838 | 0.75 |
ENSMUST00000067843.3
ENSMUST00000176489.1 ENSMUST00000175905.1 ENSMUST00000022544.7 ENSMUST00000175744.1 ENSMUST00000176128.1 |
Hmbox1
|
homeobox containing 1 |
| chr7_-_45092130 | 0.75 |
ENSMUST00000148175.1
|
Rcn3
|
reticulocalbin 3, EF-hand calcium binding domain |
| chr7_-_104353328 | 0.75 |
ENSMUST00000130139.1
ENSMUST00000059037.8 |
Trim12c
|
tripartite motif-containing 12C |
| chr7_-_103827922 | 0.74 |
ENSMUST00000023934.6
ENSMUST00000153218.1 |
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr1_-_72874877 | 0.74 |
ENSMUST00000027377.8
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr1_+_88087802 | 0.74 |
ENSMUST00000113139.1
|
Ugt1a8
|
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr18_-_36695925 | 0.73 |
ENSMUST00000115682.1
|
E230025N22Rik
|
Riken cDNA E230025N22 gene |
| chr7_+_43437073 | 0.73 |
ENSMUST00000070518.2
|
Nkg7
|
natural killer cell group 7 sequence |
| chr17_-_35979237 | 0.73 |
ENSMUST00000165613.2
ENSMUST00000173872.1 |
Prr3
|
proline-rich polypeptide 3 |
| chrX_-_9469288 | 0.73 |
ENSMUST00000015484.3
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr12_-_112860886 | 0.73 |
ENSMUST00000021729.7
|
Gpr132
|
G protein-coupled receptor 132 |
| chr5_+_77004055 | 0.72 |
ENSMUST00000071199.2
|
Arl9
|
ADP-ribosylation factor-like 9 |
| chr13_-_19824234 | 0.72 |
ENSMUST00000065335.2
|
Gpr141
|
G protein-coupled receptor 141 |
| chr3_-_95687846 | 0.72 |
ENSMUST00000015994.3
ENSMUST00000148854.1 ENSMUST00000117782.1 |
Adamtsl4
|
ADAMTS-like 4 |
| chr16_+_29209695 | 0.71 |
ENSMUST00000089824.4
|
Hrasls
|
HRAS-like suppressor |
| chr11_+_114851814 | 0.70 |
ENSMUST00000053361.5
ENSMUST00000021071.7 ENSMUST00000136785.1 |
Gprc5c
|
G protein-coupled receptor, family C, group 5, member C |
| chr1_+_170277376 | 0.70 |
ENSMUST00000179976.1
|
Sh2d1b1
|
SH2 domain protein 1B1 |
| chr17_-_56717681 | 0.70 |
ENSMUST00000164907.1
|
Vmac
|
vimentin-type intermediate filament associated coiled-coil protein |
| chrX_-_97377190 | 0.69 |
ENSMUST00000037353.3
|
Eda2r
|
ectodysplasin A2 receptor |
| chr6_-_82774448 | 0.69 |
ENSMUST00000000642.4
|
Hk2
|
hexokinase 2 |
| chr5_-_24351604 | 0.69 |
ENSMUST00000036092.7
|
Kcnh2
|
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr11_-_121039400 | 0.69 |
ENSMUST00000026159.5
|
Cd7
|
CD7 antigen |
| chr3_+_27317028 | 0.69 |
ENSMUST00000046383.5
ENSMUST00000174840.1 |
Tnfsf10
|
tumor necrosis factor (ligand) superfamily, member 10 |
| chr1_-_172297989 | 0.68 |
ENSMUST00000085913.4
ENSMUST00000097464.2 ENSMUST00000137679.1 |
Atp1a2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide |
| chr11_-_3931789 | 0.68 |
ENSMUST00000109992.1
ENSMUST00000109988.1 |
Tcn2
|
transcobalamin 2 |
| chr11_+_101246960 | 0.68 |
ENSMUST00000107282.3
|
Ramp2
|
receptor (calcitonin) activity modifying protein 2 |
| chr11_-_31824518 | 0.68 |
ENSMUST00000134944.1
|
D630024D03Rik
|
RIKEN cDNA D630024D03 gene |
| chr6_-_122602345 | 0.68 |
ENSMUST00000147760.1
ENSMUST00000112585.1 |
Apobec1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr15_+_103272893 | 0.68 |
ENSMUST00000100162.3
|
Copz1
|
coatomer protein complex, subunit zeta 1 |
| chr4_-_141825997 | 0.67 |
ENSMUST00000102481.3
|
Cela2a
|
chymotrypsin-like elastase family, member 2A |
| chr7_-_141016892 | 0.67 |
ENSMUST00000081924.3
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr15_-_3995708 | 0.67 |
ENSMUST00000046633.8
|
AW549877
|
expressed sequence AW549877 |
| chr10_+_87058043 | 0.67 |
ENSMUST00000169849.1
|
1700113H08Rik
|
RIKEN cDNA 1700113H08 gene |
| chr13_+_38151324 | 0.66 |
ENSMUST00000127906.1
|
Dsp
|
desmoplakin |
| chr8_-_3694167 | 0.66 |
ENSMUST00000005678.4
|
Fcer2a
|
Fc receptor, IgE, low affinity II, alpha polypeptide |
| chr9_-_62510498 | 0.66 |
ENSMUST00000164246.2
|
Coro2b
|
coronin, actin binding protein, 2B |
| chrX_-_102908672 | 0.65 |
ENSMUST00000119624.1
ENSMUST00000033686.1 |
Dmrtc1a
|
DMRT-like family C1a |
| chr3_-_54915867 | 0.65 |
ENSMUST00000070342.3
|
Sertm1
|
serine rich and transmembrane domain containing 1 |
| chr7_-_143074561 | 0.65 |
ENSMUST00000148715.1
|
Trpm5
|
transient receptor potential cation channel, subfamily M, member 5 |
| chr6_-_122602404 | 0.65 |
ENSMUST00000112586.1
|
Apobec1
|
apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 |
| chr19_+_56461629 | 0.65 |
ENSMUST00000178590.1
ENSMUST00000039666.6 |
Plekhs1
|
pleckstrin homology domain containing, family S member 1 |
| chr17_+_75178911 | 0.64 |
ENSMUST00000112514.1
|
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr5_-_105139539 | 0.63 |
ENSMUST00000100961.4
ENSMUST00000031235.6 ENSMUST00000100962.3 |
Gbp9
Gbp8
Gbp4
|
guanylate-binding protein 9 guanylate-binding protein 8 guanylate binding protein 4 |
| chr7_+_143005638 | 0.63 |
ENSMUST00000075172.5
ENSMUST00000105923.1 |
Tspan32
|
tetraspanin 32 |
| chr8_+_21776567 | 0.63 |
ENSMUST00000051017.8
|
Defb1
|
defensin beta 1 |
| chr11_+_49076584 | 0.63 |
ENSMUST00000109202.1
|
Ifi47
|
interferon gamma inducible protein 47 |
| chr1_+_16688405 | 0.63 |
ENSMUST00000026881.4
|
Ly96
|
lymphocyte antigen 96 |
| chr11_-_49113757 | 0.62 |
ENSMUST00000060398.1
|
Olfr1396
|
olfactory receptor 1396 |
| chr3_-_88378699 | 0.62 |
ENSMUST00000098956.2
|
Bglap2
|
bone gamma-carboxyglutamate protein 2 |
| chr14_-_51146757 | 0.62 |
ENSMUST00000080126.2
|
Rnase1
|
ribonuclease, RNase A family, 1 (pancreatic) |
| chr10_+_42502030 | 0.62 |
ENSMUST00000105500.1
ENSMUST00000019939.5 |
Snx3
|
sorting nexin 3 |
| chr17_+_37045980 | 0.62 |
ENSMUST00000174456.1
|
Gabbr1
|
gamma-aminobutyric acid (GABA) B receptor, 1 |
| chrX_-_101269023 | 0.62 |
ENSMUST00000117736.1
|
Gm20489
|
predicted gene 20489 |
| chr2_+_154436437 | 0.62 |
ENSMUST00000109725.1
ENSMUST00000099178.3 ENSMUST00000045270.8 ENSMUST00000109724.1 |
Cbfa2t2
|
core-binding factor, runt domain, alpha subunit 2, translocated to, 2 (human) |
| chr6_-_126645784 | 0.62 |
ENSMUST00000055168.3
|
Kcna1
|
potassium voltage-gated channel, shaker-related subfamily, member 1 |
| chr7_+_30776394 | 0.61 |
ENSMUST00000041703.7
|
Dmkn
|
dermokine |
| chr18_+_65800543 | 0.61 |
ENSMUST00000025394.6
ENSMUST00000153193.1 |
Sec11c
|
SEC11 homolog C (S. cerevisiae) |
| chr9_-_103219823 | 0.61 |
ENSMUST00000168142.1
|
Trf
|
transferrin |
| chr15_-_102722120 | 0.61 |
ENSMUST00000171838.1
|
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr17_-_28560704 | 0.61 |
ENSMUST00000114785.1
ENSMUST00000025062.3 |
Clps
|
colipase, pancreatic |
| chr6_-_126698192 | 0.61 |
ENSMUST00000060972.3
|
Kcna5
|
potassium voltage-gated channel, shaker-related subfamily, member 5 |
| chr17_+_75178797 | 0.61 |
ENSMUST00000112516.1
ENSMUST00000135447.1 |
Ltbp1
|
latent transforming growth factor beta binding protein 1 |
| chr15_-_97844254 | 0.60 |
ENSMUST00000119670.1
ENSMUST00000116409.2 |
Hdac7
|
histone deacetylase 7 |
| chr19_+_3935186 | 0.60 |
ENSMUST00000162708.1
ENSMUST00000165711.1 |
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr1_+_125676969 | 0.60 |
ENSMUST00000027581.6
|
Gpr39
|
G protein-coupled receptor 39 |
| chr7_-_140082246 | 0.60 |
ENSMUST00000166758.2
|
Caly
|
calcyon neuron-specific vesicular protein |
| chr7_+_30169861 | 0.60 |
ENSMUST00000085668.4
|
Gm5113
|
predicted gene 5113 |
| chr4_+_152199805 | 0.60 |
ENSMUST00000105652.2
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr12_+_10390756 | 0.60 |
ENSMUST00000020947.5
|
Rdh14
|
retinol dehydrogenase 14 (all-trans and 9-cis) |
| chr3_+_28263205 | 0.59 |
ENSMUST00000159236.2
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr6_+_56017489 | 0.59 |
ENSMUST00000052827.4
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chrX_-_97377150 | 0.59 |
ENSMUST00000113832.1
|
Eda2r
|
ectodysplasin A2 receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx2_Bcl11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.5 | 4.5 | GO:2000295 | regulation of hydrogen peroxide catabolic process(GO:2000295) |
| 1.0 | 2.9 | GO:0071661 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.8 | 2.3 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.5 | 3.3 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.4 | 1.3 | GO:0061033 | secretion by lung epithelial cell involved in lung growth(GO:0061033) |
| 0.4 | 1.1 | GO:0035483 | gastric emptying(GO:0035483) |
| 0.4 | 1.1 | GO:2000642 | negative regulation of early endosome to late endosome transport(GO:2000642) |
| 0.4 | 1.8 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.3 | 1.0 | GO:0071846 | actin filament debranching(GO:0071846) |
| 0.3 | 1.3 | GO:0090365 | regulation of mRNA modification(GO:0090365) |
| 0.3 | 1.0 | GO:2000864 | androgen catabolic process(GO:0006710) estradiol secretion(GO:0035938) regulation of estradiol secretion(GO:2000864) |
| 0.3 | 2.5 | GO:0015074 | DNA integration(GO:0015074) |
| 0.3 | 0.9 | GO:0001997 | positive regulation of the force of heart contraction by epinephrine-norepinephrine(GO:0001997) positive regulation of the force of heart contraction by chemical signal(GO:0003099) |
| 0.3 | 5.1 | GO:0051923 | sulfation(GO:0051923) |
| 0.3 | 2.6 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.3 | 0.9 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) |
| 0.3 | 1.1 | GO:0042414 | epinephrine metabolic process(GO:0042414) |
| 0.3 | 0.8 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.3 | 0.8 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.3 | 0.8 | GO:0050975 | sensory perception of touch(GO:0050975) |
| 0.3 | 1.4 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.3 | 2.4 | GO:0014052 | regulation of gamma-aminobutyric acid secretion(GO:0014052) |
| 0.3 | 1.3 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.3 | 0.8 | GO:2000016 | negative regulation of determination of dorsal identity(GO:2000016) |
| 0.3 | 0.8 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) malonyl-CoA metabolic process(GO:2001293) |
| 0.2 | 1.5 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.2 | 0.9 | GO:0061010 | gall bladder development(GO:0061010) |
| 0.2 | 1.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.2 | 0.7 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.2 | 2.9 | GO:1900118 | negative regulation of execution phase of apoptosis(GO:1900118) |
| 0.2 | 1.1 | GO:0031437 | regulation of mRNA cleavage(GO:0031437) negative regulation of mRNA cleavage(GO:0031438) regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904720) negative regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904721) |
| 0.2 | 0.4 | GO:1904252 | negative regulation of bile acid biosynthetic process(GO:0070858) negative regulation of bile acid metabolic process(GO:1904252) |
| 0.2 | 1.1 | GO:0035752 | lysosomal lumen pH elevation(GO:0035752) |
| 0.2 | 0.8 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.2 | 1.6 | GO:1901387 | positive regulation of voltage-gated calcium channel activity(GO:1901387) |
| 0.2 | 1.2 | GO:0090383 | phagosome acidification(GO:0090383) |
| 0.2 | 0.8 | GO:0016115 | terpenoid catabolic process(GO:0016115) |
| 0.2 | 0.4 | GO:0015866 | ADP transport(GO:0015866) |
| 0.2 | 0.6 | GO:0002355 | detection of tumor cell(GO:0002355) |
| 0.2 | 1.1 | GO:0060214 | endocardium formation(GO:0060214) |
| 0.2 | 0.6 | GO:1903660 | negative regulation of NAD(P)H oxidase activity(GO:0033861) pancreatic stellate cell proliferation(GO:0072343) regulation of complement-dependent cytotoxicity(GO:1903659) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of pancreatic stellate cell proliferation(GO:2000229) |
| 0.2 | 0.7 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.2 | 0.6 | GO:0002305 | gamma-delta intraepithelial T cell differentiation(GO:0002304) CD8-positive, gamma-delta intraepithelial T cell differentiation(GO:0002305) |
| 0.2 | 1.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.2 | 0.9 | GO:0019244 | lactate biosynthetic process from pyruvate(GO:0019244) |
| 0.2 | 0.7 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.2 | 0.7 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.2 | 0.5 | GO:0061090 | positive regulation of sequestering of zinc ion(GO:0061090) |
| 0.2 | 1.2 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.2 | 1.6 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.2 | 0.7 | GO:0035795 | negative regulation of mitochondrial membrane permeability(GO:0035795) |
| 0.2 | 0.7 | GO:1902303 | negative regulation of potassium ion export(GO:1902303) |
| 0.2 | 1.0 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.2 | 1.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.2 | 0.8 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.2 | 2.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.2 | 0.8 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.2 | 1.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 1.0 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.2 | 0.5 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.2 | 0.6 | GO:2000321 | positive regulation of T-helper 17 cell differentiation(GO:2000321) |
| 0.2 | 0.3 | GO:0036466 | synaptic vesicle recycling via endosome(GO:0036466) |
| 0.2 | 1.8 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.2 | 0.8 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.2 | 0.6 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.2 | 0.5 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.1 | 0.6 | GO:1900535 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) fatty-acyl-CoA catabolic process(GO:0036115) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.1 | 0.4 | GO:0021837 | motogenic signaling involved in postnatal olfactory bulb interneuron migration(GO:0021837) positive regulation of mitotic cell cycle DNA replication(GO:1903465) |
| 0.1 | 0.6 | GO:0002838 | negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) |
| 0.1 | 1.0 | GO:0061621 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.1 | 0.4 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.1 | 0.6 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 1.1 | GO:0042737 | drug catabolic process(GO:0042737) |
| 0.1 | 0.4 | GO:0009804 | coumarin metabolic process(GO:0009804) phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.8 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.8 | GO:0072675 | osteoclast fusion(GO:0072675) |
| 0.1 | 0.5 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 2.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.1 | 0.4 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.1 | 0.4 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.1 | 0.4 | GO:2000402 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 0.6 | GO:0086043 | bundle of His cell to Purkinje myocyte signaling(GO:0086028) bundle of His cell action potential(GO:0086043) |
| 0.1 | 0.6 | GO:0034154 | toll-like receptor 7 signaling pathway(GO:0034154) |
| 0.1 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 1.0 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.1 | 0.5 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.1 | 1.1 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.1 | 1.1 | GO:0045779 | negative regulation of bone resorption(GO:0045779) |
| 0.1 | 1.3 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 0.4 | GO:1900004 | regulation of serine-type endopeptidase activity(GO:1900003) negative regulation of serine-type endopeptidase activity(GO:1900004) regulation of serine-type peptidase activity(GO:1902571) negative regulation of serine-type peptidase activity(GO:1902572) |
| 0.1 | 0.3 | GO:0072244 | metanephric glomerular epithelium development(GO:0072244) |
| 0.1 | 0.3 | GO:0061402 | glycerol biosynthetic process(GO:0006114) positive regulation of transcription from RNA polymerase II promoter in response to acidic pH(GO:0061402) |
| 0.1 | 0.5 | GO:0009597 | detection of virus(GO:0009597) |
| 0.1 | 0.7 | GO:0010968 | regulation of microtubule nucleation(GO:0010968) |
| 0.1 | 0.3 | GO:0071211 | protein targeting to vacuole involved in autophagy(GO:0071211) |
| 0.1 | 0.4 | GO:0097026 | dendritic cell dendrite assembly(GO:0097026) |
| 0.1 | 0.4 | GO:0033368 | protein localization to secretory granule(GO:0033366) protein localization to mast cell secretory granule(GO:0033367) protease localization to mast cell secretory granule(GO:0033368) maintenance of protein location in mast cell secretory granule(GO:0033370) T cell secretory granule organization(GO:0033371) maintenance of protease location in mast cell secretory granule(GO:0033373) protein localization to T cell secretory granule(GO:0033374) protease localization to T cell secretory granule(GO:0033375) maintenance of protein location in T cell secretory granule(GO:0033377) maintenance of protease location in T cell secretory granule(GO:0033379) granzyme B localization to T cell secretory granule(GO:0033380) maintenance of granzyme B location in T cell secretory granule(GO:0033382) |
| 0.1 | 0.6 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.1 | 0.4 | GO:1904048 | negative regulation of synaptic vesicle recycling(GO:1903422) regulation of spontaneous neurotransmitter secretion(GO:1904048) |
| 0.1 | 0.5 | GO:0051611 | regulation of serotonin uptake(GO:0051611) |
| 0.1 | 0.4 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.7 | GO:0097647 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 0.3 | GO:0071895 | negative regulation of interleukin-13 production(GO:0032696) odontoblast differentiation(GO:0071895) |
| 0.1 | 0.3 | GO:2000847 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.1 | 1.2 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.1 | 0.3 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) |
| 0.1 | 0.7 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.6 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.1 | 1.0 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.6 | GO:0031666 | positive regulation of lipopolysaccharide-mediated signaling pathway(GO:0031666) |
| 0.1 | 0.3 | GO:0033685 | negative regulation of luteinizing hormone secretion(GO:0033685) |
| 0.1 | 1.0 | GO:0072378 | blood coagulation, fibrin clot formation(GO:0072378) |
| 0.1 | 0.6 | GO:0097460 | positive regulation of oligodendrocyte progenitor proliferation(GO:0070447) ferrous iron import into cell(GO:0097460) ferrous iron import across plasma membrane(GO:0098707) |
| 0.1 | 0.3 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 1.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.1 | 2.2 | GO:1903831 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.1 | 0.4 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.1 | 0.2 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.1 | 1.2 | GO:0071361 | cellular response to ethanol(GO:0071361) |
| 0.1 | 0.3 | GO:0061187 | regulation of chromatin silencing at rDNA(GO:0061187) negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.5 | GO:0035624 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 | 0.2 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.1 | 0.6 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 0.9 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.1 | 0.4 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.1 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.4 | GO:0002291 | T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:0002291) |
| 0.1 | 0.3 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.5 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.7 | GO:0002925 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) |
| 0.1 | 0.4 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.1 | 0.3 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.1 | 0.4 | GO:0035022 | positive regulation of Rac protein signal transduction(GO:0035022) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.8 | GO:1902916 | positive regulation of protein polyubiquitination(GO:1902916) |
| 0.1 | 0.5 | GO:0042148 | strand invasion(GO:0042148) |
| 0.1 | 0.3 | GO:0044330 | canonical Wnt signaling pathway involved in positive regulation of wound healing(GO:0044330) lactic acid secretion(GO:0046722) regulation of metanephric cap mesenchymal cell proliferation(GO:0090095) positive regulation of metanephric cap mesenchymal cell proliferation(GO:0090096) |
| 0.1 | 0.7 | GO:0052697 | flavonoid metabolic process(GO:0009812) flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.1 | 1.9 | GO:0035313 | wound healing, spreading of epidermal cells(GO:0035313) |
| 0.1 | 0.6 | GO:0061366 | behavioral response to chemical pain(GO:0061366) behavioral response to formalin induced pain(GO:0061368) |
| 0.1 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.2 | GO:0031284 | positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.1 | 0.1 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 1.2 | GO:0030149 | sphingolipid catabolic process(GO:0030149) |
| 0.1 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.1 | 0.8 | GO:0021940 | positive regulation of cerebellar granule cell precursor proliferation(GO:0021940) |
| 0.1 | 0.2 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.1 | 0.2 | GO:0061428 | peptidyl-aspartic acid hydroxylation(GO:0042264) negative regulation of transcription from RNA polymerase II promoter in response to hypoxia(GO:0061428) |
| 0.1 | 0.3 | GO:0061589 | calcium activated phosphatidylserine scrambling(GO:0061589) |
| 0.1 | 0.1 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.1 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.1 | 0.2 | GO:0018211 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.1 | 0.3 | GO:0060398 | regulation of growth hormone receptor signaling pathway(GO:0060398) |
| 0.1 | 0.3 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.3 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.1 | 0.4 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.1 | 0.2 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.1 | 0.8 | GO:0042448 | progesterone metabolic process(GO:0042448) |
| 0.1 | 0.2 | GO:0070460 | thyroid-stimulating hormone secretion(GO:0070460) metanephric nephron tubule formation(GO:0072289) |
| 0.1 | 0.8 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.1 | 0.3 | GO:0071985 | multivesicular body sorting pathway(GO:0071985) |
| 0.1 | 0.9 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.2 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 1.0 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.0 | 1.2 | GO:0032967 | positive regulation of collagen biosynthetic process(GO:0032967) |
| 0.0 | 0.1 | GO:0003127 | detection of nodal flow(GO:0003127) detection of endogenous stimulus(GO:0009726) |
| 0.0 | 1.4 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.1 | GO:0036092 | phosphatidylinositol-3-phosphate biosynthetic process(GO:0036092) |
| 0.0 | 0.6 | GO:0035278 | miRNA mediated inhibition of translation(GO:0035278) negative regulation of translation, ncRNA-mediated(GO:0040033) regulation of translation, ncRNA-mediated(GO:0045974) regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060211) positive regulation of nuclear-transcribed mRNA poly(A) tail shortening(GO:0060213) |
| 0.0 | 0.3 | GO:2001015 | negative regulation of skeletal muscle cell differentiation(GO:2001015) |
| 0.0 | 0.8 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:2000721 | positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.0 | 1.8 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.6 | GO:0001865 | NK T cell differentiation(GO:0001865) |
| 0.0 | 0.7 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.6 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.3 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.2 | GO:0032468 | cellular manganese ion homeostasis(GO:0030026) Golgi calcium ion homeostasis(GO:0032468) manganese ion homeostasis(GO:0055071) |
| 0.0 | 0.3 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0010693 | negative regulation of alkaline phosphatase activity(GO:0010693) |
| 0.0 | 0.2 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
| 0.0 | 0.1 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.6 | GO:0060575 | intestinal epithelial cell differentiation(GO:0060575) |
| 0.0 | 0.1 | GO:0032829 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.3 | GO:0002756 | MyD88-independent toll-like receptor signaling pathway(GO:0002756) |
| 0.0 | 0.2 | GO:0002414 | immunoglobulin transcytosis in epithelial cells(GO:0002414) |
| 0.0 | 1.8 | GO:0006687 | glycosphingolipid metabolic process(GO:0006687) |
| 0.0 | 0.2 | GO:1903564 | regulation of protein localization to cilium(GO:1903564) |
| 0.0 | 0.8 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 1.6 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.3 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.4 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:2000501 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.0 | 1.5 | GO:0035458 | cellular response to interferon-beta(GO:0035458) |
| 0.0 | 0.1 | GO:0090126 | protein complex assembly involved in synapse maturation(GO:0090126) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.1 | GO:0010752 | regulation of cGMP-mediated signaling(GO:0010752) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.4 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.2 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.3 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.3 | GO:1901724 | miRNA catabolic process(GO:0010587) positive regulation of cell proliferation involved in kidney development(GO:1901724) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.3 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) negative regulation of oocyte development(GO:0060283) negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.2 | GO:1904659 | glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0021666 | rhombomere formation(GO:0021594) rhombomere 3 formation(GO:0021660) rhombomere 5 morphogenesis(GO:0021664) rhombomere 5 formation(GO:0021666) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.4 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0097117 | guanylate kinase-associated protein clustering(GO:0097117) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.6 | GO:1902187 | negative regulation of viral release from host cell(GO:1902187) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.2 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 3.5 | GO:2000045 | regulation of G1/S transition of mitotic cell cycle(GO:2000045) |
| 0.0 | 0.3 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.7 | GO:0048266 | behavioral response to pain(GO:0048266) |
| 0.0 | 0.9 | GO:0044062 | regulation of excretion(GO:0044062) |
| 0.0 | 0.3 | GO:0021780 | oligodendrocyte cell fate specification(GO:0021778) oligodendrocyte cell fate commitment(GO:0021779) glial cell fate specification(GO:0021780) |
| 0.0 | 0.8 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.8 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0001802 | antibody-dependent cellular cytotoxicity(GO:0001788) type III hypersensitivity(GO:0001802) regulation of type III hypersensitivity(GO:0001803) positive regulation of type III hypersensitivity(GO:0001805) |
| 0.0 | 0.4 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.3 | GO:0035456 | response to interferon-beta(GO:0035456) |
| 0.0 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.4 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.4 | GO:0090141 | positive regulation of mitochondrial fission(GO:0090141) |
| 0.0 | 0.2 | GO:0000042 | protein targeting to Golgi(GO:0000042) |
| 0.0 | 0.7 | GO:0090200 | positive regulation of release of cytochrome c from mitochondria(GO:0090200) |
| 0.0 | 0.1 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.0 | 0.3 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.5 | GO:0010663 | positive regulation of striated muscle cell apoptotic process(GO:0010663) positive regulation of cardiac muscle cell apoptotic process(GO:0010666) |
| 0.0 | 0.1 | GO:0090649 | rRNA export from nucleus(GO:0006407) response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.6 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:0061635 | regulation of protein complex stability(GO:0061635) |
| 0.0 | 0.6 | GO:0032094 | response to food(GO:0032094) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.3 | GO:0071378 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.9 | GO:0010972 | negative regulation of G2/M transition of mitotic cell cycle(GO:0010972) |
| 0.0 | 0.3 | GO:0000083 | regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0000083) |
| 0.0 | 1.1 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.7 | GO:0051482 | positive regulation of cytosolic calcium ion concentration involved in phospholipase C-activating G-protein coupled signaling pathway(GO:0051482) |
| 0.0 | 0.7 | GO:0032012 | ARF protein signal transduction(GO:0032011) regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0003376 | sphingosine-1-phosphate signaling pathway(GO:0003376) |
| 0.0 | 0.4 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.3 | GO:1990118 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.2 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.0 | 0.1 | GO:0070560 | negative regulation of inositol phosphate biosynthetic process(GO:0010920) protein secretion by platelet(GO:0070560) |
| 0.0 | 0.1 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.3 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.1 | GO:0018992 | germ-line sex determination(GO:0018992) |
| 0.0 | 0.8 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.3 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0097033 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.7 | GO:0045668 | negative regulation of osteoblast differentiation(GO:0045668) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.4 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 2.0 | GO:0010921 | regulation of phosphatase activity(GO:0010921) |
| 0.0 | 0.1 | GO:1900095 | regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.3 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.3 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.0 | 0.4 | GO:0010667 | negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.5 | GO:0009409 | response to cold(GO:0009409) |
| 0.0 | 0.0 | GO:0098501 | polynucleotide dephosphorylation(GO:0098501) |
| 0.0 | 0.1 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.2 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.7 | GO:0033120 | positive regulation of RNA splicing(GO:0033120) |
| 0.0 | 0.0 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.2 | GO:0010574 | regulation of vascular endothelial growth factor production(GO:0010574) |
| 0.0 | 0.2 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.1 | GO:0051095 | regulation of helicase activity(GO:0051095) positive regulation of helicase activity(GO:0051096) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.3 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0043144 | snoRNA processing(GO:0043144) |
| 0.0 | 0.2 | GO:0036295 | cellular response to increased oxygen levels(GO:0036295) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0050913 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) sensory perception of bitter taste(GO:0050913) |
| 0.0 | 0.1 | GO:0033683 | nucleotide-excision repair, DNA incision(GO:0033683) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.3 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.2 | GO:0050909 | sensory perception of taste(GO:0050909) |
| 0.0 | 0.7 | GO:0006757 | glycolytic process(GO:0006096) ATP generation from ADP(GO:0006757) |
| 0.0 | 0.2 | GO:0097345 | mitochondrial outer membrane permeabilization(GO:0097345) |
| 0.0 | 0.1 | GO:0035887 | aortic smooth muscle cell differentiation(GO:0035887) |
| 0.0 | 0.8 | GO:0030032 | lamellipodium assembly(GO:0030032) |
| 0.0 | 0.1 | GO:0043312 | neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.2 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.6 | 1.8 | GO:0070442 | integrin alphaIIb-beta3 complex(GO:0070442) |
| 0.4 | 1.2 | GO:0038045 | large latent transforming growth factor-beta complex(GO:0038045) |
| 0.3 | 1.3 | GO:0044194 | cytolytic granule(GO:0044194) |
| 0.2 | 2.3 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.2 | 0.8 | GO:0005914 | spot adherens junction(GO:0005914) |
| 0.2 | 0.6 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.2 | 2.1 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.2 | 2.5 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.2 | 0.7 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 0.8 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.2 | 0.5 | GO:0005608 | laminin-3 complex(GO:0005608) |
| 0.1 | 1.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.1 | 1.8 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.1 | 0.9 | GO:1990462 | omegasome(GO:1990462) |
| 0.1 | 1.2 | GO:0002177 | manchette(GO:0002177) |
| 0.1 | 1.8 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 1.4 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 5.7 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.1 | 0.7 | GO:1903439 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 1.1 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.1 | 1.5 | GO:0032009 | early phagosome(GO:0032009) |
| 0.1 | 0.3 | GO:0098830 | presynaptic endosome(GO:0098830) |
| 0.1 | 0.8 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.1 | 1.5 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.3 | GO:0031088 | platelet dense granule membrane(GO:0031088) intrinsic component of autophagosome membrane(GO:0097636) integral component of autophagosome membrane(GO:0097637) |
| 0.1 | 0.4 | GO:0034687 | integrin alphaL-beta2 complex(GO:0034687) |
| 0.1 | 0.7 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 1.5 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.9 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.7 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.1 | 0.9 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.1 | 4.1 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.1 | 0.6 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.1 | 0.2 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.1 | 0.3 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.1 | 0.3 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 0.3 | GO:0043293 | apoptosome(GO:0043293) |
| 0.1 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.4 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.1 | 0.8 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 1.8 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.1 | 0.4 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.1 | 0.2 | GO:0044753 | amphisome(GO:0044753) |
| 0.1 | 1.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.1 | 0.4 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) DNA ligase IV complex(GO:0032807) |
| 0.1 | 0.7 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.1 | 0.2 | GO:0030312 | external encapsulating structure(GO:0030312) tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.3 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 1.1 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.2 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.7 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 1.4 | GO:0045335 | phagocytic vesicle(GO:0045335) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.2 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.6 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 1.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 1.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.3 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.5 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.8 | GO:0042470 | melanosome(GO:0042470) pigment granule(GO:0048770) |
| 0.0 | 0.8 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.6 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.5 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.3 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.7 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 1.0 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.2 | GO:0030137 | COPI-coated vesicle(GO:0030137) |
| 0.0 | 2.2 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.0 | 0.4 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.3 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) |
| 0.0 | 0.2 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 1.7 | GO:0031225 | anchored component of membrane(GO:0031225) |
| 0.0 | 4.4 | GO:0009897 | external side of plasma membrane(GO:0009897) |
| 0.0 | 0.2 | GO:0000974 | Prp19 complex(GO:0000974) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 0.2 | GO:0060091 | stereocilium tip(GO:0032426) kinocilium(GO:0060091) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 1.1 | GO:0005795 | Golgi stack(GO:0005795) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0005798 | Golgi-associated vesicle(GO:0005798) |
| 0.0 | 0.1 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 16.5 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.2 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.0 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) |
| 0.0 | 0.7 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.6 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.3 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 1.1 | 5.6 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.5 | 3.3 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.4 | 1.1 | GO:0005118 | sevenless binding(GO:0005118) |
| 0.4 | 1.5 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.4 | 2.1 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.3 | 0.9 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.3 | 2.5 | GO:0019158 | fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.3 | 2.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.3 | 0.9 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.3 | 1.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.3 | 1.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.3 | 0.8 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.2 | 2.0 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.2 | 0.7 | GO:0005152 | interleukin-1, Type II receptor binding(GO:0005151) interleukin-1 receptor antagonist activity(GO:0005152) interleukin-1 Type I receptor antagonist activity(GO:0045352) interleukin-1 Type II receptor antagonist activity(GO:0045353) |
| 0.2 | 1.3 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.2 | 0.9 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.2 | 1.8 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.2 | 0.8 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.2 | 0.8 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.2 | 1.6 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.2 | 0.8 | GO:0004008 | copper-exporting ATPase activity(GO:0004008) copper-transporting ATPase activity(GO:0043682) |
| 0.2 | 0.6 | GO:0031752 | D3 dopamine receptor binding(GO:0031750) D5 dopamine receptor binding(GO:0031752) |
| 0.2 | 1.1 | GO:0060698 | endoribonuclease inhibitor activity(GO:0060698) |
| 0.2 | 0.5 | GO:0008147 | structural constituent of bone(GO:0008147) |
| 0.2 | 0.7 | GO:0019976 | interleukin-2 binding(GO:0019976) |
| 0.2 | 0.5 | GO:0004771 | sterol esterase activity(GO:0004771) retinyl-palmitate esterase activity(GO:0050253) |
| 0.2 | 1.4 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.2 | 2.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.5 | GO:0038100 | nodal binding(GO:0038100) |
| 0.2 | 0.7 | GO:0019863 | IgE binding(GO:0019863) |
| 0.2 | 1.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.2 | 0.5 | GO:0051379 | alpha2-adrenergic receptor activity(GO:0004938) epinephrine binding(GO:0051379) |
| 0.2 | 0.5 | GO:0070615 | nucleosome-dependent ATPase activity(GO:0070615) |
| 0.2 | 2.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.2 | 1.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.2 | 0.8 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.2 | 0.8 | GO:0055056 | dehydroascorbic acid transporter activity(GO:0033300) D-glucose transmembrane transporter activity(GO:0055056) |
| 0.2 | 1.5 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.2 | 0.5 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.1 | 0.6 | GO:0019970 | interleukin-11 receptor activity(GO:0004921) interleukin-11 binding(GO:0019970) |
| 0.1 | 1.0 | GO:0001851 | complement component C3b binding(GO:0001851) |
| 0.1 | 0.6 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.7 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.1 | 0.4 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.1 | 0.5 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 1.0 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.1 | 0.5 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.9 | GO:0004118 | cGMP-stimulated cyclic-nucleotide phosphodiesterase activity(GO:0004118) |
| 0.1 | 0.8 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.1 | 0.4 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.4 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 1.4 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.9 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.4 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.4 | GO:0071077 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.1 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.2 | GO:0034875 | testosterone 16-alpha-hydroxylase activity(GO:0008390) oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.1 | 0.6 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.7 | GO:0097643 | amylin receptor activity(GO:0097643) |
| 0.1 | 0.5 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.1 | 0.8 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.9 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.1 | 3.3 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.1 | 1.4 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.1 | 1.0 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.1 | 0.9 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.1 | 0.9 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.1 | 2.2 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.4 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.6 | GO:0016892 | endoribonuclease activity, producing 3'-phosphomonoesters(GO:0016892) |
| 0.1 | 0.6 | GO:0015091 | ferric iron transmembrane transporter activity(GO:0015091) trivalent inorganic cation transmembrane transporter activity(GO:0072510) |
| 0.1 | 0.4 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.1 | 0.4 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.1 | 0.3 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.8 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 1.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.1 | 1.3 | GO:0070330 | aromatase activity(GO:0070330) |
| 0.1 | 0.8 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 0.8 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.8 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.1 | 0.5 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.1 | 0.6 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.1 | 1.0 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.1 | 0.4 | GO:0097001 | ceramide binding(GO:0097001) |
| 0.1 | 0.6 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.1 | 0.3 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.8 | GO:0016918 | retinal binding(GO:0016918) |
| 0.1 | 0.3 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.4 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.1 | 0.4 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.1 | 0.6 | GO:0008106 | alcohol dehydrogenase (NADP+) activity(GO:0008106) |
| 0.1 | 0.3 | GO:0042134 | rRNA primary transcript binding(GO:0042134) |
| 0.1 | 0.2 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.1 | 1.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.1 | 1.4 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.1 | 0.3 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.1 | 2.3 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.1 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.1 | 0.3 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.3 | GO:0004033 | aldo-keto reductase (NADP) activity(GO:0004033) |
| 0.1 | 0.3 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.1 | 1.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.1 | 0.6 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.1 | GO:0008176 | tRNA (guanine-N7-)-methyltransferase activity(GO:0008176) |
| 0.0 | 0.2 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
| 0.0 | 0.3 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 1.6 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.2 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.4 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.5 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.5 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.6 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 1.5 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.3 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.4 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.6 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.6 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.5 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.2 | GO:0015410 | manganese-transporting ATPase activity(GO:0015410) |
| 0.0 | 0.1 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.7 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.4 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.5 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 1.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.2 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 1.4 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.8 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.5 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.2 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.2 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.8 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.0 | GO:0015020 | glucuronosyltransferase activity(GO:0015020) |
| 0.0 | 0.3 | GO:0019865 | immunoglobulin binding(GO:0019865) |
| 0.0 | 0.1 | GO:0005344 | catalase activity(GO:0004096) oxygen transporter activity(GO:0005344) |
| 0.0 | 0.3 | GO:0051022 | Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.5 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0016997 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 7.5 | GO:0030246 | carbohydrate binding(GO:0030246) |
| 0.0 | 0.1 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.4 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 4.4 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.1 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.1 | GO:0097200 | cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:0097200) |
| 0.0 | 0.3 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.5 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.3 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.7 | GO:0005227 | calcium activated cation channel activity(GO:0005227) |
| 0.0 | 0.9 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.4 | GO:0015299 | solute:proton antiporter activity(GO:0015299) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0002151 | G-quadruplex RNA binding(GO:0002151) |
| 0.0 | 0.1 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.2 | GO:0051429 | corticotropin-releasing hormone receptor binding(GO:0051429) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) single guanine insertion binding(GO:0032142) |
| 0.0 | 0.9 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.3 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.7 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0030160 | GKAP/Homer scaffold activity(GO:0030160) |
| 0.0 | 0.4 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.4 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.5 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.4 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.3 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.4 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.0 | GO:0001032 | RNA polymerase III type 3 promoter DNA binding(GO:0001032) |
| 0.0 | 0.3 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 1.2 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.1 | GO:0005436 | sodium:phosphate symporter activity(GO:0005436) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.1 | GO:0004620 | phospholipase activity(GO:0004620) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.5 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0052629 | phosphatidylinositol-3,5-bisphosphate 3-phosphatase activity(GO:0052629) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 1.0 | GO:0019888 | protein phosphatase regulator activity(GO:0019888) |
| 0.0 | 1.1 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0019208 | phosphatase regulator activity(GO:0019208) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.5 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 1.1 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 1.1 | GO:0005178 | integrin binding(GO:0005178) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.3 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 2.5 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.1 | 3.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.2 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.9 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 1.1 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.4 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.9 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.5 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.9 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 1.2 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 3.0 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.6 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.9 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 3.2 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 9.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 1.6 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.6 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.6 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.5 | PID AMB2 NEUTROPHILS PATHWAY | amb2 Integrin signaling |
| 0.0 | 1.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.7 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
| 0.0 | 1.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.6 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 1.6 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.0 | 0.5 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.4 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.4 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 2.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.1 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.1 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 5.1 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.2 | 0.6 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.2 | 8.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.2 | 2.0 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.2 | 3.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.1 | 1.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.1 | 0.4 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.1 | 1.0 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.1 | 1.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.8 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.1 | 1.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.1 | 0.5 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.1 | 0.8 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.1 | 3.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.1 | 1.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.1 | 0.7 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.1 | 0.7 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.1 | 1.8 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.1 | 0.8 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.1 | 0.6 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.1 | 0.7 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.1 | 1.1 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 2.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.6 | REACTOME TRAF6 MEDIATED INDUCTION OF TAK1 COMPLEX | Genes involved in TRAF6 mediated induction of TAK1 complex |
| 0.0 | 0.4 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 1.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.6 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.9 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 1.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.8 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 1.5 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 1.0 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.4 | REACTOME POST TRANSLATIONAL MODIFICATION SYNTHESIS OF GPI ANCHORED PROTEINS | Genes involved in Post-translational modification: synthesis of GPI-anchored proteins |
| 0.0 | 0.9 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.4 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.3 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 1.1 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.6 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.6 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.4 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.9 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 1.2 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.8 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.4 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.1 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 1.2 | REACTOME VOLTAGE GATED POTASSIUM CHANNELS | Genes involved in Voltage gated Potassium channels |
| 0.0 | 0.5 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.5 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.4 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 1.1 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 3.3 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.3 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.1 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 1.0 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |