Project
2D miR_HR1_12
Navigation
Downloads
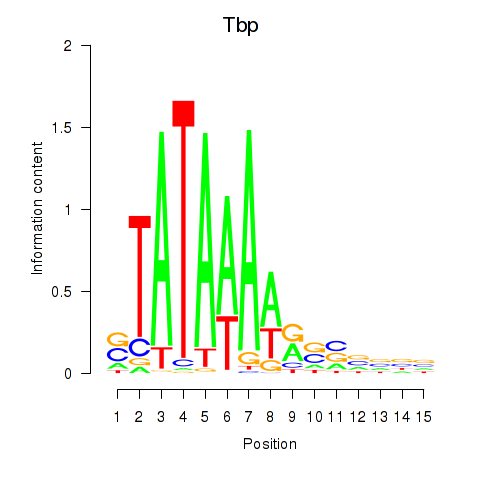
Results for Tbp
Z-value: 1.89
Transcription factors associated with Tbp
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Tbp
|
ENSMUSG00000014767.10 | TATA box binding protein |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Tbp | mm10_v2_chr17_+_15499888_15499960 | -0.31 | 3.3e-01 | Click! |
Activity profile of Tbp motif
Sorted Z-values of Tbp motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_41377604 | 3.28 |
ENSMUST00000096003.5
|
Prss3
|
protease, serine, 3 |
| chr6_+_78370877 | 3.16 |
ENSMUST00000096904.3
|
Reg3b
|
regenerating islet-derived 3 beta |
| chr3_-_90695706 | 2.82 |
ENSMUST00000069960.5
ENSMUST00000117167.1 |
S100a9
|
S100 calcium binding protein A9 (calgranulin B) |
| chr7_-_30924169 | 2.71 |
ENSMUST00000074671.6
|
Hamp2
|
hepcidin antimicrobial peptide 2 |
| chr19_-_3414464 | 2.59 |
ENSMUST00000025842.6
|
Gal
|
galanin |
| chr6_-_41314700 | 2.45 |
ENSMUST00000064324.5
|
Try5
|
trypsin 5 |
| chr3_-_106167564 | 2.16 |
ENSMUST00000063062.8
|
Chi3l3
|
chitinase 3-like 3 |
| chr6_+_41392356 | 2.08 |
ENSMUST00000049079.7
|
Gm5771
|
predicted gene 5771 |
| chr16_+_48842552 | 2.05 |
ENSMUST00000023329.4
|
Retnla
|
resistin like alpha |
| chr11_+_82101836 | 1.96 |
ENSMUST00000000194.3
|
Ccl12
|
chemokine (C-C motif) ligand 12 |
| chr5_-_135573036 | 1.95 |
ENSMUST00000004936.6
|
Ccl24
|
chemokine (C-C motif) ligand 24 |
| chr3_-_20275659 | 1.88 |
ENSMUST00000011607.5
|
Cpb1
|
carboxypeptidase B1 (tissue) |
| chr4_+_148000722 | 1.83 |
ENSMUST00000103230.4
|
Nppa
|
natriuretic peptide type A |
| chr19_+_58670358 | 1.75 |
ENSMUST00000057270.7
|
Pnlip
|
pancreatic lipase |
| chr8_-_105933832 | 1.71 |
ENSMUST00000034368.6
|
Ctrl
|
chymotrypsin-like |
| chr15_+_99591028 | 1.71 |
ENSMUST00000169082.1
|
Aqp5
|
aquaporin 5 |
| chr17_-_57228003 | 1.61 |
ENSMUST00000177046.1
ENSMUST00000024988.8 |
C3
|
complement component 3 |
| chr9_+_7445822 | 1.56 |
ENSMUST00000034497.6
|
Mmp3
|
matrix metallopeptidase 3 |
| chr6_-_41035501 | 1.54 |
ENSMUST00000031931.5
|
2210010C04Rik
|
RIKEN cDNA 2210010C04 gene |
| chr6_+_78425973 | 1.53 |
ENSMUST00000079926.5
|
Reg1
|
regenerating islet-derived 1 |
| chr6_-_78378851 | 1.46 |
ENSMUST00000089667.1
ENSMUST00000167492.1 |
Reg3d
|
regenerating islet-derived 3 delta |
| chr6_+_41354105 | 1.46 |
ENSMUST00000072103.5
|
Try10
|
trypsin 10 |
| chr2_+_164403194 | 1.42 |
ENSMUST00000017151.1
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr3_-_20242173 | 1.38 |
ENSMUST00000001921.1
|
Cpa3
|
carboxypeptidase A3, mast cell |
| chr11_+_94936224 | 1.31 |
ENSMUST00000001547.7
|
Col1a1
|
collagen, type I, alpha 1 |
| chr7_+_35119285 | 1.27 |
ENSMUST00000042985.9
|
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr4_-_103026709 | 1.26 |
ENSMUST00000084382.5
ENSMUST00000106869.2 |
Insl5
|
insulin-like 5 |
| chr5_+_90903864 | 1.23 |
ENSMUST00000075433.6
|
Cxcl2
|
chemokine (C-X-C motif) ligand 2 |
| chr11_-_83592981 | 1.20 |
ENSMUST00000019071.3
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr1_+_164048214 | 1.12 |
ENSMUST00000027874.5
|
Sele
|
selectin, endothelial cell |
| chr12_+_85473883 | 1.10 |
ENSMUST00000021674.6
|
Fos
|
FBJ osteosarcoma oncogene |
| chr18_+_65873478 | 1.08 |
ENSMUST00000025395.8
ENSMUST00000173530.1 |
Grp
|
gastrin releasing peptide |
| chr2_+_91256144 | 1.04 |
ENSMUST00000154959.1
ENSMUST00000059566.4 |
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr14_+_103046977 | 1.04 |
ENSMUST00000022722.6
|
Irg1
|
immunoresponsive gene 1 |
| chr16_+_36184082 | 1.04 |
ENSMUST00000114858.1
|
Gm5483
|
predicted gene 5483 |
| chr7_-_16259185 | 1.03 |
ENSMUST00000168818.1
|
C5ar1
|
complement component 5a receptor 1 |
| chr1_-_144249134 | 1.01 |
ENSMUST00000172388.1
|
Rgs1
|
regulator of G-protein signaling 1 |
| chr7_+_131032061 | 1.01 |
ENSMUST00000084509.3
|
Dmbt1
|
deleted in malignant brain tumors 1 |
| chr14_+_67745229 | 1.00 |
ENSMUST00000111095.2
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr11_-_83578496 | 0.99 |
ENSMUST00000019266.5
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr5_+_90759299 | 0.96 |
ENSMUST00000031318.4
|
Cxcl5
|
chemokine (C-X-C motif) ligand 5 |
| chr7_-_141266415 | 0.95 |
ENSMUST00000106023.1
ENSMUST00000097952.2 ENSMUST00000026571.4 |
Irf7
|
interferon regulatory factor 7 |
| chr2_-_164356507 | 0.93 |
ENSMUST00000109367.3
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr3_+_87948666 | 0.89 |
ENSMUST00000005019.5
|
Crabp2
|
cellular retinoic acid binding protein II |
| chr17_-_14694223 | 0.86 |
ENSMUST00000170872.1
|
Thbs2
|
thrombospondin 2 |
| chr11_-_100261021 | 0.85 |
ENSMUST00000080893.6
|
Krt17
|
keratin 17 |
| chr11_-_83530505 | 0.83 |
ENSMUST00000035938.2
|
Ccl5
|
chemokine (C-C motif) ligand 5 |
| chr6_-_67535783 | 0.81 |
ENSMUST00000058178.4
|
Tacstd2
|
tumor-associated calcium signal transducer 2 |
| chr1_+_131019843 | 0.80 |
ENSMUST00000016673.5
|
Il10
|
interleukin 10 |
| chr2_-_164356067 | 0.78 |
ENSMUST00000165980.1
|
Slpi
|
secretory leukocyte peptidase inhibitor |
| chr8_+_58912257 | 0.78 |
ENSMUST00000160055.1
|
BC030500
|
cDNA sequence BC030500 |
| chr2_+_13573927 | 0.78 |
ENSMUST00000141365.1
ENSMUST00000028062.2 |
Vim
|
vimentin |
| chr14_+_65968483 | 0.77 |
ENSMUST00000022616.6
|
Clu
|
clusterin |
| chr9_+_57697612 | 0.73 |
ENSMUST00000034865.4
|
Cyp1a1
|
cytochrome P450, family 1, subfamily a, polypeptide 1 |
| chr15_-_5063741 | 0.71 |
ENSMUST00000110689.3
|
C7
|
complement component 7 |
| chr11_+_120949053 | 0.69 |
ENSMUST00000154187.1
ENSMUST00000100130.3 ENSMUST00000129473.1 ENSMUST00000168579.1 |
Slc16a3
|
solute carrier family 16 (monocarboxylic acid transporters), member 3 |
| chr7_-_103813913 | 0.68 |
ENSMUST00000098192.3
|
Hbb-bt
|
hemoglobin, beta adult t chain |
| chr16_-_46120238 | 0.68 |
ENSMUST00000023336.9
|
Cd96
|
CD96 antigen |
| chr3_+_138217814 | 0.67 |
ENSMUST00000090171.5
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_-_89294608 | 0.66 |
ENSMUST00000107131.1
|
Cdkn2a
|
cyclin-dependent kinase inhibitor 2A |
| chr6_+_135065651 | 0.65 |
ENSMUST00000050104.7
|
Gprc5a
|
G protein-coupled receptor, family C, group 5, member A |
| chr7_-_137314394 | 0.65 |
ENSMUST00000168203.1
ENSMUST00000106118.2 ENSMUST00000169486.2 ENSMUST00000033378.5 |
Ebf3
|
early B cell factor 3 |
| chr2_+_153918391 | 0.64 |
ENSMUST00000109760.1
|
Bpifb3
|
BPI fold containing family B, member 3 |
| chr7_-_103827922 | 0.63 |
ENSMUST00000023934.6
ENSMUST00000153218.1 |
Hbb-bs
|
hemoglobin, beta adult s chain |
| chr6_-_4747157 | 0.63 |
ENSMUST00000090686.4
ENSMUST00000115579.1 ENSMUST00000115577.2 ENSMUST00000101677.3 ENSMUST00000004750.8 |
Sgce
|
sarcoglycan, epsilon |
| chr17_+_80944611 | 0.63 |
ENSMUST00000025092.4
|
Tmem178
|
transmembrane protein 178 |
| chr6_+_122707489 | 0.62 |
ENSMUST00000112581.1
ENSMUST00000112580.1 ENSMUST00000012540.4 |
Nanog
|
Nanog homeobox |
| chr13_-_23934156 | 0.62 |
ENSMUST00000052776.2
|
Hist1h2ba
|
histone cluster 1, H2ba |
| chr4_+_119637704 | 0.61 |
ENSMUST00000024015.2
|
Guca2a
|
guanylate cyclase activator 2a (guanylin) |
| chr5_+_114923234 | 0.59 |
ENSMUST00000031540.4
ENSMUST00000112143.3 |
Oasl1
|
2'-5' oligoadenylate synthetase-like 1 |
| chr11_+_94565039 | 0.57 |
ENSMUST00000040418.8
|
Chad
|
chondroadherin |
| chr14_-_51773578 | 0.56 |
ENSMUST00000073860.5
|
Ang4
|
angiogenin, ribonuclease A family, member 4 |
| chr11_-_99024179 | 0.56 |
ENSMUST00000068031.7
|
Top2a
|
topoisomerase (DNA) II alpha |
| chr6_-_4747019 | 0.54 |
ENSMUST00000126151.1
ENSMUST00000133306.1 ENSMUST00000123907.1 |
Sgce
|
sarcoglycan, epsilon |
| chr9_+_7347374 | 0.54 |
ENSMUST00000065079.5
ENSMUST00000005950.5 |
Mmp12
|
matrix metallopeptidase 12 |
| chr14_-_79771305 | 0.53 |
ENSMUST00000039568.5
|
Pcdh8
|
protocadherin 8 |
| chr9_-_67832325 | 0.53 |
ENSMUST00000054500.5
|
C2cd4a
|
C2 calcium-dependent domain containing 4A |
| chr5_+_135887988 | 0.52 |
ENSMUST00000111155.1
|
Hspb1
|
heat shock protein 1 |
| chr8_+_34115030 | 0.52 |
ENSMUST00000095345.3
|
Mboat4
|
membrane bound O-acyltransferase domain containing 4 |
| chr17_-_33955658 | 0.51 |
ENSMUST00000174609.2
ENSMUST00000008812.7 |
Rps18
|
ribosomal protein S18 |
| chr7_-_19023538 | 0.51 |
ENSMUST00000036018.5
|
Foxa3
|
forkhead box A3 |
| chr6_+_122626410 | 0.51 |
ENSMUST00000049644.2
|
Dppa3
|
developmental pluripotency-associated 3 |
| chr17_-_33824346 | 0.51 |
ENSMUST00000173879.1
ENSMUST00000166693.2 ENSMUST00000173019.1 ENSMUST00000087342.6 ENSMUST00000173844.1 |
Rps28
|
ribosomal protein S28 |
| chr6_+_90550789 | 0.51 |
ENSMUST00000130418.1
ENSMUST00000032175.8 |
Aldh1l1
|
aldehyde dehydrogenase 1 family, member L1 |
| chr9_+_7272514 | 0.50 |
ENSMUST00000015394.8
|
Mmp13
|
matrix metallopeptidase 13 |
| chr2_+_91255954 | 0.50 |
ENSMUST00000134699.1
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr6_-_113719880 | 0.50 |
ENSMUST00000064993.5
|
Ghrl
|
ghrelin |
| chr11_-_99993992 | 0.48 |
ENSMUST00000105049.1
|
Krtap17-1
|
keratin associated protein 17-1 |
| chr7_-_30612731 | 0.48 |
ENSMUST00000006476.4
|
Upk1a
|
uroplakin 1A |
| chr6_+_42286676 | 0.47 |
ENSMUST00000031894.6
|
Clcn1
|
chloride channel 1 |
| chr10_+_88091070 | 0.46 |
ENSMUST00000048621.7
|
Pmch
|
pro-melanin-concentrating hormone |
| chr1_+_120006980 | 0.45 |
ENSMUST00000072886.4
|
Sctr
|
secretin receptor |
| chr15_+_101473472 | 0.45 |
ENSMUST00000088049.3
|
Krt86
|
keratin 86 |
| chr11_-_69948145 | 0.45 |
ENSMUST00000179298.1
ENSMUST00000018710.6 ENSMUST00000135437.1 ENSMUST00000141837.2 ENSMUST00000142500.1 |
Slc2a4
|
solute carrier family 2 (facilitated glucose transporter), member 4 |
| chr5_+_135887905 | 0.44 |
ENSMUST00000005077.6
|
Hspb1
|
heat shock protein 1 |
| chr11_-_102897123 | 0.44 |
ENSMUST00000067444.3
|
Gfap
|
glial fibrillary acidic protein |
| chr4_+_41966058 | 0.44 |
ENSMUST00000108026.2
|
Gm20938
|
predicted gene, 20938 |
| chr10_-_93589621 | 0.44 |
ENSMUST00000020203.6
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr4_+_42318334 | 0.44 |
ENSMUST00000178192.1
|
Gm21598
|
predicted gene, 21598 |
| chr13_-_113663670 | 0.43 |
ENSMUST00000054650.4
|
Hspb3
|
heat shock protein 3 |
| chr9_+_30942541 | 0.43 |
ENSMUST00000068135.6
|
Adamts8
|
a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 8 |
| chr12_-_40199315 | 0.43 |
ENSMUST00000095760.2
|
Lsmem1
|
leucine-rich single-pass membrane protein 1 |
| chr13_-_34130345 | 0.42 |
ENSMUST00000075774.3
|
Tubb2b
|
tubulin, beta 2B class IIB |
| chr11_-_33163072 | 0.41 |
ENSMUST00000093201.6
ENSMUST00000101375.4 ENSMUST00000109354.3 ENSMUST00000075641.3 |
Npm1
|
nucleophosmin 1 |
| chr4_-_42853888 | 0.41 |
ENSMUST00000107979.1
|
Gm12429
|
predicted gene 12429 |
| chr4_-_41098174 | 0.41 |
ENSMUST00000055327.7
|
Aqp3
|
aquaporin 3 |
| chr6_+_4747306 | 0.41 |
ENSMUST00000175823.1
ENSMUST00000176204.1 ENSMUST00000166678.1 |
Peg10
|
paternally expressed 10 |
| chr11_-_115514374 | 0.41 |
ENSMUST00000021083.6
|
Hn1
|
hematological and neurological expressed sequence 1 |
| chr6_+_17694167 | 0.40 |
ENSMUST00000115418.1
|
St7
|
suppression of tumorigenicity 7 |
| chr15_-_101869695 | 0.40 |
ENSMUST00000087996.5
|
Krt77
|
keratin 77 |
| chr3_+_103832562 | 0.40 |
ENSMUST00000062945.5
|
Bcl2l15
|
BCLl2-like 15 |
| chr12_+_55598917 | 0.40 |
ENSMUST00000051857.3
|
Insm2
|
insulinoma-associated 2 |
| chr6_-_72958097 | 0.40 |
ENSMUST00000114049.1
|
Tmsb10
|
thymosin, beta 10 |
| chr15_-_80083374 | 0.40 |
ENSMUST00000081650.7
|
Rpl3
|
ribosomal protein L3 |
| chr17_-_31129602 | 0.39 |
ENSMUST00000024827.4
|
Tff3
|
trefoil factor 3, intestinal |
| chr6_+_17693905 | 0.39 |
ENSMUST00000115420.1
|
St7
|
suppression of tumorigenicity 7 |
| chr19_-_34255325 | 0.39 |
ENSMUST00000039631.8
|
Acta2
|
actin, alpha 2, smooth muscle, aorta |
| chr6_-_125494754 | 0.38 |
ENSMUST00000032492.8
|
Cd9
|
CD9 antigen |
| chr11_-_99438143 | 0.38 |
ENSMUST00000017743.2
|
Krt20
|
keratin 20 |
| chr6_+_24528144 | 0.38 |
ENSMUST00000031696.3
|
Asb15
|
ankyrin repeat and SOCS box-containing 15 |
| chr10_+_21994666 | 0.38 |
ENSMUST00000020145.5
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr2_-_164857542 | 0.38 |
ENSMUST00000109316.1
ENSMUST00000156255.1 ENSMUST00000128110.1 ENSMUST00000109317.3 |
Pltp
|
phospholipid transfer protein |
| chr10_+_52391606 | 0.37 |
ENSMUST00000067085.4
|
Nepn
|
nephrocan |
| chr13_+_23695791 | 0.37 |
ENSMUST00000041052.2
|
Hist1h1t
|
histone cluster 1, H1t |
| chr7_-_4812351 | 0.36 |
ENSMUST00000079496.7
|
Ube2s
|
ubiquitin-conjugating enzyme E2S |
| chr4_-_119658781 | 0.36 |
ENSMUST00000106309.2
ENSMUST00000044426.7 |
Guca2b
|
guanylate cyclase activator 2b (retina) |
| chr9_+_55541148 | 0.35 |
ENSMUST00000034869.4
|
Isl2
|
insulin related protein 2 (islet 2) |
| chr6_+_17694005 | 0.35 |
ENSMUST00000081635.6
ENSMUST00000052113.5 |
St7
|
suppression of tumorigenicity 7 |
| chr4_-_3835595 | 0.35 |
ENSMUST00000138502.1
|
Rps20
|
ribosomal protein S20 |
| chr6_-_113600645 | 0.34 |
ENSMUST00000035870.4
|
Fancd2os
|
Fancd2 opposite strand |
| chr2_-_93957040 | 0.34 |
ENSMUST00000148314.2
|
Gm13889
|
predicted gene 13889 |
| chr10_+_43579161 | 0.34 |
ENSMUST00000058714.8
|
Cd24a
|
CD24a antigen |
| chr19_-_40994133 | 0.34 |
ENSMUST00000117695.1
|
Blnk
|
B cell linker |
| chr17_+_24720063 | 0.34 |
ENSMUST00000170715.1
ENSMUST00000054289.6 ENSMUST00000146867.1 |
Rps2
|
ribosomal protein S2 |
| chr14_+_68083853 | 0.33 |
ENSMUST00000022639.7
|
Nefl
|
neurofilament, light polypeptide |
| chr9_+_21015960 | 0.33 |
ENSMUST00000086399.4
|
Icam1
|
intercellular adhesion molecule 1 |
| chr2_+_27165233 | 0.33 |
ENSMUST00000000910.6
|
Dbh
|
dopamine beta hydroxylase |
| chr6_-_72958465 | 0.33 |
ENSMUST00000114050.1
|
Tmsb10
|
thymosin, beta 10 |
| chr3_+_14863495 | 0.33 |
ENSMUST00000029076.4
|
Car3
|
carbonic anhydrase 3 |
| chr10_-_128626464 | 0.32 |
ENSMUST00000026420.5
|
Rps26
|
ribosomal protein S26 |
| chr18_+_34624621 | 0.32 |
ENSMUST00000167161.1
|
Kif20a
|
kinesin family member 20A |
| chr10_+_90576872 | 0.31 |
ENSMUST00000182550.1
ENSMUST00000099364.5 |
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr6_+_48895243 | 0.31 |
ENSMUST00000031835.7
|
Aoc1
|
amine oxidase, copper-containing 1 |
| chr11_-_120713725 | 0.30 |
ENSMUST00000106154.1
ENSMUST00000106155.3 ENSMUST00000055424.6 ENSMUST00000026137.7 |
Stra13
|
stimulated by retinoic acid 13 |
| chr2_+_122298898 | 0.29 |
ENSMUST00000028656.1
|
Duoxa2
|
dual oxidase maturation factor 2 |
| chr18_+_34625009 | 0.29 |
ENSMUST00000166044.1
|
Kif20a
|
kinesin family member 20A |
| chr6_+_17693965 | 0.28 |
ENSMUST00000115419.1
|
St7
|
suppression of tumorigenicity 7 |
| chr1_-_36273425 | 0.28 |
ENSMUST00000056946.6
|
Neurl3
|
neuralized homolog 3 homolog (Drosophila) |
| chr3_-_92485886 | 0.28 |
ENSMUST00000054599.7
|
Sprr1a
|
small proline-rich protein 1A |
| chr7_-_45526146 | 0.27 |
ENSMUST00000167273.1
ENSMUST00000042105.8 |
Ppp1r15a
|
protein phosphatase 1, regulatory (inhibitor) subunit 15A |
| chr17_+_33824591 | 0.27 |
ENSMUST00000048249.6
|
Ndufa7
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7 (B14.5a) |
| chr15_-_32244632 | 0.25 |
ENSMUST00000181536.1
|
0610007N19Rik
|
RIKEN cDNA 0610007N19 |
| chr6_-_112696604 | 0.25 |
ENSMUST00000113182.1
ENSMUST00000113180.1 ENSMUST00000068487.5 ENSMUST00000077088.4 |
Rad18
|
RAD18 homolog (S. cerevisiae) |
| chr5_+_115559505 | 0.25 |
ENSMUST00000156359.1
ENSMUST00000152976.1 |
Rplp0
|
ribosomal protein, large, P0 |
| chr9_+_21835506 | 0.25 |
ENSMUST00000058777.6
|
Gm6484
|
predicted gene 6484 |
| chr2_-_36104060 | 0.25 |
ENSMUST00000112961.3
ENSMUST00000112966.3 |
Lhx6
|
LIM homeobox protein 6 |
| chr15_+_80255184 | 0.25 |
ENSMUST00000109605.3
|
Atf4
|
activating transcription factor 4 |
| chr3_-_145649970 | 0.25 |
ENSMUST00000029846.3
|
Cyr61
|
cysteine rich protein 61 |
| chr5_-_99978914 | 0.25 |
ENSMUST00000112939.3
ENSMUST00000171786.1 ENSMUST00000072750.6 ENSMUST00000019128.8 ENSMUST00000172361.1 |
Hnrnpd
|
heterogeneous nuclear ribonucleoprotein D |
| chr5_+_115559467 | 0.24 |
ENSMUST00000086519.5
|
Rplp0
|
ribosomal protein, large, P0 |
| chr10_+_96616998 | 0.24 |
ENSMUST00000038377.7
|
Btg1
|
B cell translocation gene 1, anti-proliferative |
| chr10_-_69212996 | 0.24 |
ENSMUST00000170048.1
|
A930033H14Rik
|
RIKEN cDNA A930033H14 gene |
| chr19_+_9982694 | 0.24 |
ENSMUST00000025563.6
|
Fth1
|
ferritin heavy chain 1 |
| chr2_+_167062934 | 0.22 |
ENSMUST00000125674.1
|
1500012F01Rik
|
RIKEN cDNA 1500012F01 gene |
| chr4_-_132353605 | 0.21 |
ENSMUST00000155129.1
|
Rcc1
|
regulator of chromosome condensation 1 |
| chr16_+_90220742 | 0.21 |
ENSMUST00000023707.9
|
Sod1
|
superoxide dismutase 1, soluble |
| chr17_+_23726336 | 0.21 |
ENSMUST00000024701.7
|
Pkmyt1
|
protein kinase, membrane associated tyrosine/threonine 1 |
| chr19_+_34583528 | 0.21 |
ENSMUST00000102825.3
|
Ifit3
|
interferon-induced protein with tetratricopeptide repeats 3 |
| chr3_-_106406090 | 0.21 |
ENSMUST00000029510.7
|
BC051070
|
cDNA sequence BC051070 |
| chr9_+_121777607 | 0.21 |
ENSMUST00000098272.2
|
Klhl40
|
kelch-like 40 |
| chr11_+_62551676 | 0.21 |
ENSMUST00000136938.1
|
Ubb
|
ubiquitin B |
| chr7_+_28810886 | 0.21 |
ENSMUST00000038572.8
|
Hnrnpl
|
heterogeneous nuclear ribonucleoprotein L |
| chr8_-_13254068 | 0.21 |
ENSMUST00000168498.1
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr6_-_87981482 | 0.20 |
ENSMUST00000056403.5
|
H1fx
|
H1 histone family, member X |
| chr7_+_18987518 | 0.20 |
ENSMUST00000063563.7
|
Nanos2
|
nanos homolog 2 (Drosophila) |
| chr17_+_29032664 | 0.20 |
ENSMUST00000130216.1
|
Srsf3
|
serine/arginine-rich splicing factor 3 |
| chr19_+_8723478 | 0.20 |
ENSMUST00000180819.1
ENSMUST00000181422.1 |
Snhg1
|
small nucleolar RNA host gene (non-protein coding) 1 |
| chr17_+_80207460 | 0.20 |
ENSMUST00000134652.1
|
Ttc39d
|
tetratricopeptide repeat domain 39D |
| chr8_-_13254154 | 0.20 |
ENSMUST00000033825.4
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr17_-_56117577 | 0.20 |
ENSMUST00000019808.5
|
Plin5
|
perilipin 5 |
| chr8_-_13254096 | 0.19 |
ENSMUST00000171619.1
|
Adprhl1
|
ADP-ribosylhydrolase like 1 |
| chr8_-_117082449 | 0.19 |
ENSMUST00000098375.4
ENSMUST00000109093.2 |
Pkd1l2
|
polycystic kidney disease 1 like 2 |
| chr4_-_133967296 | 0.19 |
ENSMUST00000105893.1
|
Hmgn2
|
high mobility group nucleosomal binding domain 2 |
| chr11_+_120598421 | 0.19 |
ENSMUST00000026128.3
|
Anapc11
|
anaphase promoting complex subunit 11 |
| chrX_+_74270812 | 0.19 |
ENSMUST00000008826.7
ENSMUST00000151702.1 ENSMUST00000074085.5 ENSMUST00000135690.1 |
Rpl10
|
ribosomal protein L10 |
| chr10_+_79988584 | 0.18 |
ENSMUST00000004784.4
ENSMUST00000105374.1 |
Cnn2
|
calponin 2 |
| chr11_+_119942763 | 0.18 |
ENSMUST00000026436.3
ENSMUST00000106231.1 ENSMUST00000075180.5 ENSMUST00000103021.3 ENSMUST00000106233.1 |
Baiap2
|
brain-specific angiogenesis inhibitor 1-associated protein 2 |
| chr12_+_108851122 | 0.18 |
ENSMUST00000057026.8
|
Slc25a47
|
solute carrier family 25, member 47 |
| chr1_-_74893109 | 0.18 |
ENSMUST00000006721.2
|
Cryba2
|
crystallin, beta A2 |
| chr8_+_84969824 | 0.18 |
ENSMUST00000125893.1
|
Prdx2
|
peroxiredoxin 2 |
| chr3_-_144849301 | 0.18 |
ENSMUST00000159989.1
|
Clca4
|
chloride channel calcium activated 4 |
| chr2_-_51149100 | 0.18 |
ENSMUST00000154545.1
ENSMUST00000017288.2 |
Rnd3
|
Rho family GTPase 3 |
| chr17_-_56117265 | 0.17 |
ENSMUST00000113072.2
|
Plin5
|
perilipin 5 |
| chr4_+_136143497 | 0.17 |
ENSMUST00000008016.2
|
Id3
|
inhibitor of DNA binding 3 |
| chr16_-_91728599 | 0.17 |
ENSMUST00000122254.1
|
Cryzl1
|
crystallin, zeta (quinone reductase)-like 1 |
| chr15_-_76232045 | 0.17 |
ENSMUST00000167754.1
|
Plec
|
plectin |
| chr6_-_72439549 | 0.17 |
ENSMUST00000059472.8
|
Mat2a
|
methionine adenosyltransferase II, alpha |
| chr2_+_152736244 | 0.17 |
ENSMUST00000038368.8
ENSMUST00000109824.1 |
Id1
|
inhibitor of DNA binding 1 |
| chr19_-_31764963 | 0.17 |
ENSMUST00000182685.1
|
Prkg1
|
protein kinase, cGMP-dependent, type I |
| chr4_+_123282778 | 0.16 |
ENSMUST00000106243.1
ENSMUST00000106241.1 ENSMUST00000080178.6 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Tbp
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.0 | 3.1 | GO:0051464 | positive regulation of cortisol secretion(GO:0051464) |
| 0.9 | 2.8 | GO:0070488 | neutrophil aggregation(GO:0070488) |
| 0.7 | 2.8 | GO:0034757 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) |
| 0.6 | 1.9 | GO:2000418 | positive regulation of eosinophil migration(GO:2000418) |
| 0.6 | 2.8 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.4 | 1.8 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.4 | 3.2 | GO:1903207 | neuron death in response to hydrogen peroxide(GO:0036476) regulation of hydrogen peroxide-induced neuron death(GO:1903207) negative regulation of hydrogen peroxide-induced neuron death(GO:1903208) |
| 0.3 | 1.6 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.3 | 1.0 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.3 | 0.8 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.3 | 1.1 | GO:0036343 | psychomotor behavior(GO:0036343) positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.3 | 0.8 | GO:0060300 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) regulation of cytokine activity(GO:0060300) |
| 0.3 | 0.5 | GO:0070368 | positive regulation of hepatocyte differentiation(GO:0070368) |
| 0.2 | 0.7 | GO:0018894 | coumarin metabolic process(GO:0009804) dibenzo-p-dioxin metabolic process(GO:0018894) |
| 0.2 | 1.7 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) |
| 0.2 | 0.7 | GO:0002370 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.2 | 1.7 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.2 | 0.6 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 0.4 | GO:0090650 | response to oxygen-glucose deprivation(GO:0090649) cellular response to oxygen-glucose deprivation(GO:0090650) |
| 0.2 | 1.0 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.2 | 1.3 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.2 | 0.6 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.2 | 1.3 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.2 | 1.0 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.2 | 0.5 | GO:1901535 | regulation of DNA demethylation(GO:1901535) negative regulation of DNA demethylation(GO:1901536) |
| 0.2 | 1.7 | GO:0010727 | negative regulation of hydrogen peroxide metabolic process(GO:0010727) |
| 0.2 | 0.5 | GO:0042560 | 10-formyltetrahydrofolate catabolic process(GO:0009258) folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.2 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.2 | 0.3 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.2 | 0.8 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.2 | 0.5 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.1 | 0.4 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) rRNA export from nucleus(GO:0006407) |
| 0.1 | 1.6 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.1 | 0.8 | GO:1902998 | regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.5 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.1 | 0.4 | GO:0035519 | protein K29-linked ubiquitination(GO:0035519) |
| 0.1 | 0.4 | GO:0021524 | visceral motor neuron differentiation(GO:0021524) |
| 0.1 | 1.2 | GO:0002002 | regulation of angiotensin levels in blood(GO:0002002) |
| 0.1 | 1.3 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.1 | 0.3 | GO:0002842 | positive regulation of T cell mediated immune response to tumor cell(GO:0002842) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.1 | 0.3 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 1.0 | GO:0099640 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) axo-dendritic protein transport(GO:0099640) |
| 0.1 | 0.9 | GO:0051798 | positive regulation of hair follicle development(GO:0051798) |
| 0.1 | 0.5 | GO:0060054 | positive regulation of epithelial cell proliferation involved in wound healing(GO:0060054) |
| 0.1 | 0.3 | GO:1902310 | positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.6 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.2 | GO:0003278 | apoptotic process involved in heart morphogenesis(GO:0003278) |
| 0.1 | 0.2 | GO:1901355 | response to rapamycin(GO:1901355) |
| 0.1 | 0.6 | GO:0060346 | bone trabecula formation(GO:0060346) |
| 0.1 | 0.6 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.1 | 0.4 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.1 | 0.4 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.1 | 1.5 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.1 | 3.5 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.1 | 0.5 | GO:1902669 | positive regulation of axon guidance(GO:1902669) |
| 0.1 | 0.1 | GO:2000852 | corticosterone secretion(GO:0035934) regulation of corticosterone secretion(GO:2000852) |
| 0.1 | 2.2 | GO:0019731 | antibacterial humoral response(GO:0019731) |
| 0.1 | 0.9 | GO:0061365 | positive regulation of triglyceride lipase activity(GO:0061365) |
| 0.1 | 0.7 | GO:0035879 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.1 | 0.4 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.1 | 0.4 | GO:0030913 | paranodal junction assembly(GO:0030913) |
| 0.1 | 0.3 | GO:0036091 | positive regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0036091) |
| 0.1 | 7.6 | GO:0007586 | digestion(GO:0007586) |
| 0.1 | 0.9 | GO:0042573 | retinoic acid metabolic process(GO:0042573) |
| 0.1 | 0.4 | GO:0010624 | regulation of Schwann cell proliferation(GO:0010624) |
| 0.1 | 0.3 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.1 | 1.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) |
| 0.1 | 0.3 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.1 | 0.2 | GO:0009992 | cellular water homeostasis(GO:0009992) |
| 0.1 | 1.0 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.1 | 0.2 | GO:0098528 | skeletal muscle fiber differentiation(GO:0098528) |
| 0.1 | 0.3 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 1.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.5 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.3 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.2 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.3 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.2 | GO:1902606 | regulation of large conductance calcium-activated potassium channel activity(GO:1902606) positive regulation of large conductance calcium-activated potassium channel activity(GO:1902608) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.8 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.6 | GO:0000920 | cell separation after cytokinesis(GO:0000920) |
| 0.0 | 0.1 | GO:0031509 | telomeric heterochromatin assembly(GO:0031509) negative regulation of chromosome condensation(GO:1902340) |
| 0.0 | 0.3 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.3 | GO:0071280 | cellular response to copper ion(GO:0071280) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.3 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.7 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0002485 | antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway(GO:0002484) antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent(GO:0002485) |
| 0.0 | 0.1 | GO:0021888 | hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) |
| 0.0 | 0.9 | GO:0071353 | cellular response to interleukin-4(GO:0071353) |
| 0.0 | 0.6 | GO:0045109 | intermediate filament organization(GO:0045109) |
| 0.0 | 0.3 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.1 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.2 | GO:0044838 | cell quiescence(GO:0044838) |
| 0.0 | 0.2 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.3 | GO:0051984 | positive regulation of chromosome segregation(GO:0051984) |
| 0.0 | 0.2 | GO:1902416 | positive regulation of mRNA binding(GO:1902416) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:2000510 | positive regulation of dendritic cell chemotaxis(GO:2000510) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.5 | GO:0044381 | glucose import in response to insulin stimulus(GO:0044381) |
| 0.0 | 0.3 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.0 | GO:0070384 | Harderian gland development(GO:0070384) |
| 0.0 | 0.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.4 | GO:0000387 | spliceosomal snRNP assembly(GO:0000387) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) |
| 0.0 | 0.0 | GO:0035524 | regulation of amino acid import(GO:0010958) proline transmembrane transport(GO:0035524) glycine import(GO:0036233) |
| 0.0 | 0.3 | GO:0045671 | negative regulation of osteoclast differentiation(GO:0045671) |
| 0.0 | 0.0 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.8 | GO:0030512 | negative regulation of transforming growth factor beta receptor signaling pathway(GO:0030512) |
| 0.0 | 0.2 | GO:0051457 | maintenance of protein location in nucleus(GO:0051457) |
| 0.0 | 0.4 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0005584 | collagen type I trimer(GO:0005584) |
| 0.4 | 1.3 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.3 | 1.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.3 | 1.8 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.2 | 1.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.2 | 5.7 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.2 | 1.2 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 1.0 | GO:0001652 | granular component(GO:0001652) |
| 0.1 | 1.0 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.8 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.1 | 0.3 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.1 | 0.3 | GO:0060205 | secretory granule lumen(GO:0034774) cytoplasmic membrane-bounded vesicle lumen(GO:0060205) |
| 0.1 | 1.0 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.1 | 0.4 | GO:0097450 | astrocyte end-foot(GO:0097450) |
| 0.1 | 0.7 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.4 | GO:0005683 | U7 snRNP(GO:0005683) pICln-Sm protein complex(GO:0034715) |
| 0.1 | 0.4 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.1 | 0.5 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.3 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.1 | 0.8 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.3 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 2.0 | GO:0022627 | cytosolic small ribosomal subunit(GO:0022627) |
| 0.0 | 0.5 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.9 | GO:0031091 | platelet alpha granule(GO:0031091) |
| 0.0 | 0.4 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.5 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.7 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 1.4 | GO:0005882 | intermediate filament(GO:0005882) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 40.8 | GO:0005615 | extracellular space(GO:0005615) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.3 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 1.7 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 1.1 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.1 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.2 | GO:0045120 | pronucleus(GO:0045120) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.6 | 2.6 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.5 | 2.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.4 | 2.8 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) RAGE receptor binding(GO:0050786) |
| 0.3 | 1.3 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.3 | 0.8 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.3 | 1.0 | GO:0001847 | opsonin receptor activity(GO:0001847) |
| 0.2 | 0.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.2 | 2.3 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.2 | 1.0 | GO:0010853 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.2 | 0.5 | GO:0016155 | formyltetrahydrofolate dehydrogenase activity(GO:0016155) |
| 0.2 | 0.7 | GO:0055105 | ubiquitin-protein transferase inhibitor activity(GO:0055105) |
| 0.2 | 2.1 | GO:0015250 | water channel activity(GO:0015250) |
| 0.2 | 0.8 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.6 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.1 | 1.5 | GO:0019855 | calcium channel inhibitor activity(GO:0019855) |
| 0.1 | 1.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.1 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 0.6 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.1 | 0.9 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.7 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.1 | 0.6 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.1 | 3.0 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.1 | 1.8 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.1 | 1.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 2.1 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.1 | 1.8 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.1 | 0.5 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.1 | 0.3 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.1 | 0.7 | GO:0016679 | oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) |
| 0.1 | 1.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.1 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 6.3 | GO:0005179 | hormone activity(GO:0005179) |
| 0.1 | 10.6 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.1 | 0.6 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.1 | 0.1 | GO:0005372 | water transmembrane transporter activity(GO:0005372) |
| 0.1 | 1.7 | GO:0008236 | serine-type peptidase activity(GO:0008236) |
| 0.1 | 0.9 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.4 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.4 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.1 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.0 | 0.2 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) gamma-aminobutyric acid transmembrane transporter activity(GO:0015185) |
| 0.0 | 0.3 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0000403 | Y-form DNA binding(GO:0000403) |
| 0.0 | 0.3 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.2 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.2 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.8 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.4 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.0 | 0.2 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 1.0 | GO:0044183 | protein binding involved in protein folding(GO:0044183) |
| 0.0 | 0.3 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 2.8 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 1.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.3 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.3 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.3 | GO:0035925 | mRNA 3'-UTR AU-rich region binding(GO:0035925) |
| 0.0 | 1.0 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 3.3 | GO:0019838 | growth factor binding(GO:0019838) |
| 0.0 | 0.3 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.7 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 1.7 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.3 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.5 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 1.5 | GO:0004866 | endopeptidase inhibitor activity(GO:0004866) |
| 0.0 | 0.2 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 2.6 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
| 0.0 | 0.0 | GO:0005302 | hydrogen:amino acid symporter activity(GO:0005280) L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.6 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 1.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 1.5 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.5 | GO:0008374 | O-acyltransferase activity(GO:0008374) |
| 0.0 | 0.2 | GO:0097602 | cullin family protein binding(GO:0097602) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.6 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.1 | 2.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 2.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.1 | 2.6 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.7 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 1.8 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 2.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 2.1 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 1.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.1 | PID ATF2 PATHWAY | ATF-2 transcription factor network |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.9 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 2.0 | PID AP1 PATHWAY | AP-1 transcription factor network |
| 0.0 | 4.8 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.3 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.5 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
| 0.0 | 0.6 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.2 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.6 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 3.8 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.7 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 1.0 | PID SMAD2 3NUCLEAR PATHWAY | Regulation of nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 1.0 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 0.7 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.2 | 2.1 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.1 | 1.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.1 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.1 | 1.3 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.1 | 2.1 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.1 | 3.4 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.1 | 1.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.1 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.1 | 1.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 4.8 | REACTOME PEPTIDE LIGAND BINDING RECEPTORS | Genes involved in Peptide ligand-binding receptors |
| 0.0 | 1.8 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.7 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.7 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.4 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 2.0 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.4 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME HIV INFECTION | Genes involved in HIV Infection |
| 0.0 | 0.5 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.6 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.3 | REACTOME AUTODEGRADATION OF CDH1 BY CDH1 APC C | Genes involved in Autodegradation of Cdh1 by Cdh1:APC/C |
| 0.0 | 1.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.6 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 1.2 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 1.2 | REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT | Genes involved in Lipid digestion, mobilization, and transport |
| 0.0 | 0.2 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 1.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.3 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 1.4 | REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | Genes involved in Response to elevated platelet cytosolic Ca2+ |
| 0.0 | 0.7 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |