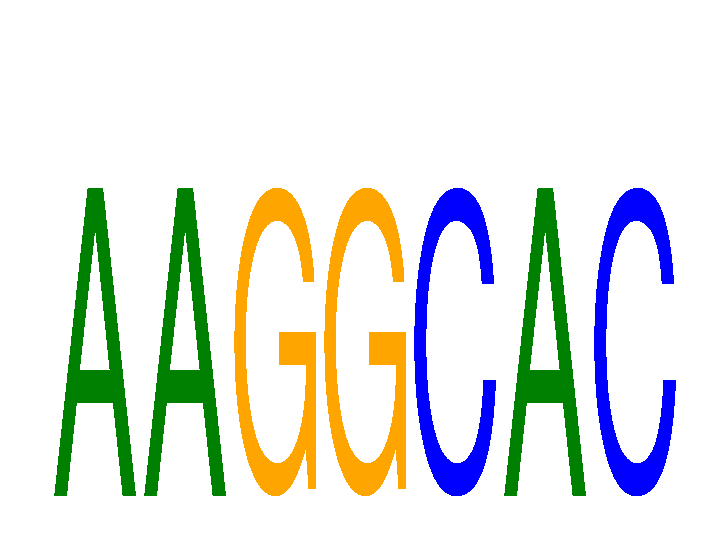Project
avrg: GFI1 WT vs 36n/n vs KD
Navigation
Downloads
Results for AAGGCAC
Z-value: 1.11
miRNA associated with seed AAGGCAC
| Name | miRBASE accession |
|---|---|
|
mmu-miR-124-3p.1
|
MIMAT0000134 |
Activity profile of AAGGCAC motif
Sorted Z-values of AAGGCAC motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_87651359 | 0.85 |
ENSMUST00000039627.12
ENSMUST00000100644.10 |
Tspoap1
|
TSPO associated protein 1 |
| chr6_-_127128007 | 0.80 |
ENSMUST00000000188.12
|
Ccnd2
|
cyclin D2 |
| chr11_+_3280401 | 0.61 |
ENSMUST00000045153.11
|
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr3_+_41510160 | 0.60 |
ENSMUST00000026865.15
ENSMUST00000194181.6 ENSMUST00000195846.6 |
Jade1
|
jade family PHD finger 1 |
| chr16_+_16714333 | 0.53 |
ENSMUST00000027373.12
ENSMUST00000232247.2 |
Ppm1f
|
protein phosphatase 1F (PP2C domain containing) |
| chr16_+_32427738 | 0.52 |
ENSMUST00000023486.15
|
Tfrc
|
transferrin receptor |
| chr3_+_121220146 | 0.50 |
ENSMUST00000029773.13
|
Cnn3
|
calponin 3, acidic |
| chr5_-_21260878 | 0.49 |
ENSMUST00000030556.8
|
Ptpn12
|
protein tyrosine phosphatase, non-receptor type 12 |
| chr3_+_104545974 | 0.48 |
ENSMUST00000046212.2
|
Slc16a1
|
solute carrier family 16 (monocarboxylic acid transporters), member 1 |
| chrX_+_65692924 | 0.44 |
ENSMUST00000166241.2
|
Slitrk2
|
SLIT and NTRK-like family, member 2 |
| chr10_+_59159118 | 0.42 |
ENSMUST00000009789.15
ENSMUST00000092512.11 ENSMUST00000105466.3 |
P4ha1
|
procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide |
| chr17_+_29709723 | 0.41 |
ENSMUST00000024811.9
|
Pim1
|
proviral integration site 1 |
| chr2_-_84481058 | 0.40 |
ENSMUST00000111670.9
ENSMUST00000111697.9 ENSMUST00000111696.8 ENSMUST00000111678.8 ENSMUST00000111690.8 ENSMUST00000111695.8 ENSMUST00000111677.8 ENSMUST00000111698.8 ENSMUST00000099941.9 ENSMUST00000111676.8 ENSMUST00000111694.8 ENSMUST00000111675.8 ENSMUST00000111689.8 ENSMUST00000111687.8 ENSMUST00000111692.8 ENSMUST00000111685.8 ENSMUST00000111686.8 ENSMUST00000111688.8 ENSMUST00000111693.8 ENSMUST00000111684.8 |
Ctnnd1
|
catenin (cadherin associated protein), delta 1 |
| chr4_+_140427799 | 0.40 |
ENSMUST00000071169.9
|
Rcc2
|
regulator of chromosome condensation 2 |
| chr17_+_87270504 | 0.40 |
ENSMUST00000024956.15
|
Rhoq
|
ras homolog family member Q |
| chr10_+_99099084 | 0.39 |
ENSMUST00000020118.5
ENSMUST00000220291.2 |
Dusp6
|
dual specificity phosphatase 6 |
| chr2_+_127112127 | 0.39 |
ENSMUST00000110375.9
|
Stard7
|
START domain containing 7 |
| chr4_+_106418224 | 0.37 |
ENSMUST00000047973.4
|
Dhcr24
|
24-dehydrocholesterol reductase |
| chr3_-_19217174 | 0.37 |
ENSMUST00000029125.10
|
Armc1
|
armadillo repeat containing 1 |
| chr5_-_121329385 | 0.36 |
ENSMUST00000054547.9
ENSMUST00000100770.9 |
Ptpn11
|
protein tyrosine phosphatase, non-receptor type 11 |
| chr4_-_87951565 | 0.35 |
ENSMUST00000078090.12
|
Mllt3
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 3 |
| chr12_-_69771604 | 0.35 |
ENSMUST00000021370.10
|
L2hgdh
|
L-2-hydroxyglutarate dehydrogenase |
| chr8_+_93581946 | 0.34 |
ENSMUST00000046290.3
ENSMUST00000210099.2 |
Lpcat2
|
lysophosphatidylcholine acyltransferase 2 |
| chr17_-_44416665 | 0.34 |
ENSMUST00000024757.14
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr18_-_10030017 | 0.33 |
ENSMUST00000116669.2
ENSMUST00000092096.14 |
Usp14
|
ubiquitin specific peptidase 14 |
| chr11_-_76289888 | 0.32 |
ENSMUST00000021204.4
|
Nxn
|
nucleoredoxin |
| chr12_-_112893382 | 0.32 |
ENSMUST00000075827.5
|
Jag2
|
jagged 2 |
| chr9_+_56902172 | 0.32 |
ENSMUST00000034832.8
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr12_-_107969853 | 0.32 |
ENSMUST00000066060.11
|
Bcl11b
|
B cell leukemia/lymphoma 11B |
| chr8_+_86219191 | 0.32 |
ENSMUST00000034136.12
|
Gpt2
|
glutamic pyruvate transaminase (alanine aminotransferase) 2 |
| chr17_+_8384333 | 0.32 |
ENSMUST00000097419.10
ENSMUST00000024636.15 |
Cep43
|
centrosomal protein 43 |
| chr11_+_103857541 | 0.31 |
ENSMUST00000057921.10
ENSMUST00000063347.12 |
Arf2
|
ADP-ribosylation factor 2 |
| chr7_-_98831916 | 0.31 |
ENSMUST00000033001.6
|
Dgat2
|
diacylglycerol O-acyltransferase 2 |
| chr4_+_139350152 | 0.31 |
ENSMUST00000039818.10
|
Aldh4a1
|
aldehyde dehydrogenase 4 family, member A1 |
| chr19_+_42135812 | 0.30 |
ENSMUST00000061111.10
|
Marveld1
|
MARVEL (membrane-associating) domain containing 1 |
| chr11_+_23615612 | 0.30 |
ENSMUST00000109525.8
ENSMUST00000020520.11 |
Pus10
|
pseudouridylate synthase 10 |
| chr7_+_27290969 | 0.30 |
ENSMUST00000108344.9
|
Akt2
|
thymoma viral proto-oncogene 2 |
| chr9_-_14292453 | 0.30 |
ENSMUST00000167549.2
|
Endod1
|
endonuclease domain containing 1 |
| chr4_-_156319232 | 0.30 |
ENSMUST00000105569.5
|
Klhl17
|
kelch-like 17 |
| chr9_+_108539296 | 0.29 |
ENSMUST00000035222.6
|
Slc25a20
|
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase), member 20 |
| chr1_-_153208477 | 0.29 |
ENSMUST00000027752.15
|
Lamc1
|
laminin, gamma 1 |
| chr18_+_69478790 | 0.29 |
ENSMUST00000202116.4
ENSMUST00000114982.8 ENSMUST00000078486.13 ENSMUST00000202772.4 ENSMUST00000201288.4 |
Tcf4
|
transcription factor 4 |
| chr1_+_16175998 | 0.28 |
ENSMUST00000027053.8
|
Rdh10
|
retinol dehydrogenase 10 (all-trans) |
| chr7_-_144837719 | 0.28 |
ENSMUST00000058022.6
|
Tpcn2
|
two pore segment channel 2 |
| chr6_-_137626207 | 0.28 |
ENSMUST00000134630.6
ENSMUST00000058210.13 ENSMUST00000111878.8 |
Eps8
|
epidermal growth factor receptor pathway substrate 8 |
| chr8_-_105169621 | 0.28 |
ENSMUST00000041769.8
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr6_+_116314975 | 0.28 |
ENSMUST00000079012.13
ENSMUST00000101032.10 ENSMUST00000123405.8 ENSMUST00000204657.3 ENSMUST00000203116.2 ENSMUST00000203193.3 ENSMUST00000126376.8 |
Marchf8
|
membrane associated ring-CH-type finger 8 |
| chr8_-_69636825 | 0.27 |
ENSMUST00000185176.8
|
Lzts1
|
leucine zipper, putative tumor suppressor 1 |
| chr14_-_106134253 | 0.27 |
ENSMUST00000022709.6
|
Spry2
|
sprouty RTK signaling antagonist 2 |
| chr15_+_44291470 | 0.27 |
ENSMUST00000226827.2
ENSMUST00000060652.5 |
Eny2
|
ENY2 transcription and export complex 2 subunit |
| chr14_-_54855446 | 0.27 |
ENSMUST00000227257.2
ENSMUST00000022803.6 |
Psmb5
|
proteasome (prosome, macropain) subunit, beta type 5 |
| chr4_-_6990774 | 0.26 |
ENSMUST00000039987.4
|
Tox
|
thymocyte selection-associated high mobility group box |
| chr10_+_61531282 | 0.26 |
ENSMUST00000020284.5
|
Tysnd1
|
trypsin domain containing 1 |
| chr18_+_84106188 | 0.26 |
ENSMUST00000060223.4
|
Zadh2
|
zinc binding alcohol dehydrogenase, domain containing 2 |
| chr9_-_21421458 | 0.26 |
ENSMUST00000213167.2
ENSMUST00000034698.9 |
Tmed1
|
transmembrane p24 trafficking protein 1 |
| chr4_+_97665843 | 0.26 |
ENSMUST00000075448.13
ENSMUST00000092532.13 |
Nfia
|
nuclear factor I/A |
| chrX_+_74425990 | 0.26 |
ENSMUST00000033541.5
|
Fundc2
|
FUN14 domain containing 2 |
| chr2_-_34645241 | 0.26 |
ENSMUST00000102800.9
|
Gapvd1
|
GTPase activating protein and VPS9 domains 1 |
| chr7_+_34818709 | 0.26 |
ENSMUST00000205391.2
ENSMUST00000042985.11 |
Cebpa
|
CCAAT/enhancer binding protein (C/EBP), alpha |
| chr7_-_121620366 | 0.26 |
ENSMUST00000033160.15
|
Gga2
|
golgi associated, gamma adaptin ear containing, ARF binding protein 2 |
| chr12_+_102724223 | 0.26 |
ENSMUST00000046404.8
|
Ubr7
|
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
| chr17_-_74601769 | 0.25 |
ENSMUST00000078459.8
ENSMUST00000232989.2 |
Memo1
|
mediator of cell motility 1 |
| chr15_+_100659622 | 0.25 |
ENSMUST00000023776.13
|
Slc4a8
|
solute carrier family 4 (anion exchanger), member 8 |
| chr4_-_120955387 | 0.25 |
ENSMUST00000058754.9
|
Zmpste24
|
zinc metallopeptidase, STE24 |
| chr2_+_162773440 | 0.25 |
ENSMUST00000130411.7
ENSMUST00000126163.3 |
Srsf6
|
serine and arginine-rich splicing factor 6 |
| chr8_+_80366247 | 0.25 |
ENSMUST00000173078.8
ENSMUST00000173286.8 |
Otud4
|
OTU domain containing 4 |
| chr5_+_141227245 | 0.25 |
ENSMUST00000085774.11
|
Sdk1
|
sidekick cell adhesion molecule 1 |
| chr10_+_53473032 | 0.25 |
ENSMUST00000020004.8
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chrX_+_138511360 | 0.25 |
ENSMUST00000113026.2
|
Rnf128
|
ring finger protein 128 |
| chr11_+_55360502 | 0.25 |
ENSMUST00000018727.4
|
G3bp1
|
GTPase activating protein (SH3 domain) binding protein 1 |
| chr4_-_151142351 | 0.24 |
ENSMUST00000030797.4
|
Vamp3
|
vesicle-associated membrane protein 3 |
| chr10_-_128640232 | 0.24 |
ENSMUST00000051011.14
|
Tmem198b
|
transmembrane protein 198b |
| chr10_+_11157047 | 0.24 |
ENSMUST00000129456.8
|
Fbxo30
|
F-box protein 30 |
| chr2_+_155223728 | 0.24 |
ENSMUST00000043237.14
ENSMUST00000174685.8 |
Trp53inp2
|
transformation related protein 53 inducible nuclear protein 2 |
| chr2_+_128907854 | 0.24 |
ENSMUST00000035812.14
|
Ttl
|
tubulin tyrosine ligase |
| chr14_+_56091454 | 0.24 |
ENSMUST00000227465.2
ENSMUST00000168479.3 ENSMUST00000100529.10 |
Nynrin
|
NYN domain and retroviral integrase containing |
| chr13_-_74465353 | 0.24 |
ENSMUST00000022060.7
|
Pdcd6
|
programmed cell death 6 |
| chr9_+_44516140 | 0.24 |
ENSMUST00000170489.2
ENSMUST00000217034.2 |
Ddx6
|
DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 |
| chr13_+_92981257 | 0.24 |
ENSMUST00000076169.4
|
Mtx3
|
metaxin 3 |
| chr19_+_3373285 | 0.24 |
ENSMUST00000025835.6
|
Cpt1a
|
carnitine palmitoyltransferase 1a, liver |
| chr2_-_156921135 | 0.24 |
ENSMUST00000069098.7
|
Soga1
|
suppressor of glucose, autophagy associated 1 |
| chr1_-_37580084 | 0.23 |
ENSMUST00000151952.8
|
Mgat4a
|
mannoside acetylglucosaminyltransferase 4, isoenzyme A |
| chr8_-_105122397 | 0.23 |
ENSMUST00000179802.2
|
Cmtm4
|
CKLF-like MARVEL transmembrane domain containing 4 |
| chr1_+_91468409 | 0.23 |
ENSMUST00000027538.9
ENSMUST00000190484.7 ENSMUST00000186068.2 |
Asb1
|
ankyrin repeat and SOCS box-containing 1 |
| chr2_-_32584132 | 0.22 |
ENSMUST00000028148.11
|
Fpgs
|
folylpolyglutamyl synthetase |
| chr6_+_7844759 | 0.22 |
ENSMUST00000040159.6
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr6_-_119394634 | 0.22 |
ENSMUST00000032272.13
|
Adipor2
|
adiponectin receptor 2 |
| chr4_+_148025316 | 0.22 |
ENSMUST00000103232.2
|
2510039O18Rik
|
RIKEN cDNA 2510039O18 gene |
| chr12_-_65010124 | 0.22 |
ENSMUST00000021331.9
|
Klhl28
|
kelch-like 28 |
| chr17_+_28910302 | 0.22 |
ENSMUST00000004990.14
ENSMUST00000114754.8 ENSMUST00000062694.16 |
Mapk14
|
mitogen-activated protein kinase 14 |
| chr11_+_58221538 | 0.22 |
ENSMUST00000116376.9
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr18_-_80194682 | 0.22 |
ENSMUST00000066743.11
|
Adnp2
|
ADNP homeobox 2 |
| chrX_-_35909011 | 0.22 |
ENSMUST00000051906.13
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr4_-_141412910 | 0.22 |
ENSMUST00000105782.2
|
Rsc1a1
|
regulatory solute carrier protein, family 1, member 1 |
| chr1_-_58544105 | 0.22 |
ENSMUST00000191206.2
ENSMUST00000027198.12 |
Orc2
|
origin recognition complex, subunit 2 |
| chr8_-_85620537 | 0.21 |
ENSMUST00000003907.14
ENSMUST00000109745.8 ENSMUST00000142748.2 |
Gcdh
|
glutaryl-Coenzyme A dehydrogenase |
| chr10_+_82465633 | 0.21 |
ENSMUST00000092266.11
ENSMUST00000151390.8 |
Tdg
|
thymine DNA glycosylase |
| chr15_+_88635852 | 0.21 |
ENSMUST00000041297.15
|
Zbed4
|
zinc finger, BED type containing 4 |
| chr2_-_144112700 | 0.21 |
ENSMUST00000110030.10
|
Snx5
|
sorting nexin 5 |
| chr7_-_55669702 | 0.21 |
ENSMUST00000052204.6
|
Nipa1
|
non imprinted in Prader-Willi/Angelman syndrome 1 homolog (human) |
| chr1_-_165021879 | 0.21 |
ENSMUST00000043338.10
|
Sft2d2
|
SFT2 domain containing 2 |
| chr12_-_85386120 | 0.21 |
ENSMUST00000040992.8
|
Nek9
|
NIMA (never in mitosis gene a)-related expressed kinase 9 |
| chr2_-_34803988 | 0.21 |
ENSMUST00000028232.7
ENSMUST00000202907.2 |
Phf19
|
PHD finger protein 19 |
| chr14_-_52258158 | 0.21 |
ENSMUST00000228580.2
ENSMUST00000226554.2 ENSMUST00000067549.15 |
Zfp219
|
zinc finger protein 219 |
| chr6_-_148732946 | 0.21 |
ENSMUST00000048418.14
|
Ipo8
|
importin 8 |
| chr12_+_111005768 | 0.21 |
ENSMUST00000084968.14
|
Rcor1
|
REST corepressor 1 |
| chr14_-_69522431 | 0.21 |
ENSMUST00000183882.2
ENSMUST00000037064.5 |
Slc25a37
|
solute carrier family 25, member 37 |
| chr19_-_29025233 | 0.20 |
ENSMUST00000025696.5
|
Ak3
|
adenylate kinase 3 |
| chr3_-_107603778 | 0.20 |
ENSMUST00000029490.15
|
Ahcyl1
|
S-adenosylhomocysteine hydrolase-like 1 |
| chr3_+_51323383 | 0.20 |
ENSMUST00000029303.13
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr17_+_5891582 | 0.20 |
ENSMUST00000002436.11
|
Snx9
|
sorting nexin 9 |
| chr14_+_54491637 | 0.20 |
ENSMUST00000180359.8
ENSMUST00000199338.2 |
Abhd4
|
abhydrolase domain containing 4 |
| chr9_+_7184514 | 0.20 |
ENSMUST00000215683.2
ENSMUST00000034499.10 |
Dcun1d5
|
DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) |
| chr11_-_116472272 | 0.20 |
ENSMUST00000082152.5
|
Ube2o
|
ubiquitin-conjugating enzyme E2O |
| chr12_-_73160181 | 0.20 |
ENSMUST00000043208.8
ENSMUST00000175693.3 |
Six4
|
sine oculis-related homeobox 4 |
| chr5_-_88823989 | 0.20 |
ENSMUST00000078945.12
|
Grsf1
|
G-rich RNA sequence binding factor 1 |
| chr7_-_119319965 | 0.20 |
ENSMUST00000033236.9
|
Thumpd1
|
THUMP domain containing 1 |
| chr17_+_81251997 | 0.20 |
ENSMUST00000025092.5
|
Tmem178
|
transmembrane protein 178 |
| chr5_+_115479284 | 0.20 |
ENSMUST00000031508.5
|
Triap1
|
TP53 regulated inhibitor of apoptosis 1 |
| chr11_-_101357046 | 0.20 |
ENSMUST00000040430.8
|
Vat1
|
vesicle amine transport 1 |
| chr11_+_20493306 | 0.20 |
ENSMUST00000093292.11
|
Sertad2
|
SERTA domain containing 2 |
| chr13_+_21663077 | 0.20 |
ENSMUST00000062609.6
ENSMUST00000225845.2 |
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr17_-_24292453 | 0.19 |
ENSMUST00000017090.6
|
Kctd5
|
potassium channel tetramerisation domain containing 5 |
| chr18_+_38552011 | 0.19 |
ENSMUST00000025293.5
|
Ndfip1
|
Nedd4 family interacting protein 1 |
| chr12_+_116244792 | 0.19 |
ENSMUST00000100986.4
ENSMUST00000220816.2 |
Esyt2
|
extended synaptotagmin-like protein 2 |
| chr12_+_33003882 | 0.19 |
ENSMUST00000076698.13
|
Sypl
|
synaptophysin-like protein |
| chr3_+_32490300 | 0.19 |
ENSMUST00000029201.14
|
Pik3ca
|
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| chr17_-_26314461 | 0.19 |
ENSMUST00000236128.2
ENSMUST00000025007.7 |
Nme4
|
NME/NM23 nucleoside diphosphate kinase 4 |
| chr4_+_149569717 | 0.19 |
ENSMUST00000030842.8
|
Lzic
|
leucine zipper and CTNNBIP1 domain containing |
| chr6_-_113717689 | 0.19 |
ENSMUST00000032440.6
|
Sec13
|
SEC13 homolog, nuclear pore and COPII coat complex component |
| chr11_-_74615496 | 0.19 |
ENSMUST00000021091.15
|
Pafah1b1
|
platelet-activating factor acetylhydrolase, isoform 1b, subunit 1 |
| chr18_-_35064906 | 0.19 |
ENSMUST00000025218.8
|
Etf1
|
eukaryotic translation termination factor 1 |
| chr2_+_73102269 | 0.19 |
ENSMUST00000090813.6
|
Sp9
|
trans-acting transcription factor 9 |
| chr9_-_82857528 | 0.18 |
ENSMUST00000034787.12
|
Phip
|
pleckstrin homology domain interacting protein |
| chr9_-_94420128 | 0.18 |
ENSMUST00000113028.2
|
Dipk2a
|
divergent protein kinase domain 2A |
| chr2_-_26823793 | 0.18 |
ENSMUST00000154651.2
ENSMUST00000015011.10 |
Surf4
|
surfeit gene 4 |
| chr5_+_140404997 | 0.18 |
ENSMUST00000100507.8
|
Eif3b
|
eukaryotic translation initiation factor 3, subunit B |
| chr5_+_64317550 | 0.18 |
ENSMUST00000101195.9
|
Tbc1d1
|
TBC1 domain family, member 1 |
| chr7_-_51655542 | 0.18 |
ENSMUST00000098414.5
|
Svip
|
small VCP/p97-interacting protein |
| chr13_+_69760346 | 0.18 |
ENSMUST00000022087.7
|
Nsun2
|
NOL1/NOP2/Sun domain family member 2 |
| chr11_-_5492175 | 0.18 |
ENSMUST00000020776.5
|
Ccdc117
|
coiled-coil domain containing 117 |
| chr13_-_12272962 | 0.18 |
ENSMUST00000099856.6
|
Mtr
|
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr4_+_152381662 | 0.18 |
ENSMUST00000048892.14
|
Icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr13_-_30170031 | 0.18 |
ENSMUST00000102948.11
|
E2f3
|
E2F transcription factor 3 |
| chr10_-_60983438 | 0.17 |
ENSMUST00000092498.12
ENSMUST00000137833.2 ENSMUST00000155919.8 |
Sgpl1
|
sphingosine phosphate lyase 1 |
| chr11_-_76400245 | 0.17 |
ENSMUST00000094012.11
|
Abr
|
active BCR-related gene |
| chr9_+_32607301 | 0.17 |
ENSMUST00000034534.13
ENSMUST00000050797.14 ENSMUST00000184887.2 |
Ets1
|
E26 avian leukemia oncogene 1, 5' domain |
| chr9_-_56151334 | 0.17 |
ENSMUST00000188142.7
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr9_+_77824646 | 0.17 |
ENSMUST00000034904.14
|
Elovl5
|
ELOVL family member 5, elongation of long chain fatty acids (yeast) |
| chr11_+_95925711 | 0.17 |
ENSMUST00000006217.10
ENSMUST00000107700.4 |
Snf8
|
SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) |
| chr14_-_47059694 | 0.17 |
ENSMUST00000111817.8
ENSMUST00000079314.12 |
Gmfb
|
glia maturation factor, beta |
| chr18_+_36812246 | 0.17 |
ENSMUST00000186538.7
|
Slc35a4
|
solute carrier family 35, member A4 |
| chr6_+_117840031 | 0.17 |
ENSMUST00000172088.8
ENSMUST00000079405.15 |
Zfp239
|
zinc finger protein 239 |
| chr11_-_70111796 | 0.17 |
ENSMUST00000060010.3
ENSMUST00000190533.2 |
Slc16a13
|
solute carrier family 16 (monocarboxylic acid transporters), member 13 |
| chr8_-_86026385 | 0.17 |
ENSMUST00000034131.10
|
Vps35
|
VPS35 retromer complex component |
| chr1_-_161078723 | 0.17 |
ENSMUST00000051925.5
ENSMUST00000071718.12 |
Prdx6
|
peroxiredoxin 6 |
| chr9_-_119151428 | 0.17 |
ENSMUST00000040853.11
|
Oxsr1
|
oxidative-stress responsive 1 |
| chr13_-_120252337 | 0.17 |
ENSMUST00000177916.8
ENSMUST00000178271.3 ENSMUST00000223722.2 |
Zfp131
|
zinc finger protein 131 |
| chr19_+_44282113 | 0.16 |
ENSMUST00000026221.7
|
Scd2
|
stearoyl-Coenzyme A desaturase 2 |
| chr2_+_152135748 | 0.16 |
ENSMUST00000028963.14
ENSMUST00000144252.8 |
Tbc1d20
|
TBC1 domain family, member 20 |
| chr18_-_77652820 | 0.16 |
ENSMUST00000026494.14
ENSMUST00000182024.2 |
Rnf165
|
ring finger protein 165 |
| chr6_-_83752749 | 0.16 |
ENSMUST00000014892.8
|
Tex261
|
testis expressed gene 261 |
| chr19_-_24202344 | 0.16 |
ENSMUST00000099558.5
ENSMUST00000232956.2 |
Tjp2
|
tight junction protein 2 |
| chr2_+_118731860 | 0.16 |
ENSMUST00000036578.7
|
Bahd1
|
bromo adjacent homology domain containing 1 |
| chr7_-_110443557 | 0.16 |
ENSMUST00000177462.8
ENSMUST00000176716.3 ENSMUST00000176746.8 ENSMUST00000177236.8 |
Rnf141
|
ring finger protein 141 |
| chr4_+_108477117 | 0.16 |
ENSMUST00000030320.13
|
Cc2d1b
|
coiled-coil and C2 domain containing 1B |
| chr3_-_95125002 | 0.16 |
ENSMUST00000107209.8
|
Gabpb2
|
GA repeat binding protein, beta 2 |
| chr18_+_67221287 | 0.16 |
ENSMUST00000025402.15
|
Gnal
|
guanine nucleotide binding protein, alpha stimulating, olfactory type |
| chr9_-_62029877 | 0.16 |
ENSMUST00000185675.7
|
Glce
|
glucuronyl C5-epimerase |
| chr19_-_29625755 | 0.16 |
ENSMUST00000159692.8
|
Ermp1
|
endoplasmic reticulum metallopeptidase 1 |
| chr2_-_152185901 | 0.16 |
ENSMUST00000040312.7
|
Trib3
|
tribbles pseudokinase 3 |
| chr19_-_47452840 | 0.15 |
ENSMUST00000081619.10
|
Sh3pxd2a
|
SH3 and PX domains 2A |
| chr11_-_20282684 | 0.15 |
ENSMUST00000004634.7
ENSMUST00000109594.8 |
Slc1a4
|
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr12_+_32428691 | 0.15 |
ENSMUST00000172332.4
|
Ccdc71l
|
coiled-coil domain containing 71 like |
| chr2_+_35172392 | 0.15 |
ENSMUST00000028239.8
|
Gsn
|
gelsolin |
| chr6_-_72416531 | 0.15 |
ENSMUST00000205335.2
ENSMUST00000206692.2 ENSMUST00000059472.10 |
Mat2a
|
methionine adenosyltransferase II, alpha |
| chr14_+_48683797 | 0.15 |
ENSMUST00000111735.10
|
Tmem260
|
transmembrane protein 260 |
| chrX_-_101687813 | 0.15 |
ENSMUST00000052012.14
ENSMUST00000043596.12 ENSMUST00000119229.8 ENSMUST00000122022.8 ENSMUST00000120270.8 ENSMUST00000113611.3 |
Phka1
|
phosphorylase kinase alpha 1 |
| chr15_+_82031382 | 0.15 |
ENSMUST00000023100.8
ENSMUST00000229336.2 |
Srebf2
|
sterol regulatory element binding factor 2 |
| chr12_+_111132779 | 0.15 |
ENSMUST00000117269.8
|
Traf3
|
TNF receptor-associated factor 3 |
| chr12_-_102844537 | 0.15 |
ENSMUST00000045652.8
ENSMUST00000223554.2 |
Btbd7
|
BTB (POZ) domain containing 7 |
| chr1_-_165462020 | 0.15 |
ENSMUST00000194437.6
ENSMUST00000068705.13 ENSMUST00000111435.9 ENSMUST00000193023.2 |
Mpzl1
|
myelin protein zero-like 1 |
| chr3_-_108053396 | 0.15 |
ENSMUST00000000001.5
|
Gnai3
|
guanine nucleotide binding protein (G protein), alpha inhibiting 3 |
| chr17_-_27423438 | 0.15 |
ENSMUST00000055117.9
|
Lemd2
|
LEM domain containing 2 |
| chr4_+_106173384 | 0.15 |
ENSMUST00000165709.8
ENSMUST00000094933.5 |
Usp24
|
ubiquitin specific peptidase 24 |
| chr2_+_167345009 | 0.15 |
ENSMUST00000078050.7
|
Rnf114
|
ring finger protein 114 |
| chr15_-_98505508 | 0.15 |
ENSMUST00000096224.6
|
Adcy6
|
adenylate cyclase 6 |
| chr11_+_115705550 | 0.15 |
ENSMUST00000021134.10
ENSMUST00000106481.9 |
Tsen54
|
tRNA splicing endonuclease subunit 54 |
| chrY_-_1286623 | 0.15 |
ENSMUST00000091190.12
|
Ddx3y
|
DEAD box helicase 3, Y-linked |
| chr9_+_122180673 | 0.15 |
ENSMUST00000156520.8
ENSMUST00000111497.5 |
Abhd5
|
abhydrolase domain containing 5 |
| chr7_-_112968533 | 0.14 |
ENSMUST00000047091.14
ENSMUST00000119278.8 |
Btbd10
|
BTB (POZ) domain containing 10 |
| chr16_-_18052937 | 0.14 |
ENSMUST00000076957.7
|
Zdhhc8
|
zinc finger, DHHC domain containing 8 |
| chr2_-_68302612 | 0.14 |
ENSMUST00000102715.4
|
Stk39
|
serine/threonine kinase 39 |
| chr19_+_56536685 | 0.14 |
ENSMUST00000071423.7
|
Nhlrc2
|
NHL repeat containing 2 |
| chr12_+_108758871 | 0.14 |
ENSMUST00000021692.9
|
Yy1
|
YY1 transcription factor |
| chr2_+_128659997 | 0.14 |
ENSMUST00000110325.8
|
Tmem87b
|
transmembrane protein 87B |
| chr5_+_122422428 | 0.14 |
ENSMUST00000053426.15
|
Pptc7
|
PTC7 protein phosphatase homolog |
| chr6_-_86710250 | 0.14 |
ENSMUST00000001185.14
|
Gmcl1
|
germ cell-less, spermatogenesis associated 1 |
| chr6_+_108805594 | 0.14 |
ENSMUST00000089162.5
|
Edem1
|
ER degradation enhancer, mannosidase alpha-like 1 |
| chr3_+_34074048 | 0.14 |
ENSMUST00000001620.13
|
Fxr1
|
fragile X mental retardation gene 1, autosomal homolog |
Network of associatons between targets according to the STRING database.
First level regulatory network of AAGGCAC
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072356 | chromosome passenger complex localization to kinetochore(GO:0072356) |
| 0.1 | 0.5 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.1 | 0.4 | GO:2000850 | negative regulation of corticosteroid hormone secretion(GO:2000847) negative regulation of glucocorticoid secretion(GO:2000850) |
| 0.1 | 0.3 | GO:2000686 | regulation of rubidium ion transmembrane transporter activity(GO:2000686) |
| 0.1 | 0.3 | GO:0097535 | lymphoid lineage cell migration(GO:0097534) lymphoid lineage cell migration into thymus(GO:0097535) |
| 0.1 | 0.3 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.3 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.3 | GO:0006285 | base-excision repair, AP site formation(GO:0006285) |
| 0.1 | 0.2 | GO:0072095 | regulation of branch elongation involved in ureteric bud branching(GO:0072095) |
| 0.1 | 0.3 | GO:0030327 | prenylated protein catabolic process(GO:0030327) |
| 0.1 | 0.3 | GO:0042851 | L-alanine metabolic process(GO:0042851) |
| 0.1 | 0.2 | GO:1902527 | positive regulation of protein monoubiquitination(GO:1902527) |
| 0.1 | 0.2 | GO:0071707 | immunoglobulin heavy chain V-D-J recombination(GO:0071707) |
| 0.1 | 0.2 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.1 | 0.7 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.1 | 0.2 | GO:0046949 | fatty-acyl-CoA biosynthetic process(GO:0046949) |
| 0.1 | 0.3 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.1 | 0.3 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.1 | 0.4 | GO:0060690 | epithelial cell differentiation involved in salivary gland development(GO:0060690) |
| 0.1 | 0.2 | GO:0044029 | DNA hypomethylation(GO:0044028) hypomethylation of CpG island(GO:0044029) |
| 0.1 | 0.2 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.1 | 0.2 | GO:2000331 | positive regulation of Wnt protein secretion(GO:0061357) regulation of terminal button organization(GO:2000331) |
| 0.1 | 0.4 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.6 | GO:0043553 | negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:0006172 | ADP biosynthetic process(GO:0006172) |
| 0.1 | 0.2 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.1 | 0.2 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.3 | GO:0071486 | cellular response to light intensity(GO:0071484) cellular response to high light intensity(GO:0071486) retinal rod cell apoptotic process(GO:0097473) |
| 0.0 | 0.8 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.0 | 0.0 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0016062 | adaptation of rhodopsin mediated signaling(GO:0016062) light adaption(GO:0036367) |
| 0.0 | 0.5 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.2 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.1 | GO:0006059 | hexitol metabolic process(GO:0006059) alditol catabolic process(GO:0019405) |
| 0.0 | 0.1 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0060448 | dichotomous subdivision of terminal units involved in lung branching(GO:0060448) |
| 0.0 | 0.5 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.1 | GO:0003365 | establishment of cell polarity involved in ameboidal cell migration(GO:0003365) |
| 0.0 | 0.5 | GO:0015727 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.4 | GO:0018401 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) |
| 0.0 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:1903433 | regulation of constitutive secretory pathway(GO:1903433) |
| 0.0 | 0.1 | GO:0018312 | peptidyl-serine ADP-ribosylation(GO:0018312) |
| 0.0 | 0.3 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.6 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.1 | GO:0000451 | rRNA 2'-O-methylation(GO:0000451) |
| 0.0 | 0.2 | GO:0006481 | C-terminal protein methylation(GO:0006481) |
| 0.0 | 0.1 | GO:0009188 | ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.4 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.5 | GO:0033572 | transferrin transport(GO:0033572) |
| 0.0 | 0.2 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.4 | GO:0031659 | positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.1 | GO:0048320 | axial mesoderm formation(GO:0048320) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.3 | GO:0031119 | tRNA pseudouridine synthesis(GO:0031119) |
| 0.0 | 0.2 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.1 | GO:0072368 | regulation of lipid transport by negative regulation of transcription from RNA polymerase II promoter(GO:0072368) |
| 0.0 | 0.5 | GO:0031998 | regulation of fatty acid beta-oxidation(GO:0031998) |
| 0.0 | 0.2 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:1903031 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.0 | 0.1 | GO:1903173 | phytol metabolic process(GO:0033306) fatty alcohol metabolic process(GO:1903173) |
| 0.0 | 0.2 | GO:1903966 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0033577 | protein glycosylation in endoplasmic reticulum(GO:0033577) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0006014 | D-ribose metabolic process(GO:0006014) |
| 0.0 | 0.2 | GO:0061087 | positive regulation of histone H3-K27 methylation(GO:0061087) |
| 0.0 | 0.1 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.3 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.2 | GO:0075525 | viral translational termination-reinitiation(GO:0075525) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.2 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.1 | GO:1903416 | response to glycoside(GO:1903416) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.2 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.1 | GO:1901187 | regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.3 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.2 | GO:0035280 | miRNA loading onto RISC involved in gene silencing by miRNA(GO:0035280) |
| 0.0 | 0.2 | GO:2001205 | negative regulation of osteoclast development(GO:2001205) |
| 0.0 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome assembly(GO:0006335) DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:0070345 | negative regulation of fat cell proliferation(GO:0070345) |
| 0.0 | 0.3 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.2 | GO:0060313 | negative regulation of neutrophil degranulation(GO:0043314) negative regulation of blood vessel remodeling(GO:0060313) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0036451 | cap mRNA methylation(GO:0036451) |
| 0.0 | 0.1 | GO:0030421 | defecation(GO:0030421) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.3 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transport(GO:0006848) mitochondrial pyruvate transport(GO:0006850) pyruvate transmembrane transport(GO:1901475) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.0 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.2 | GO:1904996 | positive regulation of leukocyte adhesion to vascular endothelial cell(GO:1904996) |
| 0.0 | 0.1 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.0 | 0.4 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.4 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.1 | GO:0010710 | regulation of collagen catabolic process(GO:0010710) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0046021 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.1 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.2 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:1903553 | positive regulation of extracellular exosome assembly(GO:1903553) |
| 0.0 | 0.4 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0035811 | negative regulation of urine volume(GO:0035811) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
| 0.0 | 0.2 | GO:1902166 | negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902166) |
| 0.0 | 0.1 | GO:0097494 | regulation of vesicle size(GO:0097494) |
| 0.0 | 0.1 | GO:0017187 | peptidyl-glutamic acid carboxylation(GO:0017187) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.6 | GO:0030539 | male genitalia development(GO:0030539) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.1 | GO:0030382 | sperm mitochondrion organization(GO:0030382) |
| 0.0 | 0.2 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:0072015 | glomerular visceral epithelial cell development(GO:0072015) glomerular epithelial cell development(GO:0072310) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.0 | GO:0043686 | co-translational protein modification(GO:0043686) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0032056 | positive regulation of translation in response to stress(GO:0032056) |
| 0.0 | 0.3 | GO:0006152 | purine nucleoside catabolic process(GO:0006152) purine ribonucleoside catabolic process(GO:0046130) |
| 0.0 | 0.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.2 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.2 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.2 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:1903003 | positive regulation of protein deubiquitination(GO:1903003) |
| 0.0 | 0.1 | GO:1905161 | protein localization to phagocytic vesicle(GO:1905161) regulation of protein localization to phagocytic vesicle(GO:1905169) positive regulation of protein localization to phagocytic vesicle(GO:1905171) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:2000346 | negative regulation of hepatocyte proliferation(GO:2000346) |
| 0.0 | 0.0 | GO:2000794 | regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0015862 | uridine transport(GO:0015862) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.4 | GO:0051443 | positive regulation of ubiquitin-protein transferase activity(GO:0051443) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.2 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0072178 | nephric duct development(GO:0072176) nephric duct morphogenesis(GO:0072178) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.2 | GO:0006415 | translational termination(GO:0006415) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.0 | GO:0042275 | error-free postreplication DNA repair(GO:0042275) |
| 0.0 | 0.2 | GO:0035493 | SNARE complex assembly(GO:0035493) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.3 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.3 | GO:0043259 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.1 | 0.4 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.1 | 0.2 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.1 | 0.2 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.1 | 0.4 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.1 | 0.2 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.1 | 0.2 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0000235 | astral microtubule(GO:0000235) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.2 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.5 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.5 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0071595 | Nem1-Spo7 phosphatase complex(GO:0071595) |
| 0.0 | 0.4 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0035363 | histone locus body(GO:0035363) |
| 0.0 | 0.2 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.1 | GO:0097226 | sperm mitochondrial sheath(GO:0097226) |
| 0.0 | 0.1 | GO:0032783 | ELL-EAF complex(GO:0032783) |
| 0.0 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0030981 | cortical microtubule cytoskeleton(GO:0030981) |
| 0.0 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.2 | GO:0071541 | eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.3 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.1 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.5 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.3 | GO:1990023 | mitotic spindle midzone(GO:1990023) |
| 0.0 | 0.2 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.1 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.3 | GO:0030134 | ER to Golgi transport vesicle(GO:0030134) |
| 0.0 | 0.1 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.0 | GO:0035068 | micro-ribonucleoprotein complex(GO:0035068) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0031211 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.6 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0005839 | proteasome core complex(GO:0005839) |
| 0.0 | 0.3 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.2 | 0.6 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.1 | 0.3 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.1 | 0.5 | GO:0004998 | transferrin receptor activity(GO:0004998) |
| 0.1 | 0.5 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.1 | 0.3 | GO:0047710 | bis(5'-adenosyl)-triphosphatase activity(GO:0047710) |
| 0.1 | 0.2 | GO:1990698 | palmitoleoyltransferase activity(GO:1990698) |
| 0.1 | 0.3 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.3 | GO:0031208 | POZ domain binding(GO:0031208) |
| 0.1 | 0.2 | GO:0008263 | pyrimidine-specific mismatch base pair DNA N-glycosylase activity(GO:0008263) |
| 0.1 | 0.3 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.1 | 0.3 | GO:0047635 | L-alanine:2-oxoglutarate aminotransferase activity(GO:0004021) alanine-oxo-acid transaminase activity(GO:0047635) |
| 0.1 | 0.4 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.1 | 0.4 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.1 | 0.4 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) |
| 0.1 | 0.2 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.5 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0017099 | very-long-chain-acyl-CoA dehydrogenase activity(GO:0017099) |
| 0.0 | 0.2 | GO:0097003 | adipokinetic hormone receptor activity(GO:0097003) |
| 0.0 | 0.4 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.2 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.3 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.2 | GO:0004478 | methionine adenosyltransferase activity(GO:0004478) |
| 0.0 | 0.1 | GO:0046964 | 3'-phosphoadenosine 5'-phosphosulfate transmembrane transporter activity(GO:0046964) |
| 0.0 | 0.5 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0070039 | rRNA (guanosine-2'-O-)-methyltransferase activity(GO:0070039) |
| 0.0 | 0.2 | GO:0003880 | protein C-terminal carboxyl O-methyltransferase activity(GO:0003880) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0043532 | angiostatin binding(GO:0043532) |
| 0.0 | 0.2 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0032896 | palmitoyl-CoA 9-desaturase activity(GO:0032896) |
| 0.0 | 0.3 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0009041 | cytidylate kinase activity(GO:0004127) uridylate kinase activity(GO:0009041) nucleoside phosphate kinase activity(GO:0050145) |
| 0.0 | 0.2 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0000702 | oxidized base lesion DNA N-glycosylase activity(GO:0000702) |
| 0.0 | 0.1 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.2 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.2 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004483 | mRNA (nucleoside-2'-O-)-methyltransferase activity(GO:0004483) |
| 0.0 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.3 | GO:1990239 | steroid hormone binding(GO:1990239) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.2 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.3 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0061649 | ubiquitinated histone binding(GO:0061649) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.3 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.3 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.4 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.1 | GO:0032296 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.2 | GO:0015193 | L-alanine transmembrane transporter activity(GO:0015180) L-proline transmembrane transporter activity(GO:0015193) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.0 | GO:0004420 | hydroxymethylglutaryl-CoA reductase (NADPH) activity(GO:0004420) hydroxymethylglutaryl-CoA reductase activity(GO:0042282) |
| 0.0 | 0.0 | GO:0005290 | L-histidine transmembrane transporter activity(GO:0005290) |
| 0.0 | 0.2 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.5 | GO:0016881 | acid-amino acid ligase activity(GO:0016881) |
| 0.0 | 0.4 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0102337 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.3 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.4 | GO:0097602 | cullin family protein binding(GO:0097602) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.2 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.3 | GO:0016646 | oxidoreductase activity, acting on the CH-NH group of donors, NAD or NADP as acceptor(GO:0016646) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.1 | GO:0015181 | arginine transmembrane transporter activity(GO:0015181) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.0 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) BLOC-2 complex binding(GO:0036461) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0005519 | cytoskeletal regulatory protein binding(GO:0005519) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.3 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.3 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.2 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.3 | PID HNF3B PATHWAY | FOXA2 and FOXA3 transcription factor networks |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.6 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.3 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.5 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME BASE FREE SUGAR PHOSPHATE REMOVAL VIA THE SINGLE NUCLEOTIDE REPLACEMENT PATHWAY | Genes involved in Base-free sugar-phosphate removal via the single-nucleotide replacement pathway |
| 0.0 | 0.4 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.9 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.6 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.3 | REACTOME MRNA DECAY BY 5 TO 3 EXORIBONUCLEASE | Genes involved in mRNA Decay by 5' to 3' Exoribonuclease |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.3 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.1 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PC | Genes involved in Acyl chain remodelling of PC |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |