Project
2D miR_HR1_12
Navigation
Downloads
Results for Zbtb16
Z-value: 1.27
Transcription factors associated with Zbtb16
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Zbtb16
|
ENSMUSG00000066687.4 | zinc finger and BTB domain containing 16 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Zbtb16 | mm10_v2_chr9_-_48835932_48835962 | 0.30 | 3.5e-01 | Click! |
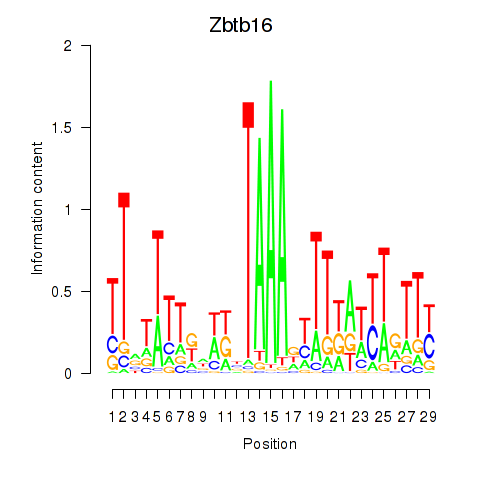
Activity profile of Zbtb16 motif
Sorted Z-values of Zbtb16 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrY_+_90785442 | 4.03 |
ENSMUST00000177591.1
ENSMUST00000177671.1 ENSMUST00000179077.1 |
Erdr1
|
erythroid differentiation regulator 1 |
| chr4_+_44300876 | 3.52 |
ENSMUST00000045607.5
|
Melk
|
maternal embryonic leucine zipper kinase |
| chr4_-_117178726 | 3.42 |
ENSMUST00000153953.1
ENSMUST00000106436.1 |
Kif2c
|
kinesin family member 2C |
| chrX_-_97377190 | 2.67 |
ENSMUST00000037353.3
|
Eda2r
|
ectodysplasin A2 receptor |
| chr11_+_69045640 | 2.61 |
ENSMUST00000108666.1
ENSMUST00000021277.5 |
Aurkb
|
aurora kinase B |
| chrX_-_97377150 | 2.07 |
ENSMUST00000113832.1
|
Eda2r
|
ectodysplasin A2 receptor |
| chr14_-_65833963 | 1.92 |
ENSMUST00000022613.9
|
Esco2
|
establishment of cohesion 1 homolog 2 (S. cerevisiae) |
| chr5_+_110330697 | 1.89 |
ENSMUST00000112481.1
|
Pole
|
polymerase (DNA directed), epsilon |
| chr4_-_116821501 | 1.47 |
ENSMUST00000055436.3
|
Hpdl
|
4-hydroxyphenylpyruvate dioxygenase-like |
| chr4_+_107367757 | 1.20 |
ENSMUST00000139560.1
|
Ndc1
|
NDC1 transmembrane nucleoporin |
| chr3_+_138143846 | 0.99 |
ENSMUST00000159481.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr4_-_119320417 | 0.98 |
ENSMUST00000147077.1
ENSMUST00000056458.7 ENSMUST00000106321.2 ENSMUST00000106319.1 ENSMUST00000106317.1 ENSMUST00000106318.1 |
Ppih
|
peptidyl prolyl isomerase H |
| chr8_+_70616816 | 0.98 |
ENSMUST00000052437.3
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr19_-_47171134 | 0.94 |
ENSMUST00000169692.1
|
Gm6970
|
predicted gene 6970 |
| chr5_+_8798139 | 0.89 |
ENSMUST00000009058.5
|
Abcb1b
|
ATP-binding cassette, sub-family B (MDR/TAP), member 1B |
| chr13_+_52583437 | 0.88 |
ENSMUST00000118756.1
|
Syk
|
spleen tyrosine kinase |
| chr11_-_69008422 | 0.86 |
ENSMUST00000021282.5
|
Pfas
|
phosphoribosylformylglycinamidine synthase (FGAR amidotransferase) |
| chr6_-_99632376 | 0.86 |
ENSMUST00000176255.1
|
Gm20696
|
predicted gene 20696 |
| chr5_-_104982656 | 0.86 |
ENSMUST00000031239.6
|
Abcg3
|
ATP-binding cassette, sub-family G (WHITE), member 3 |
| chr17_-_45685973 | 0.85 |
ENSMUST00000145873.1
|
Tmem63b
|
transmembrane protein 63b |
| chr6_-_142322941 | 0.82 |
ENSMUST00000128446.1
|
Slco1a5
|
solute carrier organic anion transporter family, member 1a5 |
| chr17_-_35175995 | 0.81 |
ENSMUST00000173324.1
|
Aif1
|
allograft inflammatory factor 1 |
| chr11_-_106779483 | 0.80 |
ENSMUST00000021060.5
|
Polg2
|
polymerase (DNA directed), gamma 2, accessory subunit |
| chr3_-_33082004 | 0.79 |
ENSMUST00000108225.3
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chrY_+_90784738 | 0.78 |
ENSMUST00000179483.1
|
Erdr1
|
erythroid differentiation regulator 1 |
| chr3_+_138143888 | 0.77 |
ENSMUST00000161141.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr2_+_15049395 | 0.74 |
ENSMUST00000017562.6
|
Arl5b
|
ADP-ribosylation factor-like 5B |
| chr11_-_115048372 | 0.72 |
ENSMUST00000092459.3
|
Cd300lh
|
CD300 antigen like family member H |
| chr9_+_57827284 | 0.71 |
ENSMUST00000163186.1
|
Gm17231
|
predicted gene 17231 |
| chr7_+_140967221 | 0.70 |
ENSMUST00000106042.2
|
Ifitm1
|
interferon induced transmembrane protein 1 |
| chr17_+_46496753 | 0.70 |
ENSMUST00000046497.6
|
Dnph1
|
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr15_-_36164872 | 0.69 |
ENSMUST00000058643.3
|
Fbxo43
|
F-box protein 43 |
| chr4_-_136053343 | 0.66 |
ENSMUST00000102536.4
|
Rpl11
|
ribosomal protein L11 |
| chr6_+_137754529 | 0.65 |
ENSMUST00000087675.6
|
Dera
|
2-deoxyribose-5-phosphate aldolase homolog (C. elegans) |
| chr7_+_28169744 | 0.64 |
ENSMUST00000042405.6
|
Fbl
|
fibrillarin |
| chr13_-_74376566 | 0.63 |
ENSMUST00000091481.2
|
Zfp72
|
zinc finger protein 72 |
| chr2_-_6130117 | 0.63 |
ENSMUST00000126551.1
ENSMUST00000054254.5 ENSMUST00000114942.2 |
Proser2
|
proline and serine rich 2 |
| chr8_-_41215146 | 0.62 |
ENSMUST00000034003.4
|
Fgl1
|
fibrinogen-like protein 1 |
| chr8_+_95534078 | 0.60 |
ENSMUST00000041569.3
|
Ccdc113
|
coiled-coil domain containing 113 |
| chr9_+_20888175 | 0.60 |
ENSMUST00000004203.5
|
Ppan
|
peter pan homolog (Drosophila) |
| chr11_+_114668524 | 0.60 |
ENSMUST00000106602.3
ENSMUST00000077915.3 ENSMUST00000106599.1 ENSMUST00000082092.4 |
Rpl38
|
ribosomal protein L38 |
| chr9_+_123806468 | 0.59 |
ENSMUST00000049810.7
|
Cxcr6
|
chemokine (C-X-C motif) receptor 6 |
| chr7_+_4792874 | 0.58 |
ENSMUST00000032597.5
ENSMUST00000078432.4 |
Rpl28
|
ribosomal protein L28 |
| chr2_+_30281043 | 0.58 |
ENSMUST00000143119.2
|
RP23-395P6.9
|
RP23-395P6.9 |
| chr9_-_79977782 | 0.58 |
ENSMUST00000093811.4
|
Filip1
|
filamin A interacting protein 1 |
| chr13_+_54071815 | 0.56 |
ENSMUST00000021930.8
|
Sfxn1
|
sideroflexin 1 |
| chr6_+_113333304 | 0.55 |
ENSMUST00000147945.1
|
Ogg1
|
8-oxoguanine DNA-glycosylase 1 |
| chr17_+_23853519 | 0.55 |
ENSMUST00000061725.7
|
Prss32
|
protease, serine, 32 |
| chr8_+_106683052 | 0.54 |
ENSMUST00000048359.4
|
Tango6
|
transport and golgi organization 6 |
| chr4_+_123282778 | 0.53 |
ENSMUST00000106243.1
ENSMUST00000106241.1 ENSMUST00000080178.6 |
Pabpc4
|
poly(A) binding protein, cytoplasmic 4 |
| chr11_-_78183551 | 0.53 |
ENSMUST00000102483.4
|
Rpl23a
|
ribosomal protein L23A |
| chr4_-_19922599 | 0.52 |
ENSMUST00000029900.5
|
Atp6v0d2
|
ATPase, H+ transporting, lysosomal V0 subunit D2 |
| chr11_+_23665615 | 0.51 |
ENSMUST00000109525.1
ENSMUST00000020520.4 |
Pus10
|
pseudouridylate synthase 10 |
| chr19_+_53310495 | 0.51 |
ENSMUST00000003870.7
|
Mxi1
|
Max interacting protein 1 |
| chr2_+_19344317 | 0.48 |
ENSMUST00000141289.1
|
4930447M23Rik
|
RIKEN cDNA 4930447M23 gene |
| chr11_-_106998483 | 0.47 |
ENSMUST00000124541.1
|
Kpna2
|
karyopherin (importin) alpha 2 |
| chr2_-_15049254 | 0.47 |
ENSMUST00000028034.8
ENSMUST00000114713.1 |
Nsun6
|
NOL1/NOP2/Sun domain family member 6 |
| chr1_+_165788746 | 0.46 |
ENSMUST00000161559.2
|
Cd247
|
CD247 antigen |
| chr4_+_146514920 | 0.46 |
ENSMUST00000140089.1
ENSMUST00000179175.1 |
Gm13247
|
predicted gene 13247 |
| chr9_-_20952838 | 0.46 |
ENSMUST00000004202.9
|
Dnmt1
|
DNA methyltransferase (cytosine-5) 1 |
| chr4_-_141846359 | 0.45 |
ENSMUST00000037059.10
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr4_+_146097312 | 0.45 |
ENSMUST00000105730.1
ENSMUST00000091878.5 |
Gm13051
|
predicted gene 13051 |
| chrX_-_111537947 | 0.45 |
ENSMUST00000132319.1
ENSMUST00000123951.1 |
Rps6ka6
|
ribosomal protein S6 kinase polypeptide 6 |
| chr17_-_40880525 | 0.44 |
ENSMUST00000068258.2
|
9130008F23Rik
|
RIKEN cDNA 9130008F23 gene |
| chr6_+_29272488 | 0.43 |
ENSMUST00000115289.1
ENSMUST00000054445.8 |
Hilpda
|
hypoxia inducible lipid droplet associated |
| chr12_-_87233556 | 0.43 |
ENSMUST00000021423.7
|
Noxred1
|
NADP+ dependent oxidoreductase domain containing 1 |
| chr7_+_75643223 | 0.43 |
ENSMUST00000137959.1
|
Akap13
|
A kinase (PRKA) anchor protein 13 |
| chr18_-_67449083 | 0.43 |
ENSMUST00000025408.8
|
Afg3l2
|
AFG3(ATPase family gene 3)-like 2 (yeast) |
| chr3_+_138143799 | 0.43 |
ENSMUST00000159622.1
|
Trmt10a
|
tRNA methyltransferase 10A |
| chr15_+_3270767 | 0.43 |
ENSMUST00000082424.4
ENSMUST00000159158.1 ENSMUST00000159216.1 ENSMUST00000160311.1 |
Sepp1
|
selenoprotein P, plasma, 1 |
| chr13_-_60897439 | 0.43 |
ENSMUST00000171347.1
ENSMUST00000021884.8 |
Ctla2b
|
cytotoxic T lymphocyte-associated protein 2 beta |
| chr15_-_99705490 | 0.43 |
ENSMUST00000163472.2
|
Gm17349
|
predicted gene, 17349 |
| chr10_+_33905015 | 0.42 |
ENSMUST00000169670.1
|
Rsph4a
|
radial spoke head 4 homolog A (Chlamydomonas) |
| chr5_+_121777929 | 0.41 |
ENSMUST00000160821.1
|
Atxn2
|
ataxin 2 |
| chr13_+_29016267 | 0.40 |
ENSMUST00000140415.1
|
A330102I10Rik
|
RIKEN cDNA A330102I10 gene |
| chr1_-_139858684 | 0.40 |
ENSMUST00000094489.3
|
Cfhr2
|
complement factor H-related 2 |
| chr17_-_80563834 | 0.39 |
ENSMUST00000086545.4
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr9_-_50617428 | 0.39 |
ENSMUST00000131351.1
ENSMUST00000171462.1 |
AU019823
|
expressed sequence AU019823 |
| chrX_-_134111852 | 0.38 |
ENSMUST00000033610.6
|
Nox1
|
NADPH oxidase 1 |
| chr7_-_115846080 | 0.37 |
ENSMUST00000166207.1
|
Sox6
|
SRY-box containing gene 6 |
| chr11_-_93965957 | 0.37 |
ENSMUST00000021220.3
|
Nme1
|
NME/NM23 nucleoside diphosphate kinase 1 |
| chr6_-_30390997 | 0.36 |
ENSMUST00000152391.2
ENSMUST00000115184.1 ENSMUST00000080812.7 ENSMUST00000102992.3 |
Zc3hc1
|
zinc finger, C3HC type 1 |
| chr6_-_118419388 | 0.36 |
ENSMUST00000032237.6
|
Bms1
|
BMS1 homolog, ribosome assembly protein (yeast) |
| chr9_+_50617516 | 0.36 |
ENSMUST00000141366.1
|
Pih1d2
|
PIH1 domain containing 2 |
| chr2_+_126152141 | 0.35 |
ENSMUST00000170908.1
|
Dtwd1
|
DTW domain containing 1 |
| chr4_-_19570073 | 0.35 |
ENSMUST00000029885.4
|
Cpne3
|
copine III |
| chr4_+_33310306 | 0.35 |
ENSMUST00000108153.2
ENSMUST00000029942.7 |
Rngtt
|
RNA guanylyltransferase and 5'-phosphatase |
| chr13_-_56178864 | 0.35 |
ENSMUST00000169652.1
|
Tifab
|
TRAF-interacting protein with forkhead-associated domain, family member B |
| chr14_-_23650189 | 0.34 |
ENSMUST00000112423.3
|
Kcnma1
|
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr8_-_106573461 | 0.33 |
ENSMUST00000073722.5
|
Gm10073
|
predicted pseudogene 10073 |
| chr5_-_65335564 | 0.31 |
ENSMUST00000172780.1
|
Rfc1
|
replication factor C (activator 1) 1 |
| chr9_+_19641224 | 0.31 |
ENSMUST00000079042.6
|
Zfp317
|
zinc finger protein 317 |
| chr3_+_116878227 | 0.31 |
ENSMUST00000040260.6
|
Frrs1
|
ferric-chelate reductase 1 |
| chr11_-_32527551 | 0.31 |
ENSMUST00000054327.2
|
Efcab9
|
EF-hand calcium binding domain 9 |
| chr1_+_20917856 | 0.31 |
ENSMUST00000167119.1
|
Paqr8
|
progestin and adipoQ receptor family member VIII |
| chrX_-_164076482 | 0.30 |
ENSMUST00000134272.1
|
Siah1b
|
seven in absentia 1B |
| chr4_+_80910646 | 0.30 |
ENSMUST00000055922.3
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr5_+_138161071 | 0.28 |
ENSMUST00000019638.8
ENSMUST00000110951.1 |
Cops6
|
COP9 (constitutive photomorphogenic) homolog, subunit 6 (Arabidopsis thaliana) |
| chr1_-_105659008 | 0.28 |
ENSMUST00000070699.8
|
Pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr1_+_165788681 | 0.28 |
ENSMUST00000161971.1
ENSMUST00000178336.1 ENSMUST00000005907.5 ENSMUST00000027849.4 |
Cd247
|
CD247 antigen |
| chr1_+_157506728 | 0.28 |
ENSMUST00000086130.2
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chrX_+_164506320 | 0.28 |
ENSMUST00000033756.2
|
Asb9
|
ankyrin repeat and SOCS box-containing 9 |
| chr18_+_56562443 | 0.28 |
ENSMUST00000130163.1
ENSMUST00000132628.1 |
Phax
|
phosphorylated adaptor for RNA export |
| chr7_-_38019505 | 0.28 |
ENSMUST00000085513.4
|
Uri1
|
URI1, prefoldin-like chaperone |
| chr1_+_157506777 | 0.27 |
ENSMUST00000027881.8
|
Sec16b
|
SEC16 homolog B (S. cerevisiae) |
| chr16_-_75909272 | 0.27 |
ENSMUST00000114239.2
|
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chrX_-_9469288 | 0.27 |
ENSMUST00000015484.3
|
Cybb
|
cytochrome b-245, beta polypeptide |
| chr4_+_135911808 | 0.27 |
ENSMUST00000068830.3
|
Cnr2
|
cannabinoid receptor 2 (macrophage) |
| chr12_-_102423741 | 0.27 |
ENSMUST00000110020.1
|
Lgmn
|
legumain |
| chr11_-_83578496 | 0.27 |
ENSMUST00000019266.5
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr5_+_143548700 | 0.26 |
ENSMUST00000169329.1
ENSMUST00000067145.5 ENSMUST00000119488.1 ENSMUST00000118121.1 |
Fam220a
Fam220a
|
family with sequence similarity 220, member A family with sequence similarity 220, member A |
| chr10_+_22360552 | 0.26 |
ENSMUST00000182677.1
|
Raet1d
|
retinoic acid early transcript delta |
| chr4_-_141846277 | 0.26 |
ENSMUST00000105781.1
|
Ctrc
|
chymotrypsin C (caldecrin) |
| chr11_-_101987004 | 0.25 |
ENSMUST00000107173.2
ENSMUST00000107172.1 |
Dusp3
|
dual specificity phosphatase 3 (vaccinia virus phosphatase VH1-related) |
| chr15_-_96699698 | 0.25 |
ENSMUST00000023099.6
|
Slc38a2
|
solute carrier family 38, member 2 |
| chr10_-_93891141 | 0.24 |
ENSMUST00000180840.1
|
Metap2
|
methionine aminopeptidase 2 |
| chr4_+_133220810 | 0.24 |
ENSMUST00000105910.1
|
Cd164l2
|
CD164 sialomucin-like 2 |
| chr9_-_71896047 | 0.23 |
ENSMUST00000184448.1
|
Tcf12
|
transcription factor 12 |
| chr9_+_96259246 | 0.23 |
ENSMUST00000179065.1
ENSMUST00000165768.2 |
Tfdp2
|
transcription factor Dp 2 |
| chr4_-_132463873 | 0.23 |
ENSMUST00000102567.3
|
Med18
|
mediator of RNA polymerase II transcription, subunit 18 homolog (yeast) |
| chr11_-_5707658 | 0.23 |
ENSMUST00000154330.1
|
Mrps24
|
mitochondrial ribosomal protein S24 |
| chr17_+_46110982 | 0.23 |
ENSMUST00000024763.3
ENSMUST00000123646.1 |
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr17_+_26252903 | 0.22 |
ENSMUST00000025023.7
|
Luc7l
|
Luc7 homolog (S. cerevisiae)-like |
| chr7_-_125491397 | 0.22 |
ENSMUST00000138616.1
|
Nsmce1
|
non-SMC element 1 homolog (S. cerevisiae) |
| chr14_-_73548242 | 0.22 |
ENSMUST00000043813.1
|
Nudt15
|
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr2_-_58160495 | 0.22 |
ENSMUST00000028175.6
|
Cytip
|
cytohesin 1 interacting protein |
| chr1_+_107511489 | 0.21 |
ENSMUST00000064916.2
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr5_-_65335597 | 0.21 |
ENSMUST00000172660.1
ENSMUST00000172732.1 ENSMUST00000031092.8 |
Rfc1
|
replication factor C (activator 1) 1 |
| chr5_-_76905463 | 0.21 |
ENSMUST00000146570.1
ENSMUST00000142450.1 ENSMUST00000120963.1 |
Aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr6_-_131247342 | 0.20 |
ENSMUST00000032306.8
ENSMUST00000088867.6 |
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chr4_+_129287614 | 0.20 |
ENSMUST00000102599.3
|
Sync
|
syncoilin |
| chr2_+_11705287 | 0.20 |
ENSMUST00000135341.1
ENSMUST00000138349.1 ENSMUST00000123600.2 |
Il15ra
|
interleukin 15 receptor, alpha chain |
| chr17_-_34305715 | 0.20 |
ENSMUST00000174074.1
|
Gm20513
|
predicted gene 20513 |
| chr3_-_157925056 | 0.20 |
ENSMUST00000118539.1
|
Cth
|
cystathionase (cystathionine gamma-lyase) |
| chr15_-_83595111 | 0.19 |
ENSMUST00000016901.3
|
Ttll12
|
tubulin tyrosine ligase-like family, member 12 |
| chr8_-_72443772 | 0.19 |
ENSMUST00000019876.5
|
Calr3
|
calreticulin 3 |
| chr18_+_77773956 | 0.19 |
ENSMUST00000114748.1
|
Atp5a1
|
ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 |
| chr11_-_5950018 | 0.19 |
ENSMUST00000102920.3
|
Gck
|
glucokinase |
| chr2_-_121380940 | 0.19 |
ENSMUST00000038389.8
|
Strc
|
stereocilin |
| chr5_+_92925400 | 0.18 |
ENSMUST00000172706.1
|
Shroom3
|
shroom family member 3 |
| chr1_-_128417352 | 0.18 |
ENSMUST00000027602.8
ENSMUST00000064309.7 |
Dars
|
aspartyl-tRNA synthetase |
| chr15_+_79670856 | 0.18 |
ENSMUST00000023062.3
|
Tomm22
|
translocase of outer mitochondrial membrane 22 homolog (yeast) |
| chr7_+_27233003 | 0.18 |
ENSMUST00000003860.6
ENSMUST00000108378.3 |
Adck4
|
aarF domain containing kinase 4 |
| chr10_-_127522428 | 0.18 |
ENSMUST00000026470.4
|
Shmt2
|
serine hydroxymethyltransferase 2 (mitochondrial) |
| chr1_+_107511416 | 0.17 |
ENSMUST00000009356.4
|
Serpinb2
|
serine (or cysteine) peptidase inhibitor, clade B, member 2 |
| chr7_+_119896292 | 0.17 |
ENSMUST00000106517.1
|
Lyrm1
|
LYR motif containing 1 |
| chr11_-_100613334 | 0.17 |
ENSMUST00000146840.1
|
Dnajc7
|
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr9_-_71168657 | 0.17 |
ENSMUST00000113570.1
|
Aqp9
|
aquaporin 9 |
| chr5_-_76905390 | 0.17 |
ENSMUST00000135954.1
|
Aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr7_-_125491586 | 0.17 |
ENSMUST00000033006.7
|
Nsmce1
|
non-SMC element 1 homolog (S. cerevisiae) |
| chr9_+_96258697 | 0.17 |
ENSMUST00000179416.1
|
Tfdp2
|
transcription factor Dp 2 |
| chrX_-_95478107 | 0.17 |
ENSMUST00000033549.2
|
Asb12
|
ankyrin repeat and SOCS box-containing 12 |
| chr7_+_19282613 | 0.16 |
ENSMUST00000032559.9
|
Rtn2
|
reticulon 2 (Z-band associated protein) |
| chr6_-_39377681 | 0.16 |
ENSMUST00000090243.4
|
Slc37a3
|
solute carrier family 37 (glycerol-3-phosphate transporter), member 3 |
| chr9_+_105053239 | 0.16 |
ENSMUST00000035177.8
ENSMUST00000149243.1 |
Mrpl3
|
mitochondrial ribosomal protein L3 |
| chr5_-_149051300 | 0.15 |
ENSMUST00000110505.1
|
Hmgb1
|
high mobility group box 1 |
| chr14_-_26442824 | 0.14 |
ENSMUST00000136635.1
ENSMUST00000125437.1 |
Slmap
|
sarcolemma associated protein |
| chr9_-_107679592 | 0.14 |
ENSMUST00000010205.7
|
Gnat1
|
guanine nucleotide binding protein, alpha transducing 1 |
| chr6_+_134640940 | 0.13 |
ENSMUST00000062755.8
|
Loh12cr1
|
loss of heterozygosity, 12, chromosomal region 1 homolog (human) |
| chr19_-_30175414 | 0.13 |
ENSMUST00000025778.7
|
Gldc
|
glycine decarboxylase |
| chr6_+_116506516 | 0.13 |
ENSMUST00000075756.2
|
Olfr212
|
olfactory receptor 212 |
| chr6_-_129678908 | 0.13 |
ENSMUST00000118447.1
ENSMUST00000169545.1 ENSMUST00000032270.6 ENSMUST00000032271.6 |
Klrc1
|
killer cell lectin-like receptor subfamily C, member 1 |
| chr11_-_115627948 | 0.13 |
ENSMUST00000154623.1
ENSMUST00000106503.3 ENSMUST00000141614.1 |
Slc25a19
|
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr6_-_129622685 | 0.12 |
ENSMUST00000032252.5
|
Klrk1
|
killer cell lectin-like receptor subfamily K, member 1 |
| chr8_+_27042555 | 0.12 |
ENSMUST00000033875.8
ENSMUST00000098851.4 |
Prosc
|
proline synthetase co-transcribed |
| chr6_+_136518820 | 0.12 |
ENSMUST00000032335.6
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr10_-_81364846 | 0.12 |
ENSMUST00000131736.1
|
4930404N11Rik
|
RIKEN cDNA 4930404N11 gene |
| chr17_-_80290476 | 0.12 |
ENSMUST00000086555.3
ENSMUST00000038166.7 |
Dhx57
|
DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 |
| chr2_-_50296680 | 0.12 |
ENSMUST00000144143.1
ENSMUST00000102769.4 ENSMUST00000133768.1 |
Mmadhc
|
methylmalonic aciduria (cobalamin deficiency) cblD type, with homocystinuria |
| chr1_-_149961230 | 0.12 |
ENSMUST00000070200.8
|
Pla2g4a
|
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr10_-_116549101 | 0.12 |
ENSMUST00000164088.1
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr16_-_62786742 | 0.11 |
ENSMUST00000152553.1
ENSMUST00000063089.5 |
Nsun3
|
NOL1/NOP2/Sun domain family member 3 |
| chr4_+_155582476 | 0.11 |
ENSMUST00000105612.1
|
Nadk
|
NAD kinase |
| chr19_-_8786408 | 0.11 |
ENSMUST00000176496.1
|
Taf6l
|
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr2_+_80638798 | 0.11 |
ENSMUST00000028382.6
ENSMUST00000124377.1 |
Nup35
|
nucleoporin 35 |
| chr5_+_110110068 | 0.11 |
ENSMUST00000112528.1
|
Zfp605
|
zinc finger protein 605 |
| chr17_+_26252915 | 0.10 |
ENSMUST00000114976.2
ENSMUST00000140427.1 ENSMUST00000119928.1 |
Luc7l
|
Luc7 homolog (S. cerevisiae)-like |
| chr15_-_97910622 | 0.10 |
ENSMUST00000173104.1
ENSMUST00000174633.1 |
Vdr
|
vitamin D receptor |
| chr2_-_25627960 | 0.10 |
ENSMUST00000028307.8
|
Fcna
|
ficolin A |
| chr11_-_99438143 | 0.09 |
ENSMUST00000017743.2
|
Krt20
|
keratin 20 |
| chr4_+_86930691 | 0.09 |
ENSMUST00000164590.1
|
Acer2
|
alkaline ceramidase 2 |
| chr7_-_41448641 | 0.09 |
ENSMUST00000165029.1
|
Vmn2r57
|
vomeronasal 2, receptor 57 |
| chrX_-_157598656 | 0.09 |
ENSMUST00000149249.1
ENSMUST00000058098.8 |
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr6_-_142702259 | 0.09 |
ENSMUST00000073173.5
ENSMUST00000111771.1 ENSMUST00000087527.4 ENSMUST00000100827.2 |
Abcc9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr15_+_6422240 | 0.08 |
ENSMUST00000163082.1
|
Dab2
|
disabled 2, mitogen-responsive phosphoprotein |
| chr16_+_20097554 | 0.08 |
ENSMUST00000023509.3
|
Klhl24
|
kelch-like 24 |
| chr19_-_29367294 | 0.08 |
ENSMUST00000138051.1
|
Plgrkt
|
plasminogen receptor, C-terminal lysine transmembrane protein |
| chr2_-_120353094 | 0.08 |
ENSMUST00000028752.7
ENSMUST00000102501.3 |
Vps39
|
vacuolar protein sorting 39 (yeast) |
| chrX_-_7188713 | 0.08 |
ENSMUST00000004428.7
|
Clcn5
|
chloride channel 5 |
| chr4_+_155451570 | 0.08 |
ENSMUST00000135407.1
ENSMUST00000105619.1 |
C030017K20Rik
|
RIKEN cDNA C030017K20 gene |
| chr17_-_70998010 | 0.08 |
ENSMUST00000024846.6
|
Myl12a
|
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr18_+_61275002 | 0.08 |
ENSMUST00000135688.1
|
Pde6a
|
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr2_+_29124106 | 0.07 |
ENSMUST00000129544.1
|
Setx
|
senataxin |
| chr11_-_77787747 | 0.07 |
ENSMUST00000092883.2
|
Gm10277
|
predicted gene 10277 |
| chr16_+_43510267 | 0.07 |
ENSMUST00000114695.2
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr3_-_96452306 | 0.07 |
ENSMUST00000093126.4
ENSMUST00000098841.3 |
BC107364
|
cDNA sequence BC107364 |
| chr11_+_65807175 | 0.07 |
ENSMUST00000071465.2
ENSMUST00000018491.7 |
Zkscan6
|
zinc finger with KRAB and SCAN domains 6 |
| chr19_+_29367447 | 0.06 |
ENSMUST00000016640.7
|
Cd274
|
CD274 antigen |
| chr6_+_6863269 | 0.06 |
ENSMUST00000160937.2
ENSMUST00000171311.1 |
Dlx6
|
distal-less homeobox 6 |
| chr15_+_85736107 | 0.05 |
ENSMUST00000057979.5
|
Ppara
|
peroxisome proliferator activated receptor alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of Zbtb16
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.9 | 2.6 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.7 | 3.4 | GO:0030951 | establishment or maintenance of microtubule cytoskeleton polarity(GO:0030951) |
| 0.6 | 1.9 | GO:0045004 | DNA replication proofreading(GO:0045004) |
| 0.4 | 1.9 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.3 | 0.9 | GO:0032752 | positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.2 | 0.8 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) muscle hyperplasia(GO:0014900) |
| 0.2 | 0.9 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.2 | 0.6 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.6 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.7 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.1 | 0.6 | GO:0018364 | peptidyl-glutamine methylation(GO:0018364) |
| 0.1 | 0.9 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) |
| 0.1 | 0.4 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.1 | 0.8 | GO:0070127 | tRNA aminoacylation for mitochondrial protein translation(GO:0070127) |
| 0.1 | 0.5 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.1 | 0.8 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
| 0.1 | 0.3 | GO:0051030 | snRNA transport(GO:0051030) |
| 0.1 | 0.3 | GO:0007356 | thorax and anterior abdomen determination(GO:0007356) |
| 0.1 | 0.3 | GO:1904305 | negative regulation of gastro-intestinal system smooth muscle contraction(GO:1904305) negative regulation of small intestine smooth muscle contraction(GO:1904348) |
| 0.1 | 1.2 | GO:0051292 | nuclear pore complex assembly(GO:0051292) |
| 0.1 | 2.3 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.1 | 0.6 | GO:0044791 | modulation by host of viral release from host cell(GO:0044789) positive regulation by host of viral release from host cell(GO:0044791) |
| 0.1 | 0.6 | GO:0045002 | double-strand break repair via single-strand annealing(GO:0045002) |
| 0.1 | 0.7 | GO:0009162 | deoxyribonucleoside monophosphate metabolic process(GO:0009162) |
| 0.1 | 0.4 | GO:0019482 | beta-alanine metabolic process(GO:0019482) |
| 0.1 | 0.7 | GO:1904667 | negative regulation of ubiquitin protein ligase activity(GO:1904667) |
| 0.1 | 0.3 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.1 | 0.6 | GO:0048318 | axial mesoderm development(GO:0048318) |
| 0.1 | 4.7 | GO:0035019 | somatic stem cell population maintenance(GO:0035019) |
| 0.1 | 0.2 | GO:1905065 | cysteine biosynthetic process via cystathionine(GO:0019343) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.1 | 0.4 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 3.5 | GO:0008631 | intrinsic apoptotic signaling pathway in response to oxidative stress(GO:0008631) |
| 0.1 | 0.3 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.1 | 0.2 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.1 | 0.6 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) |
| 0.1 | 0.4 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.3 | GO:0097411 | hypoxia-inducible factor-1alpha signaling pathway(GO:0097411) |
| 0.1 | 0.4 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.1 | 0.5 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.1 | 0.2 | GO:0002270 | plasmacytoid dendritic cell activation(GO:0002270) positive regulation of glycogen catabolic process(GO:0045819) regulation of nucleotide-excision repair(GO:2000819) |
| 0.1 | 0.9 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 1.5 | GO:0009072 | aromatic amino acid family metabolic process(GO:0009072) |
| 0.0 | 1.0 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.3 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0002223 | stimulatory C-type lectin receptor signaling pathway(GO:0002223) |
| 0.0 | 0.3 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 4.5 | GO:0072332 | intrinsic apoptotic signaling pathway by p53 class mediator(GO:0072332) |
| 0.0 | 0.2 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.4 | GO:2000381 | negative regulation of mesoderm development(GO:2000381) |
| 0.0 | 0.4 | GO:2000741 | positive regulation of mesenchymal stem cell differentiation(GO:2000741) |
| 0.0 | 0.4 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:1904747 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0030974 | thiamine pyrophosphate transport(GO:0030974) |
| 0.0 | 0.1 | GO:0090285 | negative regulation of protein glycosylation in Golgi(GO:0090285) |
| 0.0 | 0.6 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.1 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.1 | GO:0036500 | ATF6-mediated unfolded protein response(GO:0036500) |
| 0.0 | 0.3 | GO:0015838 | amino-acid betaine transport(GO:0015838) |
| 0.0 | 0.1 | GO:0051342 | sensory perception of umami taste(GO:0050917) regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) |
| 0.0 | 0.3 | GO:0032229 | negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.4 | GO:0000479 | endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.6 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.0 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.9 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.1 | GO:1902774 | late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.5 | GO:0000027 | ribosomal large subunit assembly(GO:0000027) |
| 0.0 | 0.5 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.2 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.5 | GO:0001510 | RNA methylation(GO:0001510) |
| 0.0 | 0.1 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.7 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0072369 | regulation of glycolytic process by positive regulation of transcription from RNA polymerase II promoter(GO:0072363) regulation of lipid transport by positive regulation of transcription from RNA polymerase II promoter(GO:0072369) |
| 0.0 | 0.4 | GO:0046597 | negative regulation of viral entry into host cell(GO:0046597) |
| 0.0 | 0.2 | GO:0045040 | protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.3 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.4 | GO:0010884 | positive regulation of lipid storage(GO:0010884) |
| 0.0 | 0.6 | GO:1903146 | regulation of mitophagy(GO:1903146) |
| 0.0 | 0.4 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.2 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0033147 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.6 | GO:0006826 | iron ion transport(GO:0006826) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.4 | GO:1901099 | negative regulation of signal transduction in absence of ligand(GO:1901099) negative regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001240) |
| 0.0 | 0.2 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.1 | GO:0010756 | positive regulation of plasminogen activation(GO:0010756) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 2.6 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.4 | 1.9 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.3 | 1.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.1 | 0.9 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.1 | 0.7 | GO:0001651 | dense fibrillar component(GO:0001651) |
| 0.1 | 1.9 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.1 | 0.8 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.1 | 0.4 | GO:0005745 | m-AAA complex(GO:0005745) |
| 0.1 | 3.4 | GO:0035371 | microtubule plus-end(GO:0035371) |
| 0.1 | 0.5 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.1 | 0.7 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.7 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.1 | 0.9 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.1 | 0.4 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.2 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.5 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.8 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 1.8 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.2 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.5 | GO:0005721 | pericentric heterochromatin(GO:0005721) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.6 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 1.7 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.8 | GO:0009295 | nucleoid(GO:0009295) mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.3 | GO:0071004 | U2-type prespliceosome(GO:0071004) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.3 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.5 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.2 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.2 | GO:0045180 | basal cortex(GO:0045180) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 4.2 | GO:0005938 | cell cortex(GO:0005938) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.6 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.3 | 1.9 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.3 | 2.2 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.2 | 1.9 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.2 | 3.4 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.2 | 0.6 | GO:0008534 | oxidized purine nucleobase lesion DNA N-glycosylase activity(GO:0008534) |
| 0.1 | 0.8 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.1 | 0.9 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.3 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.1 | 0.7 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.4 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.1 | 0.8 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.6 | GO:0019958 | C-X-C chemokine binding(GO:0019958) |
| 0.1 | 0.9 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.4 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.1 | 0.5 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.3 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.1 | 0.6 | GO:0001094 | TFIID-class transcription factor binding(GO:0001094) |
| 0.1 | 0.7 | GO:1990948 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.1 | 0.9 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.1 | 0.3 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.1 | 0.3 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 0.6 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 1.5 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.1 | 0.4 | GO:0004043 | L-aminoadipate-semialdehyde dehydrogenase activity(GO:0004043) |
| 0.1 | 0.2 | GO:0004793 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.1 | 0.5 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 3.3 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.1 | 0.2 | GO:0000402 | open form four-way junction DNA binding(GO:0000401) crossed form four-way junction DNA binding(GO:0000402) |
| 0.0 | 0.6 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.5 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 1.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.2 | GO:0005350 | purine nucleobase transmembrane transporter activity(GO:0005345) pyrimidine nucleobase transmembrane transporter activity(GO:0005350) nucleobase transmembrane transporter activity(GO:0015205) glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:1902121 | lithocholic acid binding(GO:1902121) D3 vitamins binding(GO:1902271) |
| 0.0 | 0.3 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.5 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.8 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.4 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.7 | GO:0016799 | hydrolase activity, hydrolyzing N-glycosyl compounds(GO:0016799) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0001160 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.5 | GO:0008143 | poly(A) binding(GO:0008143) |
| 0.0 | 1.0 | GO:0003755 | peptidyl-prolyl cis-trans isomerase activity(GO:0003755) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.5 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0015234 | thiamine transmembrane transporter activity(GO:0015234) |
| 0.0 | 0.2 | GO:0015266 | protein channel activity(GO:0015266) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.3 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.4 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0035497 | cAMP response element binding(GO:0035497) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 1.1 | GO:1990782 | protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 1.8 | GO:0003735 | structural constituent of ribosome(GO:0003735) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 6.0 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.9 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.7 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.7 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.7 | PID RAC1 PATHWAY | RAC1 signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.1 | 1.9 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.1 | 0.7 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 0.9 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.1 | 0.6 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 2.6 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 2.4 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.8 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.7 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.6 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.3 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.3 | REACTOME MRNA CAPPING | Genes involved in mRNA Capping |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |